Specific Circular RNA Signature of Endothelial Cells: Potential Implications in Vascular Pathophysiology
Abstract
1. Introduction
2. Results
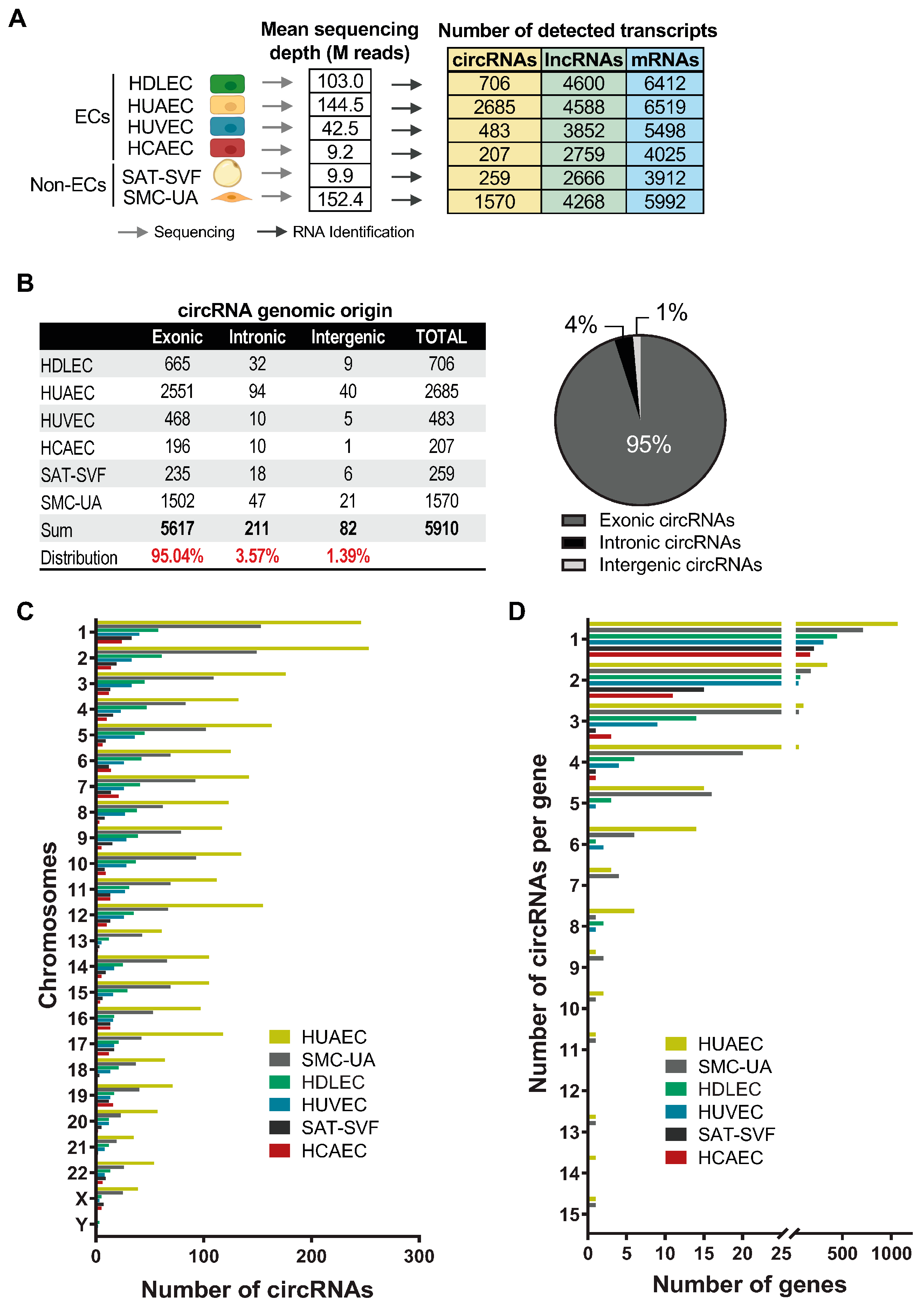
2.1. EC circRNA De Novo Identification
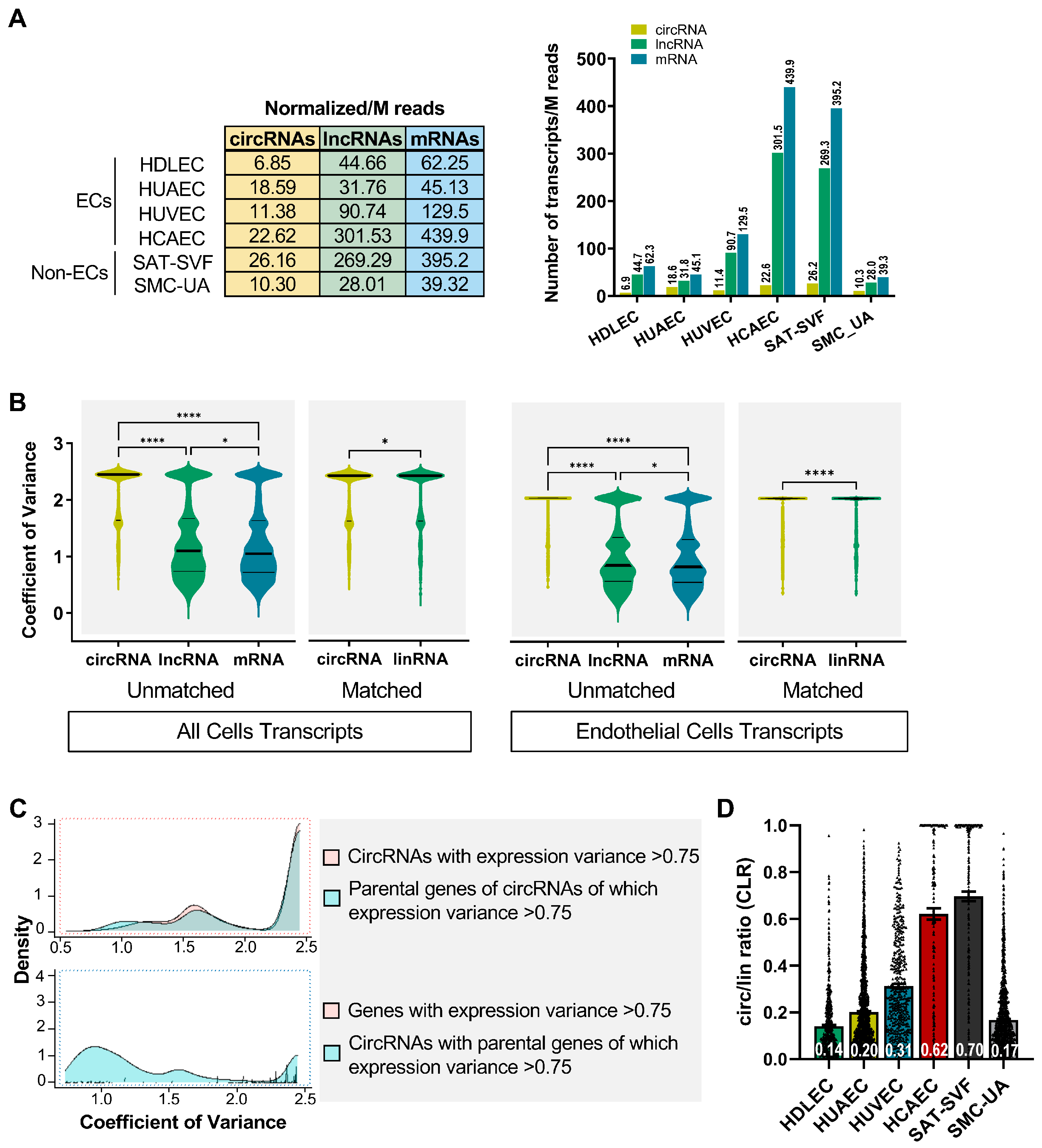
2.2. Expression Profiles of circRNAs Prove More Adept at Discerning Distinctions between Endothelial Cells (ECs) Than Their Linear RNA Counterparts
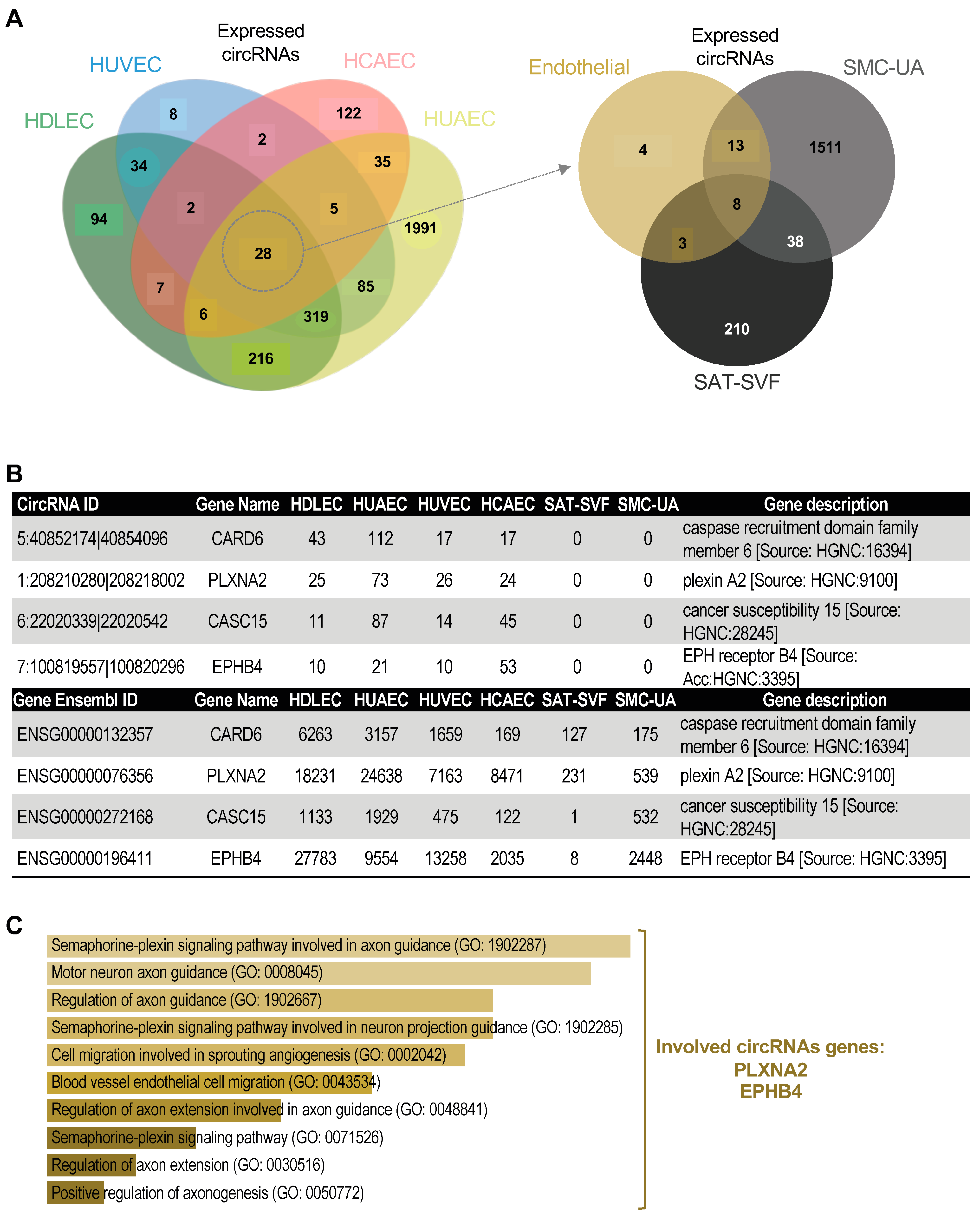
2.3. EC-Specific circRNA Identification
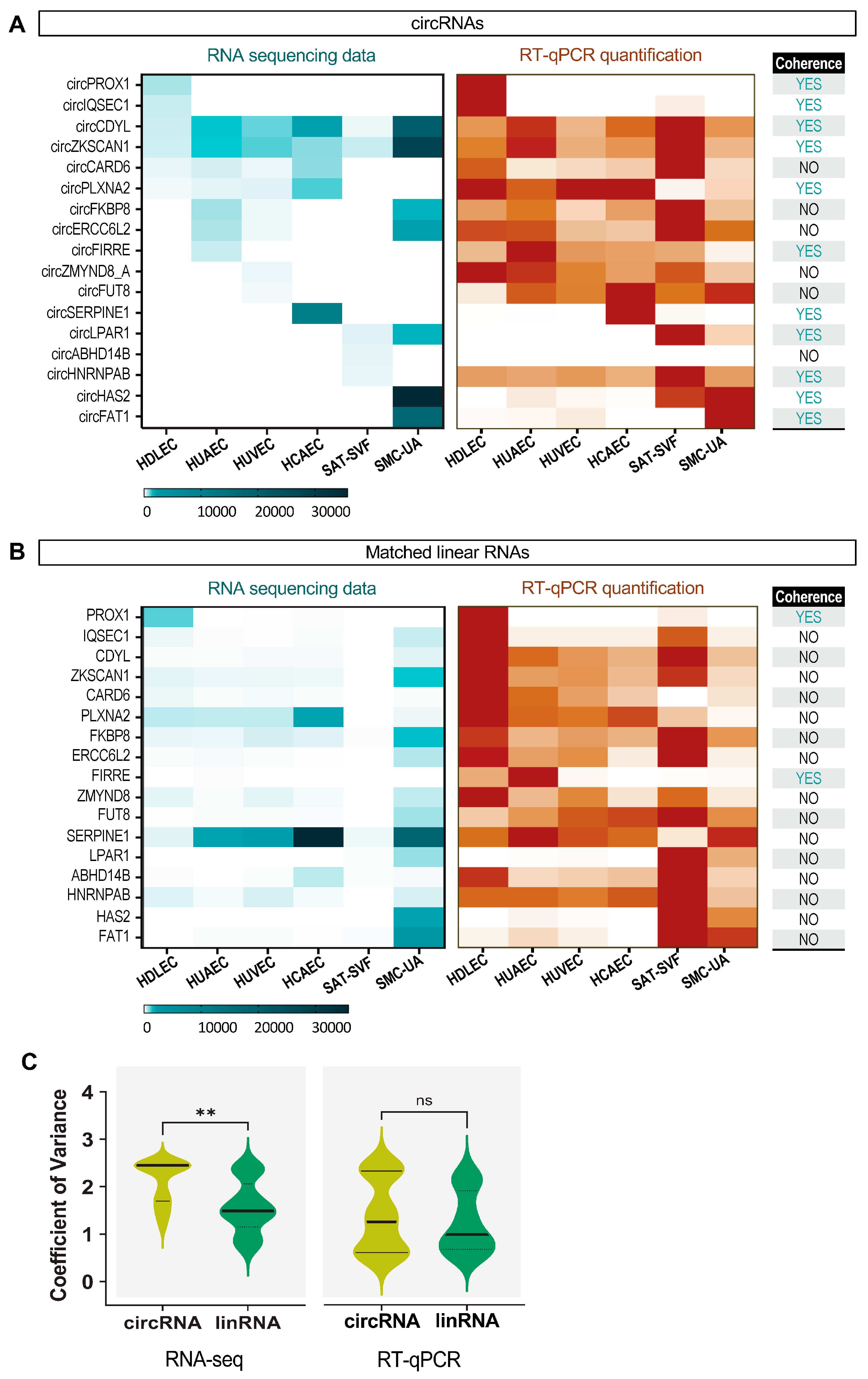
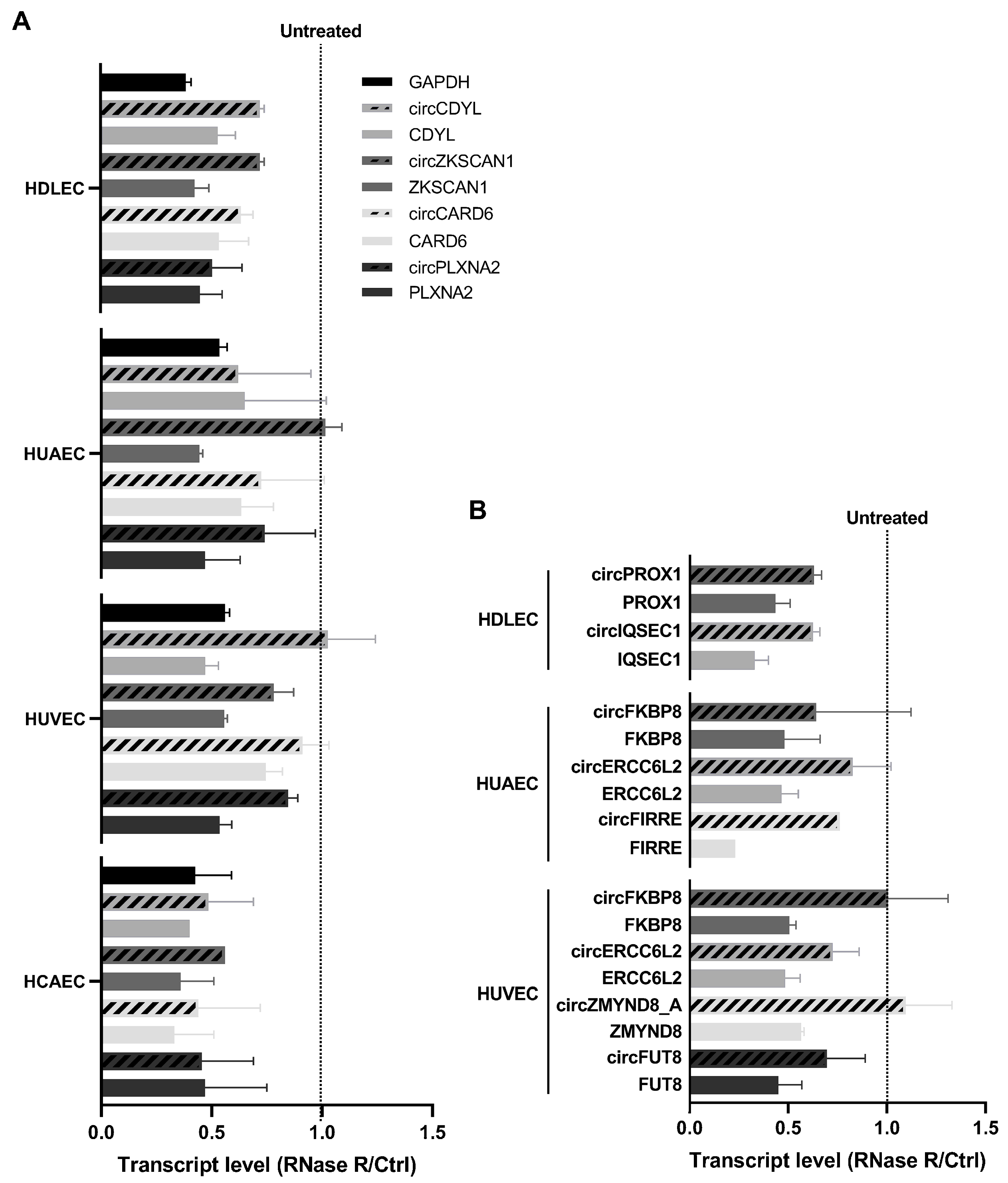
2.4. Specific circRNA Expression Validation by RT-qPCR and RNase R Treatment
3. Discussion
3.1. Endothelial Cell Specific Expression Profiles
3.2. Endothelial Cell Specific Circular RNAs
3.3. Specific Endothelial Cell circRNA Signature
3.4. Biomedical Implications
3.5. Conclusions
4. Materials and Methods
4.1. RNA Sequencing
4.2. RNA Sequencing Dataset Collection and Analysis
4.3. RNA Sample Recovery for RT-qPCR
4.4. Ribonuclease R (RNase R) Treatment
4.5. RNA Quantification by RT-qPCR Analysis
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADAR | Adenosine Deaminase Associated with RNA |
| BEC | Blood Endothelial Cell |
| circRNA | circular RNA |
| CLR | CircRNA on Linear RNA Ratio |
| EC | Endothelial Cell |
| HCAEC | Human Coronary Artery endothelial Cell |
| HDLEC | Human Dermal Lymphatic Endothelial Cell |
| hnRNP | heterogenous nuclear RiboNucleoProtein |
| HUAEC | Human Artery Endothelial Cell |
| HUVEC | Human Umbilical Vein Endothelial Cell |
| LEC | Lymphatic Endothelial Cell |
| lncRNA | long non-coding RNA |
| mRNA | messenger RNA |
| RBP | RNA-Binding Protein |
| RNA QC | RNA Quality Control |
| RNase | RiboNuclease |
| RNAseq | RNA sequencing |
| SAT-SVF | Superficial Adipose Tissue Stromal Vascular Fraction |
| SMC-UA | Smooth Muscle Cells from Umbilical Artery |
References
- Humphrey, J.D.; McCulloch, A.D. The Cardiovascular System—Anatomy, Physiology and Cell Biology. In Biomechanics of Soft Tissue in Cardiovascular Systems; Holzapfel, G.A., Ogden, R.W., Eds.; International Centre for Mechanical Sciences; Springer: Vienna, Austria, 2003; pp. 1–14. ISBN 978-3-7091-2736-0. [Google Scholar]
- Pittman, R.N. The Circulatory System and Oxygen Transport; Morgan & Claypool Life Sciences: San Rafael, CA, USA, 2011. [Google Scholar]
- Swartz, M.A. The Physiology of the Lymphatic System. Adv. Drug Deliv. Rev. 2001, 50, 3–20. [Google Scholar] [CrossRef]
- Krüger-Genge, A.; Blocki, A.; Franke, R.-P.; Jung, F. Vascular Endothelial Cell Biology: An Update. Int. J. Mol. Sci. 2019, 20, 4411. [Google Scholar] [CrossRef]
- Alitalo, K.; Tammela, T.; Petrova, T.V. Lymphangiogenesis in Development and Human Disease. Nature 2005, 438, 946–953. [Google Scholar] [CrossRef]
- Folkman, J. Angiogenesis. Annu. Rev. Med. 2006, 57, 1–18. [Google Scholar] [CrossRef]
- Libby, P.; Ridker, P.M.; Hansson, G.K. Inflammation in Atherosclerosis: From Pathophysiology to Practice. J. Am. Coll. Cardiol. 2009, 54, 2129–2138. [Google Scholar] [CrossRef]
- Kholová, I.; Dragneva, G.; Čermáková, P.; Laidinen, S.; Kaskenpää, N.; Hazes, T.; Čermáková, E.; Šteiner, I.; Ylä-Herttuala, S. Lymphatic Vasculature Is Increased in Heart Valves, Ischaemic and Inflamed Hearts and in Cholesterol-Rich and Calcified Atherosclerotic Lesions. Eur. J. Clin. Investig. 2011, 41, 487–497. [Google Scholar] [CrossRef]
- Warren, A.G.; Brorson, H.; Borud, L.J.; Slavin, S.A. Lymphedema: A Comprehensive Review. Ann. Plast. Surg. 2007, 59, 464. [Google Scholar] [CrossRef]
- Dayan, J.H.; Ly, C.L.; Kataru, R.P.; Mehrara, B.J. Lymphedema: Pathogenesis and Novel Therapies. Annu. Rev. Med. 2018, 69, 263–276. [Google Scholar] [CrossRef]
- Huggenberger, R.; Siddiqui, S.S.; Brander, D.; Ullmann, S.; Zimmermann, K.; Antsiferova, M.; Werner, S.; Alitalo, K.; Detmar, M. An Important Role of Lymphatic Vessel Activation in Limiting Acute Inflammation. Blood 2011, 117, 4667–4678. [Google Scholar] [CrossRef]
- Sánchez-Duffhues, G.; García de Vinuesa, A.; van de Pol, V.; Geerts, M.E.; de Vries, M.R.; Janson, S.G.; van Dam, H.; Lindeman, J.H.; Goumans, M.-J.; ten Dijke, P. Inflammation Induces Endothelial-to-Mesenchymal Transition and Promotes Vascular Calcification through Downregulation of BMPR2. J. Pathol. 2019, 247, 333–346. [Google Scholar] [CrossRef]
