Chromatin Organization after High-LET Irradiation Revealed by Super-Resolution STED Microscopy
Abstract
1. Introduction
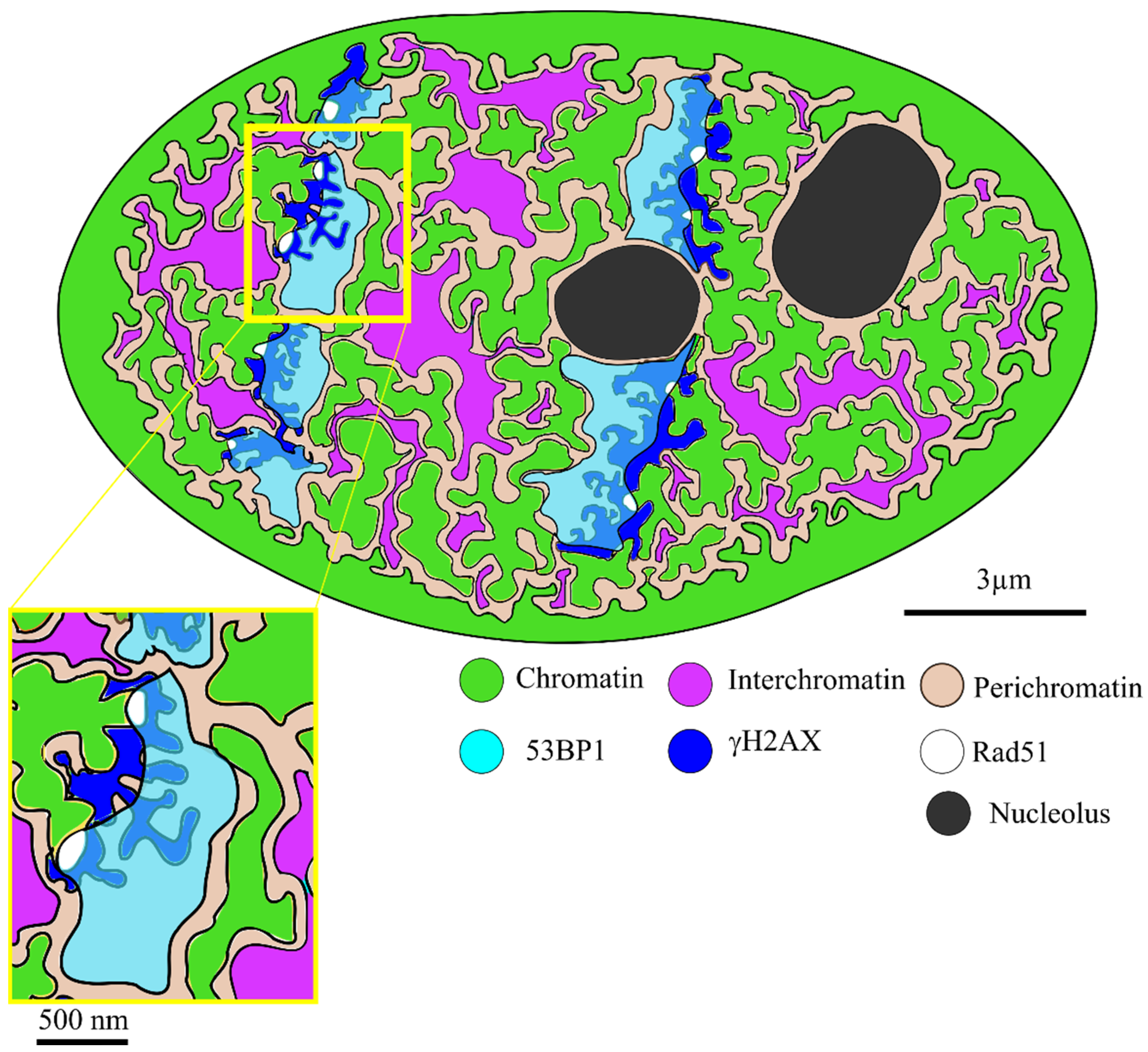
2. Results
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Irradiation
4.2. Antibodies and Immunofluorescence Detection
4.3. Microscopy
4.4. Heterochromatin Isolation
4.5. Overlap Analysis
4.6. Distillation of Chromatin Network
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rothkamm, K.; Barnard, S.; Moquet, J.; Ellender, M.; Rana, Z.; Burdak-Rothkamm, S. DNA damage foci: Meaning and significance. Environ. Mol. Mutagen. 2015, 56, 491–504. [Google Scholar] [CrossRef]
- Natarajan, A.T.; Palitti, F. DNA repair and chromosomal alterations. Mutat. Res. 2008, 657, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Pardo, B.; Gómez-González, B.; Aguilera, A. DNA repair in mammalian cells: DNA double-strand break repair: How to fix a broken relationship. Cell. Mol. Life Sci. 2009, 66, 1039–1056. [Google Scholar] [CrossRef]
- Shibata, A.; Conrad, S.; Birraux, J.; Geuting, V.; Barton, O.; Ismail, A.; Kakarougkas, A.; Meek, K.; Taucher-Scholz, G.; Löbrich, M.; et al. Factors determining DNA double-strand break repair pathway choice in G2 phase. EMBO J. 2011, 30, 1079–1092. [Google Scholar] [CrossRef] [PubMed]
- Schipler, A.; Iliakis, G. DNA double-strand-break complexity levels and their possible contributions to the probability for error-prone processing and repair pathway choice. Nucleic Acids Res. 2013, 41, 7589–7605. [Google Scholar] [CrossRef] [PubMed]
- Rothkamm, K.; Krüger, I.; Thompson, L.H.; Löbrich, M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 2003, 23, 5706–5715. [Google Scholar] [CrossRef] [PubMed]