- von der Weid, P.-Y.; Rehal, S.; Ferraz, J.G. Role of the Lymphatic System in the Pathogenesis of Crohn’s Disease. Curr. Opin. Gastroenterol. 2011, 27, 335. [Google Scholar] [CrossRef]
- Karaman, S.; Detmar, M. Mechanisms of Lymphatic Metastasis. J. Clin. Investig. 2014, 124, 922–928. [Google Scholar] [CrossRef]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular Principles of Metastasis: A Hallmark of Cancer Revisited. Signal Transduct. Target. Ther. 2020, 5, 28. [Google Scholar] [CrossRef]
- Conway, E.M.; Carmeliet, P. The Diversity of Endothelial Cells: A Challenge for Therapeutic Angiogenesis. Genome Biol. 2004, 5, 207. [Google Scholar] [CrossRef]
- Chi, J.-T.; Chang, H.Y.; Haraldsen, G.; Jahnsen, F.L.; Troyanskaya, O.G.; Chang, D.S.; Wang, Z.; Rockson, S.G.; van de Rijn, M.; Botstein, D.; et al. Endothelial Cell Diversity Revealed by Global Expression Profiling. Proc. Natl. Acad. Sci. USA 2003, 100, 10623–10628. [Google Scholar] [CrossRef]
- Ho, M.; Yang, E.; Matcuk, G.; Deng, D.; Sampas, N.; Tsalenko, A.; Tabibiazar, R.; Zhang, Y.; Chen, M.; Talbi, S.; et al. Identification of Endothelial Cell Genes by Combined Database Mining and Microarray Analysis. Physiol. Genom. 2003, 13, 249–262. [Google Scholar] [CrossRef]
- Feng, W.; Chen, L.; Nguyen, P.K.; Wu, S.M.; Li, G. Single Cell Analysis of Endothelial Cells Identified Organ-Specific Molecular Signatures and Heart-Specific Cell Populations and Molecular Features. Front. Cardiovasc. Med. 2019, 6, 165. [Google Scholar] [CrossRef]
- Huang, X.; Shen, W.; Veizades, S.; Liang, G.; Sayed, N.; Nguyen, P.K. Single-Cell Transcriptional Profiling Reveals Sex and Age Diversity of Gene Expression in Mouse Endothelial Cells. Front. Genet. 2021, 12, 590377. [Google Scholar] [CrossRef]
- Weirick, T.; Militello, G.; Uchida, S. Long Non-Coding RNAs in Endothelial Biology. Front. Physiol. 2018, 9, 522. [Google Scholar] [CrossRef]
- Jaé, N.; Dimmeler, S. Noncoding RNAs in Vascular Diseases. Circ. Res. 2020, 126, 1127–1145. [Google Scholar] [CrossRef]
- Nicolet, B.P.; Engels, S.; Aglialoro, F.; van den Akker, E.; von Lindern, M.; Wolkers, M.C. Circular RNA Expression in Human Hematopoietic Cells Is Widespread and Cell-Type Specific. Nucleic Acids Res. 2018, 46, 8168–8180. [Google Scholar] [CrossRef]
- Maass, P.G.; Glažar, P.; Memczak, S.; Dittmar, G.; Hollfinger, I.; Schreyer, L.; Sauer, A.V.; Toka, O.; Aiuti, A.; Luft, F.C.; et al. A Map of Human Circular RNAs in Clinically Relevant Tissues. J. Mol. Med. 2017, 95, 1179–1189. [Google Scholar] [CrossRef]
- Eger, N.; Schoppe, L.; Schuster, S.; Laufs, U.; Boeckel, J.-N. Circular RNA Splicing. In Circular RNAs: Biogenesis and Functions; Xiao, J., Ed.; Advances in Experimental Medicine and Biology; Springer: Singapore, 2018; pp. 41–52. ISBN 9789811314261. [Google Scholar]
- Nigro, J.M.; Cho, K.R.; Fearon, E.R.; Kern, S.E.; Ruppert, J.M.; Oliner, J.D.; Kinzler, K.W.; Vogelstein, B. Scrambled Exons. Cell 1991, 64, 607–613. [Google Scholar] [CrossRef]
- Cocquerelle, C.; Daubersies, P.; Majérus, M.A.; Kerckaert, J.P.; Bailleul, B. Splicing with Inverted Order of Exons Occurs Proximal to Large Introns. EMBO J. 1992, 11, 1095–1098. [Google Scholar] [CrossRef]
- Cocquerelle, C.; Mascrez, B.; Hétuin, D.; Bailleul, B. Mis-Splicing Yields Circular RNA Molecules. FASEB J. 1993, 7, 155–160. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs Are the Predominant Transcript Isoform from Hundreds of Human Genes in Diverse Cell Types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs Are Abundant, Conserved, and Associated with ALU Repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs Are a Large Class of Animal RNAs with Regulatory Potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Suzuki, H.; Zuo, Y.; Wang, J.; Zhang, M.Q.; Malhotra, A.; Mayeda, A. Characterization of RNase R-Digested Cellular RNA Source That Consists of Lariat and Circular RNAs from Pre-mRNA Splicing. Nucleic Acids Res. 2006, 34, e63. [Google Scholar] [CrossRef]
- Shao, T.; Pan, Y.; Xiong, X. Circular RNA: An Important Player with Multiple Facets to Regulate Its Parental Gene Expression. Mol. Ther. Nucleic Acids 2021, 23, 369–376. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA Circles Function as Efficient microRNA Sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Ragan, C.; Goodall, G.J.; Shirokikh, N.E.; Preiss, T. Insights into the Biogenesis and Potential Functions of Exonic Circular RNA. Sci. Rep. 2019, 9, 2048. [Google Scholar] [CrossRef]
- Zang, J.; Lu, D.; Xu, A. The Interaction of circRNAs and RNA Binding Proteins: An Important Part of circRNA Maintenance and Function. J. Neurosci. Res. 2020, 98, 87–97. [Google Scholar] [CrossRef]
- Diallo, L.H.; Tatin, F.; David, F.; Godet, A.-C.; Zamora, A.; Prats, A.-C.; Garmy-Susini, B.; Lacazette, E. How Are circRNAs Translated by Non-Canonical Initiation Mechanisms? Biochimie 2019, 164, 45–52. [Google Scholar] [CrossRef]
- Prats, A.-C.; David, F.; Diallo, L.H.; Roussel, E.; Tatin, F.; Garmy-Susini, B.; Lacazette, E. Circular RNA, the Key for Translation. Int. J. Mol. Sci. 2020, 21, E8591. [Google Scholar] [CrossRef]
- Wen, S.; Qadir, J.; Yang, B.B. Circular RNA Translation: Novel Protein Isoforms and Clinical Significance. Trends Mol. Med. 2022, 28, 405–420. [Google Scholar] [CrossRef]
- Boeckel, J.-N.; Jaé, N.; Heumüller, A.W.; Chen, W.; Boon, R.A.; Stellos, K.; Zeiher, A.M.; John, D.; Uchida, S.; Dimmeler, S. Identification and Characterization of Hypoxia-Regulated Endothelial Circular RNA. Circ. Res. 2015, 117, 884–890. [Google Scholar] [CrossRef]
- Heumüller, A.W.; Jones, A.N.; Mourão, A.; Klangwart, M.; Shi, C.; Wittig, I.; Fischer, A.; Muhly-Reinholz, M.; Buchmann, G.K.; Dieterich, C.; et al. Locus-Conserved Circular RNA cZNF292 Controls Endothelial Cell Flow Responses. Circ. Res. 2022, 130, 67–79. [Google Scholar] [CrossRef]
- Shan, K.; Liu, C.; Liu, B.-H.; Chen, X.; Dong, R.; Liu, X.; Zhang, Y.-Y.; Liu, B.; Zhang, S.-J.; Wang, J.-J.; et al. Circular Noncoding RNA HIPK3 Mediates Retinal Vascular Dysfunction in Diabetes Mellitus. Circulation 2017, 136, 1629–1642. [Google Scholar] [CrossRef]
- Wu, W.; Zhou, M.; Liu, D.; Min, X.; Shao, T.; Xu, Z.; Jing, X.; Cai, M.; Xu, S.; Liang, X.; et al. circGNAQ, a Circular RNA Enriched in Vascular Endothelium, Inhibits Endothelial Cell Senescence and Atherosclerosis Progression. Mol. Ther.—Nucleic Acids 2021, 26, 374–387. [Google Scholar] [CrossRef]
- Feng, J.; Chen, K.; Dong, X.; Xu, X.; Jin, Y.; Zhang, X.; Chen, W.; Han, Y.; Shao, L.; Gao, Y.; et al. Genome-Wide Identification of Cancer-Specific Alternative Splicing in circRNA. Mol. Cancer 2019, 18, 35. [Google Scholar] [CrossRef]
- Shi, X.; Wang, B.; Feng, X.; Xu, Y.; Lu, K.; Sun, M. circRNAs and Exosomes: A Mysterious Frontier for Human Cancer. Mol. Ther.—Nucleic Acids 2020, 19, 384–392. [Google Scholar] [CrossRef]
- Zhu, J.; Luo, Y.; Zhao, Y.; Kong, Y.; Zheng, H.; Li, Y.; Gao, B.; Ai, L.; Huang, H.; Huang, J.; et al. circEHBP1 Promotes Lymphangiogenesis and Lymphatic Metastasis of Bladder Cancer via miR-130a-3p/TGFβR1/VEGF-D Signaling. Mol. Ther. 2021, 29, 1838–1852. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, J.; Zhao, F. Circular RNA Identification Based on Multiple Seed Matching. Brief. Bioinform. 2018, 19, 803–810. [Google Scholar] [CrossRef]
- Ivanov, A.; Memczak, S.; Wyler, E.; Torti, F.; Porath, H.T.; Orejuela, M.R.; Piechotta, M.; Levanon, E.Y.; Landthaler, M.; Dieterich, C.; et al. Analysis of Intron Sequences Reveals Hallmarks of Circular RNA Biogenesis in Animals. Cell Rep. 2015, 10, 170–177. [Google Scholar] [CrossRef]
- Sabin, F.R. On the Origin of the Lymphatic System from the Veins and the Development of the Lymph Hearts and Thoracic Duct in the Pig. Am. J. Anat. 1902, 1, 367–389. [Google Scholar] [CrossRef]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. Jvenn: An Interactive Venn Diagram Viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef]
- Stehlik, C.; Hayashi, H.; Pio, F.; Godzik, A.; Reed, J.C. CARD6 Is a Modulator of NF-κB Activation by Nod1- and Cardiak-Mediated Pathways *. J. Biol. Chem. 2003, 278, 31941–31949. [Google Scholar] [CrossRef]
- Kim, S.S.; Ahn, C.H.; Kang, M.R.; Kim, Y.R.; Kim, H.S.; Yoo, N.J.; Lee, S.H. Expression of CARD6, an NF-kappaB Activator, in Gastric, Colorectal and Oesophageal Cancers. Pathology 2010, 42, 50–53. [Google Scholar] [CrossRef]