- Baumann, P.; West, S.C. Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem. Sci. 1998, 23, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Sung, P.; Krejci, L.; van Komen, S.; Sehorn, M.G. Rad51 recombinase and recombination mediators. J. Biol. Chem. 2003, 278, 42729–42732. [Google Scholar] [CrossRef]
- Reindl, J.; Girst, S.; Walsh, D.W.M.; Greubel, C.; Schwarz, B.; Siebenwirth, C.; Drexler, G.A.; Friedl, A.A.; Dollinger, G. Chromatin organization revealed by nanostructure of irradiation induced γH2AX, 53BP1 and Rad51 foci. Sci. Rep. 2017, 7, 40616. [Google Scholar] [CrossRef]
- Schwarz, B.; Friedl, A.A.; Girst, S.; Dollinger, G.; Reindl, J. Nanoscopic analysis of 53BP1, BRCA1 and Rad51 reveals new insights in temporal progression of DNA-repair and pathway choice. Mutat. Res. 2019, 816–818, 111675. [Google Scholar] [CrossRef]
- Adams, M.M.; Carpenter, P.B. Tying the loose ends together in DNA double strand break repair with 53BP1. Cell Div. 2006, 1, 19. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Kwak, H.-J.; Cho, I.-t.; Park, S.H.; Lee, C.-H. S1219 residue of 53BP1 is phosphorylated by ATM kinase upon DNA damage and required for proper execution of DNA damage response. Biochem. Biophys. Res. Commun. 2009, 378, 32–36. [Google Scholar] [CrossRef] [PubMed]
- Hable, V.; Drexler, G.A.; Brüning, T.; Burgdorf, C.; Greubel, C.; Derer, A.; Seel, J.; Strickfaden, H.; Cremer, T.; Friedl, A.A.; et al. Recruitment kinetics of DNA repair proteins Mdc1 and Rad52 but not 53BP1 depend on damage complexity. PLoS ONE 2012, 7, e41943. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Fong, K.-W.; Wang, J.; Wang, W.; Chen, J. RIF1 counteracts BRCA1-mediated end resection during DNA repair. J. Biol. Chem. 2013, 288, 11135–11143. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Hunt, C.R.; Chakraborty, S.; Pandita, R.K.; Yordy, J.; Ramnarain, D.B.; Horikoshi, N.; Pandita, T.K. Role of 53BP1 in the regulation of DNA double-strand break repair pathway choice. Radiat. Res. 2014, 181, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, M.; Lottersberger, F.; Buonomo, S.B.; Sfeir, A.; de Lange, T. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science 2013, 339, 700–704. [Google Scholar] [CrossRef]
- Huen, M.S.Y.; Huang, J.; Leung, J.W.C.; Sy, S.M.-H.; Leung, K.M.; Ching, Y.-P.; Tsao, S.W.; Chen, J. Regulation of chromatin architecture by the PWWP domain-containing DNA damage-responsive factor EXPAND1/MUM1. Mol. Cell 2010, 37, 854–864. [Google Scholar] [CrossRef]
- Escribano-Diaz, C.; Durocher, D. DNA repair pathway choice—A PTIP of the hat to 53BP1. EMBO Rep. 2013, 14, 665–666. [Google Scholar] [CrossRef]
- Reindl, J.; Drexler, G.A.; Girst, S.; Greubel, C.; Siebenwirth, C.; Drexler, S.E.; Dollinger, G.; Friedl, A.A. Nanoscopic exclusion between Rad51 and 53BP1 after ion irradiation in human HeLa cells. Phys. Biol. 2015, 12, 66005. [Google Scholar] [CrossRef]
- Ochs, F.; Somyajit, K.; Altmeyer, M.; Rask, M.-B.; Lukas, J.; Lukas, C. 53BP1 fosters fidelity of homology-directed DNA repair. Nat. Struct. Mol. Biol. 2016, 23, 714–721. [Google Scholar] [CrossRef]
- Klosin, A.; Hyman, A.A. Molecular biology: A liquid reservoir for silent chromatin. Nature 2017, 547, 168–170. [Google Scholar] [CrossRef] [PubMed]
- Phipps, J.; Dubrana, K. DNA Repair in Space and Time: Safeguarding the Genome with the Cohesin Complex. Genes 2022, 13, 198. [Google Scholar] [CrossRef] [PubMed]
- Lisby, M.; Mortensen, U.H.; Rothstein, R. Colocalization of multiple DNA double-strand breaks at a single Rad52 repair centre. Nat. Cell Biol. 2003, 5, 572–577. [Google Scholar] [CrossRef] [PubMed]