- Dong, X.; Xing, J.; Liu, Q.; Ye, M.; Zhou, Z.; Li, Y.; Huang, R.; Li, Z.; Nie, Q. CircPLXNA2 Affects the Proliferation and Apoptosis of Myoblast through circPLXNA2/Gga-miR-12207-5P/MDM4 Axis. Int. J. Mol. Sci. 2023, 24, 5459. [Google Scholar] [CrossRef]
- Salzman, J.; Chen, R.E.; Olsen, M.N.; Wang, P.L.; Brown, P.O. Cell-Type Specific Features of Circular RNA Expression. PLoS Genet. 2013, 9, e1003777. [Google Scholar] [CrossRef]
- Rachinger, N.; Fischer, S.; Böhme, I.; Linck-Paulus, L.; Kuphal, S.; Kappelmann-Fenzl, M.; Bosserhoff, A.K. Loss of Gene Information: Discrepancies between RNA Sequencing, cDNA Microarray, and qRT-PCR. Int. J. Mol. Sci. 2021, 22, 9349. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Li, S.; Yang, N.; Zou, Y.; Zheng, D.; Xiao, T. Recent Progress in Circular RNAs in Human Cancers. Cancer Lett. 2017, 404, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; He, D.; Peng, Z.; Peng, W.; Shi, W.; Wang, J.; Li, B.; Zhang, C.; Duan, C. Circular RNAs in Cancer: An Emerging Key Player. J. Hematol. Oncol. 2017, 10, 2. [Google Scholar] [CrossRef] [PubMed]
- Dragomir, M.; Calin, G.A. Circular RNAs in Cancer–Lessons Learned From microRNAs. Front. Oncol. 2018, 8, 179. [Google Scholar] [CrossRef] [PubMed]
- Gallogly, S.; Fujisawa, T.; Hung, J.D.; Brittan, M.; Skinner, E.M.; Mitchell, A.J.; Medine, C.; Luque, N.; Zodda, E.; Cascante, M.; et al. Generation of a Novel In Vitro Model to Study Endothelial Dysfunction from Atherothrombotic Specimens. Cardiovasc. Drugs Ther. 2021, 35, 1281–1290. [Google Scholar] [CrossRef]
- Martín de Llano, J.J.; Fuertes, G.; Torró, I.; García Vicent, C.; Fayos, J.L.; Lurbe, E. Birth Weight and Characteristics of Endothelial and Smooth Muscle Cell Cultures from Human Umbilical Cord Vessels. J. Transl. Med. 2009, 7, 30. [Google Scholar] [CrossRef]
- Van, R.L.; Bayliss, C.E.; Roncari, D.A. Cytological and Enzymological Characterization of Adult Human Adipocyte Precursors in Culture. J. Clin. Investig. 1976, 58, 699–704. [Google Scholar] [CrossRef]
- Shen, H.; An, O.; Ren, X.; Song, Y.; Tang, S.J.; Ke, X.-Y.; Han, J.; Tay, D.J.T.; Ng, V.H.E.; Molias, F.B.; et al. ADARs Act as Potent Regulators of Circular Transcriptome in Cancer. Nat. Commun. 2022, 13, 1508. [Google Scholar] [CrossRef]
- Aktaş, T.; Avşar Ilık, İ.; Maticzka, D.; Bhardwaj, V.; Pessoa Rodrigues, C.; Mittler, G.; Manke, T.; Backofen, R.; Akhtar, A. DHX9 Suppresses RNA Processing Defects Originating from the Alu Invasion of the Human Genome. Nature 2017, 544, 115–119. [Google Scholar] [CrossRef]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA Binding Protein Quaking Regulates Formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef] [PubMed]
- Errichelli, L.; Dini Modigliani, S.; Laneve, P.; Colantoni, A.; Legnini, I.; Capauto, D.; Rosa, A.; De Santis, R.; Scarfò, R.; Peruzzi, G.; et al. FUS Affects Circular RNA Expression in Murine Embryonic Stem Cell-Derived Motor Neurons. Nat. Commun. 2017, 8, 14741. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, B.; Zhao, J.; Li, Q.; Chen, S.; Guo, T.; Li, Y.; Lai, H.; Chen, Z.; Meng, Z.; et al. HNRNPL Circularizes ARHGAP35 to Produce an Oncogenic Protein. Adv. Sci. 2021, 8, 2001701. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.S.; Di Tullio, F.; Schwarz, M.; Low, D.; Incarnato, D.; Gay, F.; Tabaglio, T.; Zhang, J.; Wollmann, H.; Chen, L.; et al. HNRNPM Controls circRNA Biogenesis and Splicing Fidelity to Sustain Cancer Cell Fitness. eLife 2021, 10, e59654. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.F.; Reckman, Y.J.; Aufiero, S.; van den Hoogenhof, M.M.G.; van der Made, I.; Beqqali, A.; Koolbergen, D.R.; Rasmussen, T.B.; van der Velden, J.; Creemers, E.E.; et al. RBM20 Regulates Circular RNA Production From the Titin Gene. Circ. Res. 2016, 119, 996–1003. [Google Scholar] [CrossRef] [PubMed]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA Biogenesis Competes with Pre-mRNA Splicing. Mol. Cell 2014, 56, P55–P66. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, C.-X.; Xue, W.; Zhang, Y.; Jiang, S.; Yin, Q.-F.; Wei, J.; Yao, R.-W.; Yang, L.; Chen, L.-L. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol. Cell 2017, 67, 214–227.e7. [Google Scholar] [CrossRef]