- Caron, P.; Choudjaye, J.; Clouaire, T.; Bugler, B.; Daburon, V.; Aguirrebengoa, M.; Mangeat, T.; Iacovoni, J.S.; Álvarez-Quilón, A.; Cortés-Ledesma, F.; et al. Non-redundant Functions of ATM and DNA-PKcs in Response to DNA Double-Strand Breaks. Cell Rep. 2015, 13, 1598–1609. [Google Scholar] [CrossRef] [PubMed]
- Ortega, P.; Gómez-González, B.; Aguilera, A. Heterogeneity of DNA damage incidence and repair in different chromatin contexts. DNA Repair 2021, 107, 103210. [Google Scholar] [CrossRef] [PubMed]
- Yasuhara, T.; Zou, L. Impacts of chromatin dynamics and compartmentalization on DNA repair. DNA Repair 2021, 105, 103162. [Google Scholar] [CrossRef] [PubMed]
- Minami, K.; Iida, S.; Maeshima, K. Chromatin organization and DNA damage. Enzymes 2022, 51, 29–51. [Google Scholar] [CrossRef] [PubMed]
- Fakan, S.; van Driel, R. The perichromatin region: A functional compartment in the nucleus that determines large-scale chromatin folding. Semin. Cell Dev. Biol. 2007, 18, 676–681. [Google Scholar] [CrossRef]
- Cremer, T.; Cremer, M. Chromosome territories. Cold Spring Harb. Perspect. Biol. 2010, 2, a003889. [Google Scholar] [CrossRef]
- Albiez, H.; Cremer, M.; Tiberi, C.; Vecchio, L.; Schermelleh, L.; Dittrich, S.; Küpper, K.; Joffe, B.; Thormeyer, T.; von Hase, J.; et al. Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res. 2006, 14, 707–733. [Google Scholar] [CrossRef]
- Dion, V.; Gasser, S.M. Chromatin movement in the maintenance of genome stability. Cell 2013, 152, 1355–1364. [Google Scholar] [CrossRef] [PubMed]
- Shinkai, S.; Nozaki, T.; Maeshima, K.; Togashi, Y. Dynamic Nucleosome Movement Provides Structural Information of Topological Chromatin Domains in Living Human Cells. PLoS Comput. Biol. 2016, 12, e1005136. [Google Scholar] [CrossRef] [PubMed]