- Muniz, L.; Nicolas, E.; Trouche, D. RNA Polymerase II Speed: A Key Player in Controlling and Adapting Transcriptome Composition. EMBO J. 2021, 40, e105740. [Google Scholar] [CrossRef]
- Fossey, S.C.; Kuroda, S.; Price, J.A.; Pendleton, J.K.; Freedman, B.I.; Bowden, D.W. Identification and Characterization of PRKCBP1, a Candidate RACK-like Protein. Inc. Mouse Genome 2000, 11, 919–925. [Google Scholar] [CrossRef]
- Li, N.; Li, Y.; Lv, J.; Zheng, X.; Wen, H.; Shen, H.; Zhu, G.; Chen, T.-Y.; Dhar, S.S.; Kan, P.-Y.; et al. ZMYND8 Reads the Dual Histone Mark H3K4me1-H3K14ac to Antagonize the Expression of Metastasis-Linked Genes. Mol. Cell 2016, 63, 470–484. [Google Scholar] [CrossRef]
- Wang, D.; Guan, H.; Wang, Y.; Song, G.; Xia, Y. N6-Methyladenosine Modification in Trophoblasts Promotes circSETD2 Expression, Inhibits miR-181a-5p, and Elevates MCL1 Transcription to Reduce Apoptosis of Trophoblasts. Environ. Toxicol. 2023, 38, 422–435. [Google Scholar] [CrossRef] [PubMed]
- Wigle, J.T.; Harvey, N.; Detmar, M.; Lagutina, I.; Grosveld, G.; Gunn, M.D.; Jackson, D.G.; Oliver, G. An Essential Role for Prox1 in the Induction of the Lymphatic Endothelial Cell Phenotype. EMBO J. 2002, 21, 1505–1513. [Google Scholar] [CrossRef] [PubMed]
- Bixel, M.G.; Adams, R.H. Master and Commander: Continued Expression of Prox1 Prevents the Dedifferentiation of Lymphatic Endothelial Cells. Genes Dev. 2008, 22, 3232–3235. [Google Scholar] [CrossRef]
- Harvey, N.L.; Srinivasan, R.S.; Dillard, M.E.; Johnson, N.C.; Witte, M.H.; Boyd, K.; Sleeman, M.W.; Oliver, G. Lymphatic Vascular Defects Promoted by Prox1 Haploinsufficiency Cause Adult-Onset Obesity. Nat. Genet. 2005, 37, 1072–1081. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y. Emerging Mechanisms of Tumour Lymphangiogenesis and Lymphatic Metastasis. Nat. Rev. Cancer 2005, 5, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, R.S.; Escobedo, N.; Yang, Y.; Interiano, A.; Dillard, M.E.; Finkelstein, D.; Mukatira, S.; Gil, H.J.; Nurmi, H.; Alitalo, K.; et al. The Prox1-Vegfr3 Feedback Loop Maintains the Identity and the Number of Lymphatic Endothelial Cell Progenitors. Genes Dev. 2014, 28, 2175–2187. [Google Scholar] [CrossRef] [PubMed]
- Morfoisse, F.; De Toni, F.; Nigri, J.; Hosseini, M.; Zamora, A.; Tatin, F.; Pujol, F.; Sarry, J.-E.; Langin, D.; Lacazette, E.; et al. Coordinating Effect of VEGFC and Oleic Acid Participates to Tumor Lymphangiogenesis. Cancers 2021, 13, 2851. [Google Scholar] [CrossRef]
- Erber, R.; Eichelsbacher, U.; Powajbo, V.; Korn, T.; Djonov, V.; Lin, J.; Hammes, H.-P.; Grobholz, R.; Ullrich, A.; Vajkoczy, P. EphB4 Controls Blood Vascular Morphogenesis during Postnatal Angiogenesis. EMBO J. 2006, 25, 628–641. [Google Scholar] [CrossRef]
- Martin-Almedina, S.; Martinez-Corral, I.; Holdhus, R.; Vicente, A.; Fotiou, E.; Lin, S.; Petersen, K.; Simpson, M.A.; Hoischen, A.; Gilissen, C.; et al. EPHB4 Kinase-Inactivating Mutations Cause Autosomal Dominant Lymphatic-Related Hydrops Fetalis. J. Clin. Investig. 2016, 126, 3080–3088. [Google Scholar] [CrossRef]
- Vivanti, A.; Ozanne, A.; Grondin, C.; Saliou, G.; Quevarec, L.; Maurey, H.; Aubourg, P.; Benachi, A.; Gut, M.; Gut, I.; et al. Loss of Function Mutations in EPHB4 Are Responsible for Vein of Galen Aneurysmal Malformation. Brain 2018, 141, 979–988. [Google Scholar] [CrossRef]
- Alto, L.T.; Terman, J.R. Semaphorins and Their Signaling Mechanisms. Methods Mol. Biol. 2017, 1493, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Gabrovska, P.N.; Smith, R.A.; Tiang, T.; Weinstein, S.R.; Haupt, L.M.; Griffiths, L.R. Semaphorin–Plexin Signalling Genes Associated with Human Breast Tumourigenesis. Gene 2011, 489, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Acevedo, L.M.; Barillas, S.; Weis, S.M.; Göthert, J.R.; Cheresh, D.A. Semaphorin 3A Suppresses VEGF-Mediated Angiogenesis yet Acts as a Vascular Permeability Factor. Blood 2008, 111, 2674–2680. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-κB Signaling in Inflammation. Sig. Transduct. Target. Ther. 2017, 2, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, T.; Zheng, G. Exosome-Transmitted Circ-CARD6 Facilitates Posterior Capsule Opacification Development by miR-31/FGF7 Axis. Exp. Eye Res. 2021, 207, 108572. [Google Scholar] [CrossRef] [PubMed]