- Tacke, R.; Manley, J.L. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995, 14, 3540–3551. [Google Scholar] [CrossRef] [PubMed]
- Spector, D.L.; Lamond, A.I. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 2011, 3, a000646. [Google Scholar] [CrossRef]
- Fraser, P.; Bickmore, W. Nuclear organization of the genome and the potential for gene regulation. Nature 2007, 447, 413–417. [Google Scholar] [CrossRef]
- Morgan, G.T. Imaging the dynamics of transcription loops in living chromosomes. Chromosoma 2018, 127, 361–374. [Google Scholar] [CrossRef]
- Dabin, J.; Mori, M.; Polo, S.E. The DNA damage response in the chromatin context: A coordinated process. Curr. Opin. Cell Biol. 2023, 82, 102176. [Google Scholar] [CrossRef]
- Rouquette, J.; Genoud, C.; Vazquez-Nin, G.H.; Kraus, B.; Cremer, T.; Fakan, S. Revealing the high-resolution three-dimensional network of chromatin and interchromatin space: A novel electron-microscopic approach to reconstructing nuclear architecture. Chromosome Res. 2009, 17, 801–810. [Google Scholar] [CrossRef]
- Cremer, T.; Küpper, K.; Dietzel, S.; Fakan, S. Higher order chromatin architecture in the cell nucleus: On the way from structure to function. Biol. Cell 2004, 96, 555–567. [Google Scholar] [CrossRef]
- Fakan, S. The functional architecture of the nucleus as analysed by ultrastructural cytochemistry. Histochem. Cell Biol. 2004, 122, 83–93. [Google Scholar] [CrossRef]
- Barzel, A.; Kupiec, M. Finding a match: How do homologous sequences get together for recombination? Nat. Rev. Genet. 2008, 9, 27–37. [Google Scholar] [CrossRef]
- Renkawitz, J.; Lademann, C.A.; Jentsch, S. Mechanisms and principles of homology search during recombination. Nat. Rev. Mol. Cell Biol. 2014, 15, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Greene, E.C. DNA Sequence Alignment during Homologous Recombination. J. Biol. Chem. 2016, 291, 11572–11580. [Google Scholar] [CrossRef] [PubMed]