- Leslie Pedrioli, D.M.; Karpanen, T.; Dabouras, V.; Jurisic, G.; van de Hoek, G.; Shin, J.W.; Marino, D.; Kälin, R.E.; Leidel, S.; Cinelli, P.; et al. miR-31 Functions as a Negative Regulator of Lymphatic Vascular Lineage-Specific Differentiation In Vitro and Vascular Development In Vivo. Mol. Cell Biol. 2010, 30, 3620–3634. [Google Scholar] [CrossRef] [PubMed]
- Lessard, L.; Liu, M.; Marzese, D.M.; Wang, H.; Chong, K.; Kawas, N.; Donovan, N.C.; Kiyohara, E.; Hsu, S.; Nelson, N.; et al. The CASC15 Long Intergenic Noncoding RNA Locus Is Involved in Melanoma Progression and Phenotype Switching. J. Investig. Dermatol. 2015, 135, 2464–2474. [Google Scholar] [CrossRef]
- Wu, Q.; Xiang, S.; Ma, J.; Hui, P.; Wang, T.; Meng, W.; Shi, M.; Wang, Y. Long Non-Coding RNA CASC15 Regulates Gastric Cancer Cell Proliferation, Migration and Epithelial Mesenchymal Transition by Targeting CDKN1A and ZEB1. Mol. Oncol. 2018, 12, 799–813. [Google Scholar] [CrossRef]
- Fernando, T.R.; Contreras, J.R.; Zampini, M.; Rodriguez-Malave, N.I.; Alberti, M.O.; Anguiano, J.; Tran, T.M.; Palanichamy, J.K.; Gajeton, J.; Ung, N.M.; et al. The lncRNA CASC15 Regulates SOX4 Expression in RUNX1-Rearranged Acute Leukemia. Mol. Cancer 2017, 16, 126. [Google Scholar] [CrossRef]
- Jin, C.; Zhao, J.; Zhang, Z.-P.; Wu, M.; Li, J.; Liu, B.; Bin Liao, X.-; Liao, Y.-X.; Liu, J.-P. CircRNA EPHB4 Modulates Stem Properties and Proliferation of Gliomas via Sponging miR-637 and up-Regulating SOX10. Mol. Oncol. 2021, 15, 596–622. [Google Scholar] [CrossRef]
- Zhan, R.; Leng, X.; Liu, X.; Wang, X.; Gong, J.; Yan, L.; Wang, L.; Wang, Y.; Wang, X.; Qian, L.-J. Heat Shock Protein 70 Is Secreted from Endothelial Cells by a Non-Classical Pathway Involving Exosomes. Biochem. Biophys. Res. Commun. 2009, 387, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, H.; Heikamp, E.; Turley, H.; Dragovic, R.; Thomas, P.; Oon, C.E.; Leek, R.; Edelmann, M.; Kessler, B.; Sainson, R.C.A.; et al. New Mechanism for Notch Signaling to Endothelium at a Distance by Delta-like 4 Incorporation into Exosomes. Blood 2010, 116, 2385–2394. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA Is Enriched and Stable in Exosomes: A Promising Biomarker for Cancer Diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef] [PubMed]
- Tuo, B.; Chen, Z.; Dang, Q.; Chen, C.; Zhang, H.; Hu, S.; Sun, Z. Roles of Exosomal circRNAs in Tumour Immunity and Cancer Progression. Cell Death Dis. 2022, 13, 1–9. [Google Scholar] [CrossRef]
- Li, J.; Li, Z.; Jiang, P.; Peng, M.; Zhang, X.; Chen, K.; Liu, H.; Bi, H.; Liu, X.; Li, X. Circular RNA IARS (Circ-IARS) Secreted by Pancreatic Cancer Cells and Located within Exosomes Regulates Endothelial Monolayer Permeability to Promote Tumor Metastasis. J. Exp. Clin. Cancer Res. 2018, 37, 177. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]

| circBase ID | Gene Name | circRNA Name | circRNA Localization | Strand | Origin | Genomic Lenght (pb) | Spliced Lenght (nt) | Gene Description [Sources] | Gene Associated Pathologies | Known circRNA Functions | Sources (Pubmed ID) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| hsa_circ_0111 952 | PROX1 | circPROX1 | chr1:21399646 9-214011715 | + | exonic | 15247 | 2095 | prospero homeobox 1_protein coding transcription factor activity-cell specification [HGNC: 9459; NCBI: 5629] | lymphatic malfunctions; cancers; obesity | none | 19056879; 16170315 28024299; 22023334 |
| #N/A | IQSEC1 | circlQSEC1 | chr3:13129305 -13129525 | − | intronic | 221 | 221 | IQ Motif And Sec7 Domain ArtGEF 1 _protein coding GEF activity-endosomal protein traffic [HGNC: 29112 NCBI: 9922] | intellectual development disorder | none | 31607425 |
| hsa_dic_0008 285 | CDYL | circCDYL | chr6:4891713- 4892379 | − | exonic | 667 | 667 | Chromodomain Y Like_protein coding histone modification- transcriptional repression [HGNC: 1811; NCBI: 9425] | cancer chemoresistance | pro-myocardial regeneration in vitro | 19061646;31367252 32522972 |
| hsa_circ_0001 727 | ZKSCAN1 | circZKSCAN1 | chr7:10002341 9-100024307 | + | exonic | 889 | 668 | Zinc Finger With KRAB And SCAN Domains 1 _protein coding transcription factor activity-[HGNC: 13101; NCBI 7586] | gastric cancer, Hepatocellular cancer | anti-hepatocellular cancers progression | 7557990; 28211215 33439397 |
| hsa_dirc_0005 895 | CARD6 | circCARD6 | chr5:40852174 40854096 | + | exonic | 1923 | 1923 | Caspase Recruitment Domain Family Member 6 protein coding [HGNC: 16394; NCBI: 84674] | inflamatory bowel diseases; pro- intestinal cancers | pro-posterior capsul opacification (eye) | 12775719; 16418290 20025480; 33844960, |
| hsa_drc_0002 472 | PLXNA2 | circPLXNA2 | chr1:20821028 0-208218002 | − | exonic | 7723 | 1451 | Plexin A2 protein coding [HGNC: 9100 NCBI 5362] | axonal guidance disfunctions Breast cancer tumorigenesis | pro-profiferation and anti-apoptotic in myoblasts | 16402134;21925246 10.3390ljjms24065459 |
| hsa_drc_0000 914 | FKBP8 | circFKBP8 | chr19:1853760 1-18538436 | − | exonic | 836 | 394 | FKBP Proly Isomerase 8 _protein coding [HGNC: 3724; NCBI: 23770] | spina bifida | none | 18003640; 32969478 |
| #N/A | ERCC6L2 | circERCO6L2 | chr9:95978061 -96004701 | + | exonic | 26641 | 337 | ERCC Excision Repair 6 Like 2 _protein coding [HGNC: 26922 NCBI 375748] | bone marrow failure syndrome | none | 4507776 |
| #N/A | FIRRE | circFIRRE | chrX:13174930 6-131794466 | − | exonic | 45161 | 872 | Firre Intergenic Repeating RNA Element_IncRNA [HGNC: 49627; NCBI 286467] | various cancers | chrX inactivation and nuclear organisation? RNA stability? | 35110535;3 35988459 29678151;30124921; |
| hsa_dirc_0007 026 | ZMYND8 | dircZMYND8_A | chr20:4726228 8-47276795 | − | exonic | 14508 | 623 | Zinc Finger MYND-Type Containing 8 protein coding [HGNC: 9397; NCBI: 23613] | various cancers | none | 11003709; 27477906 |
| hsa_drc_0003 028 | FUT8 | circFUT8 | chr14:6556133 7-65561766 | + | exonic | 430 | 430 | Fucosyltransferase 8 _protein coding [HGNC: 4019 NCBI: 2530] | various cancers; congenital disorders | cancer suppression or promotion? | 19302290; 26289314; 29304374;32072011; 33500381 |
| #N/A | SERPINE 1 | circSERPINE1 | chr7:10113191 6-101135801 | + | exonic | 3886 | 541 | Serpin Family E Member 1_protein coding [HGNC: 8583 NCBI: 5054] | Thrombosis PAI1-deficiency | none | 3922531; 9207454 |
| hsa_dirc_0087 960 | LPAR1 | airdLPAR1 | chr9:11097207 3-110973558 | − | exonic | 1486 | 226 | Lysophosphatidic Receptor 1_protein coding [HGNC: 3166 NCBI: 1902] | pertusis | invasive bladder cancer biomarker | 30867795;9804623 |
| #N/A | ABHD14B | dirAAHB14B | chr3:51968816 -51969034 | − | exonic | 219 | 219 | Abhydrolase Domain Containing 14B _protein coding [HGNC: 28235 NCBI 84836] | none | none | |
| hsa_drc_0128 684 | HNRNPA B | circHNRNPAB | chr5:17821063 5-178210877 | + | exonic | 243 | 243 | Heterogeneous Nuclear Ribonudeoprotein A/B protein coding [HGNC: 5034 NCBI: 3182] | none | none | |
| hsa_circ_0005 015 | HAS2 | circHAS2 | chr8:12162871 4-121629340 | − | exonic | 627 | 627 | Hyaluronan Synthase 2 _protein coding [HGNC: 4819 NCBI: 3037] | breast cancer | diabeties retinopathy biomarker | 29288268; 22113945; 33954907 |
| hsa_drc_0001 461 | FAT1 | circFAT1 | chr4:18670656 3-186709845 | − | exonic | 3283 | 3283 | FAT Atypical Cadherin 1_protein coding [HGNC: 3595 NCBI: 2195] | various cancers | pro-cancer cell stemness; pro-breast cancer drug resistance osteoblast differentiation | 34314629; 34288822 35003269; 23354438; |
| Cell Line | Run | Spots (Millions) | Bases | Size | GC Content | Published | Access Type |
|---|---|---|---|---|---|---|---|
| HDLEC | Acc. Numb.pending | 105.3 | 15.8 G | 16.3 G | 47.00% | 15 November 2023 | public |
| HDLEC | Acc. Numb.pending | 107.7 | 16.1 G | 16.7 G | 48.00% | 15 November 2023 | public |
| HDLEC | Acc. Numb.pending | 96 | 14.4 G | 15 G | 48.00% | 15 November 2023 | public |
| HUAEC | SRR3192390 | 200.7 | 40.5 G | 26.1 G | 56.80% | 29 March 2016 | public |
| HUAEC | SRR3192391 | 88.2 | 17.8 G | 9.9 G | 53.20% | 29 March 2016 | public |
| HUVEC | SRR1959051 | 42.8 | 8.3 G | 5.3 G | 51.80% | 6 September 2016 | public |
| HUVEC | SRR1959052 | 42.1 | 8.2 G | 5.2 G | 52.10% | 6 September 2016 | public |
| HCAEC | SRR9163311 | 12.8 | 3.4 G | 1.0 G | 54.00% | 31 May 2019 | public |
| HCAEC | SRR9163312 | 9.2 | 2.4 G | 752.7 M | 54.30% | 31 May 2019 | public |
| HCAEC | SRR9163313 | 9.3 | 2.5 G | 755.0 M | 52.90% | 31 May 2019 | public |
| HCAEC | SRR9163314 | 5.3 | 1.4 G | 429.2 M | 53.20% | 31 May 2019 | public |