- Mersch, J.; Jackson, M.A.; Park, M.; Nebgen, D.; Peterson, S.K.; Singletary, C.; Arun, B.K.; Litton, J.K. Cancers associated with BRCA1 and BRCA2 mutations other than breast and ovarian. Cancer 2015, 121, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Marnef, A.; Legube, G. Organizing DNA repair in the nucleus: DSBs hit the road. Curr. Opin. Cell Biol. 2017, 46, 1–8. [Google Scholar] [CrossRef]
- Jakob, B.; Splinter, J.; Conrad, S.; Voss, K.-O.; Zink, D.; Durante, M.; Löbrich, M.; Taucher-Scholz, G. DNA double-strand breaks in heterochromatin elicit fast repair protein recruitment, histone H2AX phosphorylation and relocation to euchromatin. Nucleic Acids Res. 2011, 39, 6489–6499. [Google Scholar] [CrossRef]
- Guo, X.; Bai, Y.; Zhao, M.; Zhou, M.; Shen, Q.; Yun, C.-H.; Zhang, H.; Zhu, W.-G.; Wang, J. Acetylation of 53BP1 dictates the DNA double strand break repair pathway. Nucleic Acids Res. 2018, 46, 689–703. [Google Scholar] [CrossRef]
- Xu, Y.; Ning, S.; Wei, Z.; Xu, R.; Xu, X.; Xing, M.; Guo, R.; Xu, D. 53BP1 and BRCA1 control pathway choice for stalled replication restart. eLife 2017, 6, e30523. [Google Scholar] [CrossRef]
- Shanbhag, N.M.; Rafalska-Metcalf, I.U.; Balane-Bolivar, C.; Janicki, S.M.; Greenberg, R.A. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 2010, 141, 970–981. [Google Scholar] [CrossRef]
- Beli, P.; Lukashchuk, N.; Wagner, S.A.; Weinert, B.T.; Olsen, J.V.; Baskcomb, L.; Mann, M.; Jackson, S.P.; Choudhary, C. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol. Cell 2012, 46, 212–225. [Google Scholar] [CrossRef]
- Lopez Perez, R.; Best, G.; Nicolay, N.H.; Greubel, C.; Rossberger, S.; Reindl, J.; Dollinger, G.; Weber, K.-J.; Cremer, C.; Huber, P.E. Superresolution light microscopy shows nanostructure of carbon ion radiation-induced DNA double-strand break repair foci. FASEB J. 2016, 30, 2767–2776. [Google Scholar] [CrossRef] [PubMed]
- Varga, D.; Majoros, H.; Ujfaludi, Z.; Erdélyi, M.; Pankotai, T. Quantification of DNA damage induced repair focus formation via super-resolution dSTORM localization microscopy. Nanoscale 2019, 11, 14226–14236. [Google Scholar] [CrossRef] [PubMed]
- Liddle, P.; Jara-Wilde, J.; Lafon-Hughes, L.; Castro, I.; Härtel, S.; Folle, G. dSTORM microscopy evidences in HeLa cells clustered and scattered γH2AX nanofoci sensitive to ATM, DNA-PK, and ATR kinase inhibitors. Mol. Cell. Biochem. 2020, 473, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Hausmann, M.; Wagner, E.; Lee, J.-H.; Schrock, G.; Schaufler, W.; Krufczik, M.; Papenfuß, F.; Port, M.; Bestvater, F.; Scherthan, H. Super-resolution localization microscopy of radiation-induced histone H2AX-phosphorylation in relation to H3K9-trimethylation in HeLa cells. Nanoscale 2018, 10, 4320–4331. [Google Scholar] [CrossRef]
- Hauptner, A.; Dietzel, S.; Drexler, G.A.; Reichart, P.; Krücken, R.; Cremer, T.; Friedl, A.A.; Dollinger, G. Microirradiation of cells with energetic heavy ions. Radiat. Environ. Biophys. 2004, 42, 237–245. [Google Scholar] [CrossRef]
- Imai, R.; Nozaki, T.; Tani, T.; Kaizu, K.; Hibino, K.; Ide, S.; Tamura, S.; Takahashi, K.; Shribak, M.; Maeshima, K. Density imaging of heterochromatin in live cells using orientation-independent-DIC microscopy. Mol. Biol. Cell 2017, 28, 3349–3359. [Google Scholar] [CrossRef]
- Lee, T.C.; Kashyap, R.L.; Chu, C.N. Building Skeleton Models via 3-D Medial Surface Axis Thinning Algorithms. CVGIP Graph. Models Image Process. 1994, 56, 462–478. [Google Scholar] [CrossRef]