| SAT-SVF | SRR9163319 | 13.5 | 3.6 G | 1.1 G | 53.40% | 31 May 2019 | public |
| SAT-SVF | SRR9163320 | 7.6 | 2.0 G | 610.9 M | 54.40% | 31 May 2019 | public |
| SAT-SVF | SRR9163321 | 10.2 | 2.7 G | 824.3 M | 54.70% | 31 May 2019 | public |
| SAT-SVF | SRR9163322 | 8.3 | 2.2 G | 677.6 M | 53.90% | 31 May 2019 | public |
| SMC_UA | SRR3192392 | 129.4 | 26.1 G | 15.4 G | 56.10% | 29 March 2016 | public |
| SMC_UA | SRR3192393 | 175.4 | 35.4 G | 22.1 G | 52.60% | 29 March 2016 | public |
| Primer Sequences | |||
|---|---|---|---|
| Cell Type Specificity | Targeted Transcripts | Forward | Reverse |
| All | GAPDH | TCAAGGCTGAGAACGGGAAG | CGCCCCACTTGATTTTGGAG |
| circCDYL | GCTGTTAACGGGAAAGGTTGA | GTCCTCGCTGTCATAGCCTT | |
| CDYL | GGCTTCACCCACATCTTGTT | TACCAGCTTGCTGTCATCGG | |
| circZKSCAN1 | AAACCCCGCCTCTTACAGTC | AAACAGGGTCTGTGCTCACC | |
| ZKSCAN1 | ACATTCGTCTCGGAAACCCC | GGTCTCTGGGACTACCCTCA | |
| Endothelial | circCARD6 | GCAAGGAGTCCAGATGAAGACA | CCTTTCTGCTTCTATCCATGTTCA |
| CARD6 | CCATTTGCGGCTTAAGGCAT | TGAATTACTGCTTGCCCCCAT | |
| circPLXNA2 | CTGTGGCCTCCTACGTTTACA | GCCCTCACATGATTCTTTTTCA | |
| PLXNA2 | GTCAAGTGCTCCAACCCTCA | GCGATCTCGGAGAAGTCCAG | |
| Umbilical | circFKBP8 | ACAACATCAAGGCTCTCTTCCG | GTGAGCATCTCCAGGTCAGG |
| FKBP8 | GCCAGACAACATCAAGGCTCT | AAGGTTCCAGCTTCAGGGCT | |
| circERCC6L2 | ACCATACAAACCAGACCACCTT | CTTCCTGAACGCCATCTGCG | |
| ERCC6L2 | AGGGTGCATTCTGGGTGATG | CCTCACGAGTTCCCTTTTTATGC | |
| Non-endothelial | circLPAR1 | GGCTGCCATCTCTACTTCCAT | ACTCAGATAGGTGGATGGGGA |
| LPAR1 | GGCTGCCATCTCTACTTCCAT | AGGCAATGGACTCGTTGTAGA | |
| HDLEC | circP2 | AGACTGTGAGCTGTACAGGG | GTGCTGTCATGGTCAGGCAT |
| PROX1 | ATCAACGATGGGGTCACCAG | GGATCAACATCTTTGCCTGCG | |
| circIQSEC1 | GACACAAATTACCAATACCAGGAATGAGAA | CCACACTGATATTTTTCTTCGTCCTTTGG | |
| IQSEC1 | CTATGAGCTCTCCTCGGACCT | GTACTGGCGAAACGCCGTC | |
| HUAEC | circFIRRE | TGCAGATACGATGCTGAGTGAA | ACTGACACCTTAGTCTCCTCA |
| FIRRE | GGGAAGACTTGGTTGTGCAGAA | CCAGCCAGGATTGCTCCAGT | |
| HUVEC | circZMYND8_A | GTCAGCTCCTATCACGACGA | ATTTATTGGAACCCAGGCCCC |
| ZMYND8 | TGTGAACATGAGATGAATGAAATCG | CATCGACCTGCCCGTCTTTA | |
| circFUT8 | AGCCGAGAACTGTCCAAGAT | TATTGTCCTGTACTTCATGCGC | |
| FUT8 | CCCACAGCCTTGGCTAGAAA | CTGTGCGTCTGACATGGACT | |
| HCAEC | circSERPINE1 | AGATCGAGGTGAACGAGAGTG | GAGTCGGGGAAGGGAGTCTT |
| SERPINE1 | TCGCAAGGCACCTCTGAGAAC | GCAGACCCTTCACCAAAGACA | |
| SAT-SVF | circABHD14B | CCTCAAGCGAAGGGTCATATTTGGA | ATGAGCCTCCACACAAGCACT |
| ABHD14B | GAACCTGGGTACACTGCACA | GCTGCTGCTTCCTTGGAGT | |
| circHNRNPAB | TTAGGCAGCGTGTGGTGTCT | CCAAACAAAGCATGTGTGCGATC | |
| HNRNPAB | AACCCGTGAAGAAGGTTCTGG | ATAGACTTCTTTGGGCTGGGC | |
| SMC-UA | circHAS2 | CTTCAGAGCACTGGGACGAA | TCCAAGGAGGAGAGAGACTCC |
| HAS2 | TGTACACAGCCTTCAGAGCA | GGCTGGGTCAAGCATAGTGT | |
| circFAT1 | TGGTAATGACGGTGTCGGCT | GGCTGCCATCACTGTCTCCAA | |
| FAT1 | AACCCTTGCCAGAATGGAGG | AGGAACACGGATTGACGCTT | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diallo, L.H.; Mariette, J.; Laugero, N.; Touriol, C.; Morfoisse, F.; Prats, A.-C.; Garmy-Susini, B.; Lacazette, E. Specific Circular RNA Signature of Endothelial Cells: Potential Implications in Vascular Pathophysiology. Int. J. Mol. Sci. 2024, 25, 680. https://doi.org/10.3390/ijms25010680
Diallo LH, Mariette J, Laugero N, Touriol C, Morfoisse F, Prats A-C, Garmy-Susini B, Lacazette E. Specific Circular RNA Signature of Endothelial Cells: Potential Implications in Vascular Pathophysiology. International Journal of Molecular Sciences. 2024; 25(1):680. https://doi.org/10.3390/ijms25010680
Chicago/Turabian StyleDiallo, Leïla Halidou, Jérôme Mariette, Nathalie Laugero, Christian Touriol, Florent Morfoisse, Anne-Catherine Prats, Barbara Garmy-Susini, and Eric Lacazette. 2024. "Specific Circular RNA Signature of Endothelial Cells: Potential Implications in Vascular Pathophysiology" International Journal of Molecular Sciences 25, no. 1: 680. https://doi.org/10.3390/ijms25010680
APA StyleDiallo, L. H., Mariette, J., Laugero, N., Touriol, C., Morfoisse, F., Prats, A.-C., Garmy-Susini, B., & Lacazette, E. (2024). Specific Circular RNA Signature of Endothelial Cells: Potential Implications in Vascular Pathophysiology. International Journal of Molecular Sciences, 25(1), 680. https://doi.org/10.3390/ijms25010680