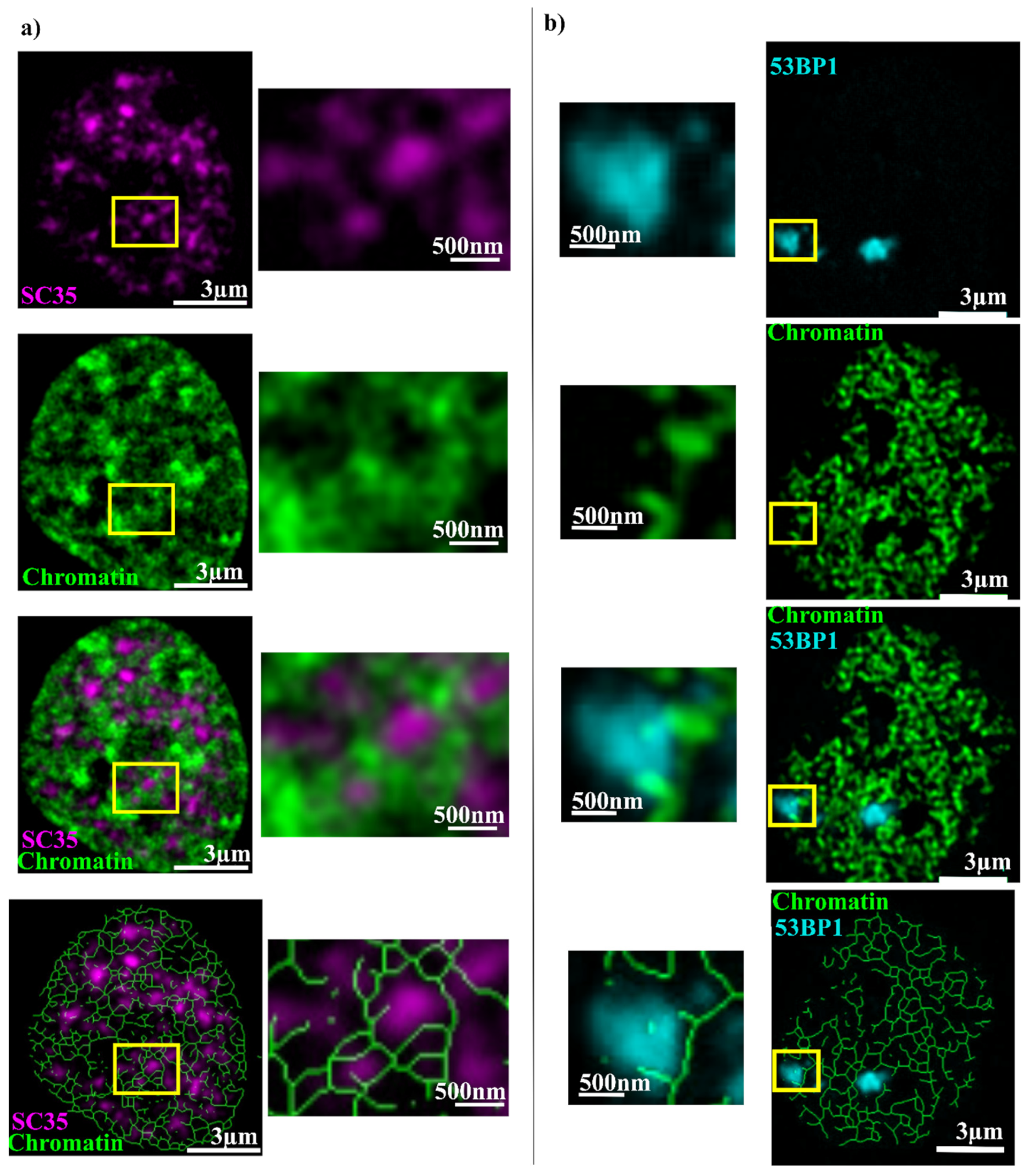
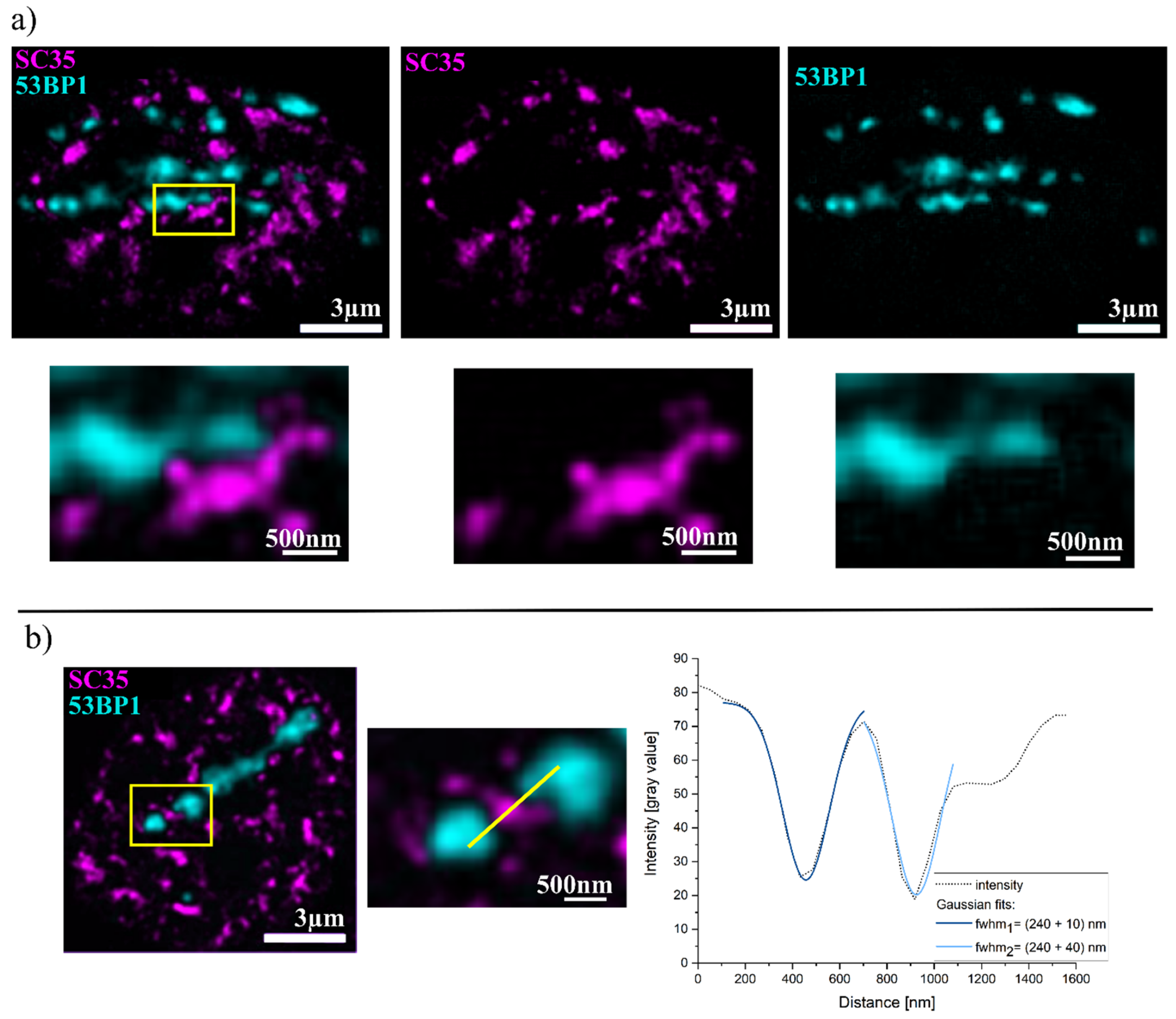
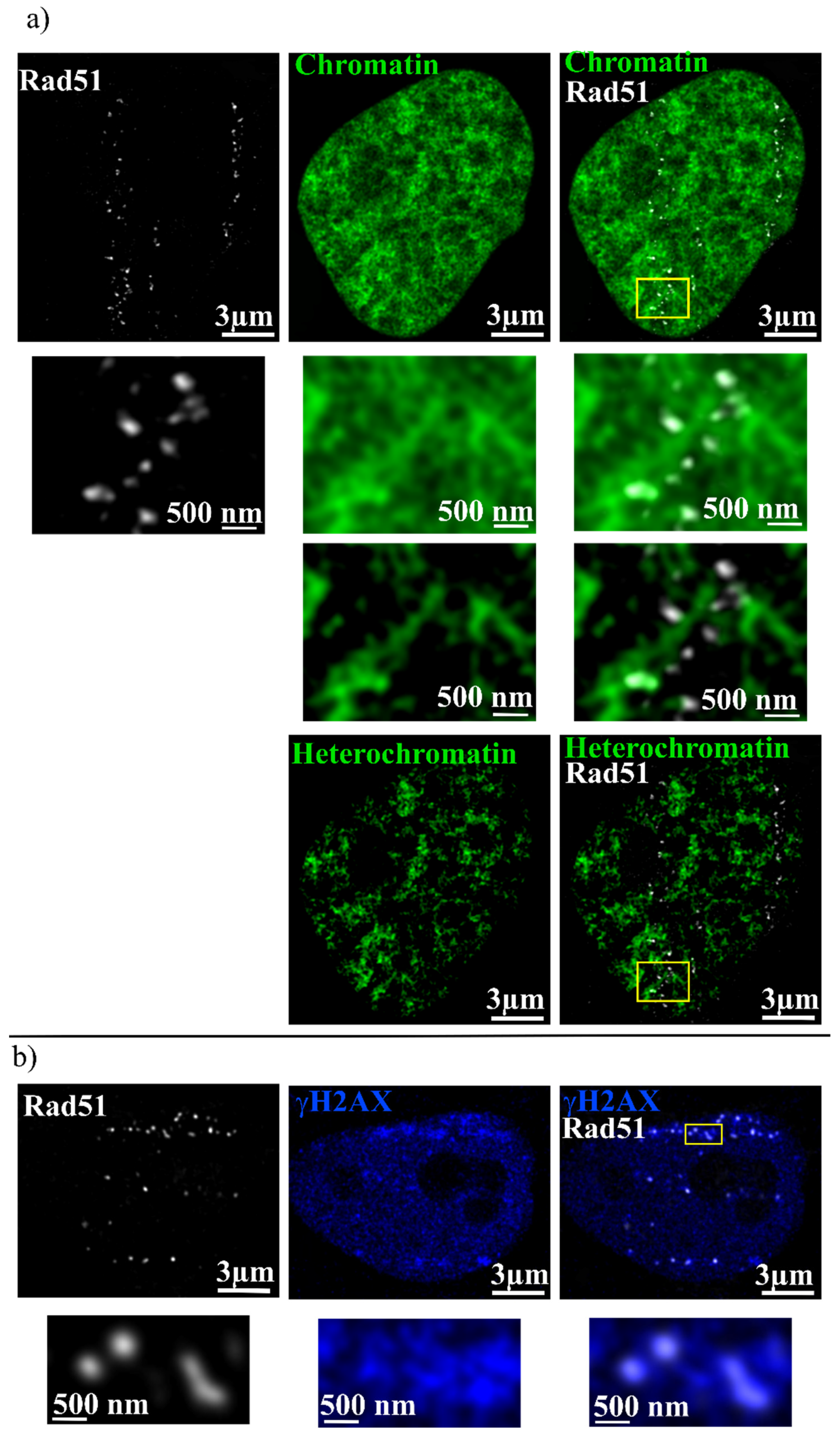
| Combination | Overlap % |
|---|---|
| SC35 + Chromatin | 29 ± 9 |
| 53BP1 + Chromatin | 38 ± 14 |
| Rad51 + Chromatin | 60 ± 15 |
| Rad51 + Heterochromatin | 10 ± 2 |
| 53BP1 + SC35 | 1.0 ± 0.2 |
| Rad51 + γH2AX | 36 ± 15 |
| Co-Staining | Dye 1 | Dye 2 |
|---|---|---|
| m-a-SC35 + SiR-DNA | gar-Alexa 488 | SiR 650 |
| r-a-53BP1 + SiR-DNA | gar-Alexa 488 | SiR 650 |
| m-a-SC35 + r-a-53BP1 | gam-Alexa 488 | gar-Alexa 532 |
| r-a-Rad51 + SiR-DNA | gar-Alexa 488 | SiR 650 |
| r-a-Rad51 + m-a-γH2AX | gam-Alexa 488 | gar-Alexa 532 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schwarz, B.; Matejka, N.; Rudigkeit, S.; Sammer, M.; Reindl, J. Chromatin Organization after High-LET Irradiation Revealed by Super-Resolution STED Microscopy. Int. J. Mol. Sci. 2024, 25, 628. https://doi.org/10.3390/ijms25010628
Schwarz B, Matejka N, Rudigkeit S, Sammer M, Reindl J. Chromatin Organization after High-LET Irradiation Revealed by Super-Resolution STED Microscopy. International Journal of Molecular Sciences. 2024; 25(1):628. https://doi.org/10.3390/ijms25010628
Chicago/Turabian StyleSchwarz, Benjamin, Nicole Matejka, Sarah Rudigkeit, Matthias Sammer, and Judith Reindl. 2024. "Chromatin Organization after High-LET Irradiation Revealed by Super-Resolution STED Microscopy" International Journal of Molecular Sciences 25, no. 1: 628. https://doi.org/10.3390/ijms25010628
APA StyleSchwarz, B., Matejka, N., Rudigkeit, S., Sammer, M., & Reindl, J. (2024). Chromatin Organization after High-LET Irradiation Revealed by Super-Resolution STED Microscopy. International Journal of Molecular Sciences, 25(1), 628. https://doi.org/10.3390/ijms25010628







