Genome-Wide Identification and Expression Profiling of Heavy Metal ATPase (HMA) Genes in Peanut: Potential Roles in Heavy Metal Transport
Abstract
1. Introduction
2. Results
2.1. Identification of the AhHMA Gene Family in Peanut
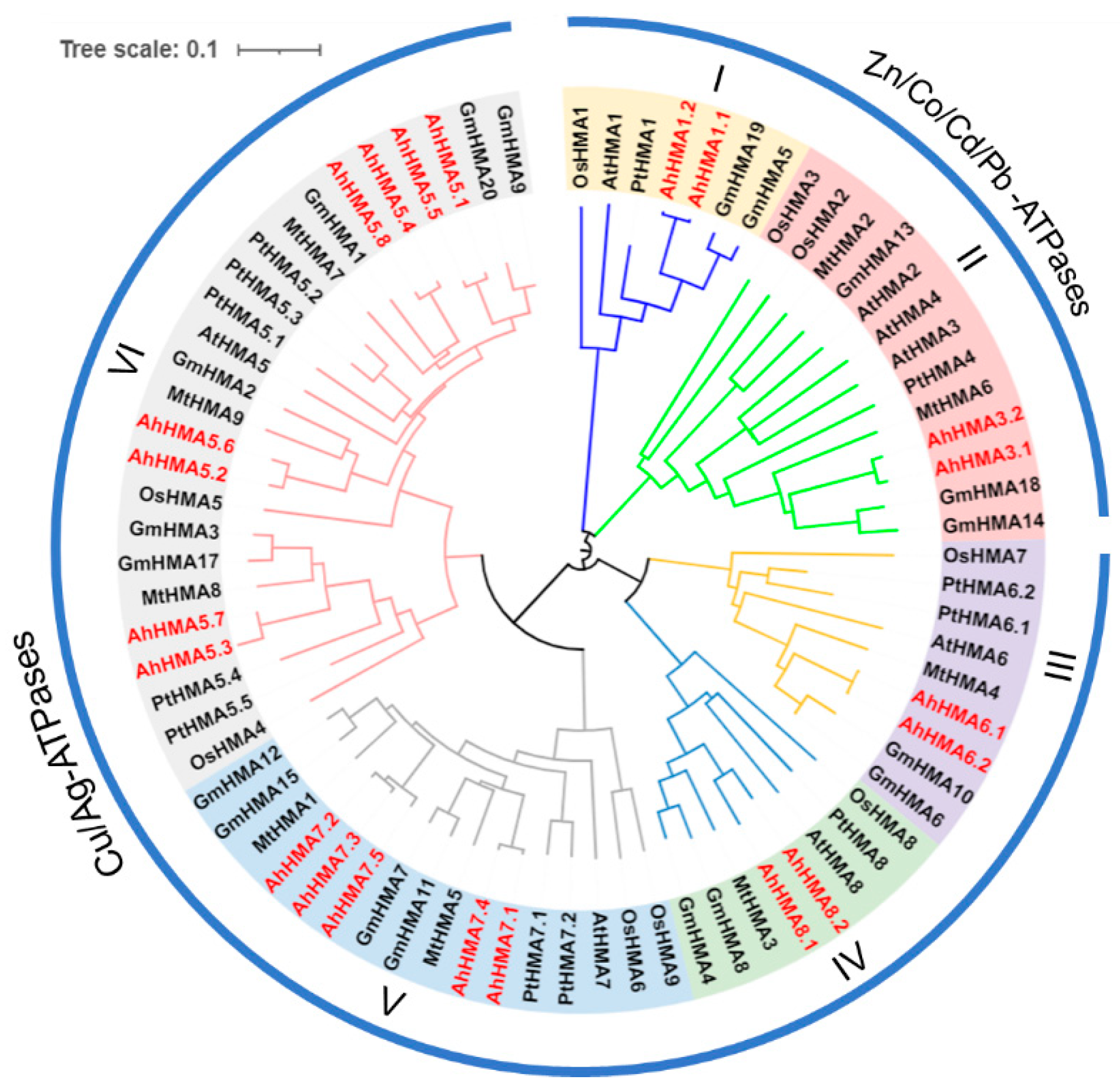
2.2. Phylogenetic Analysis of HMA Genes
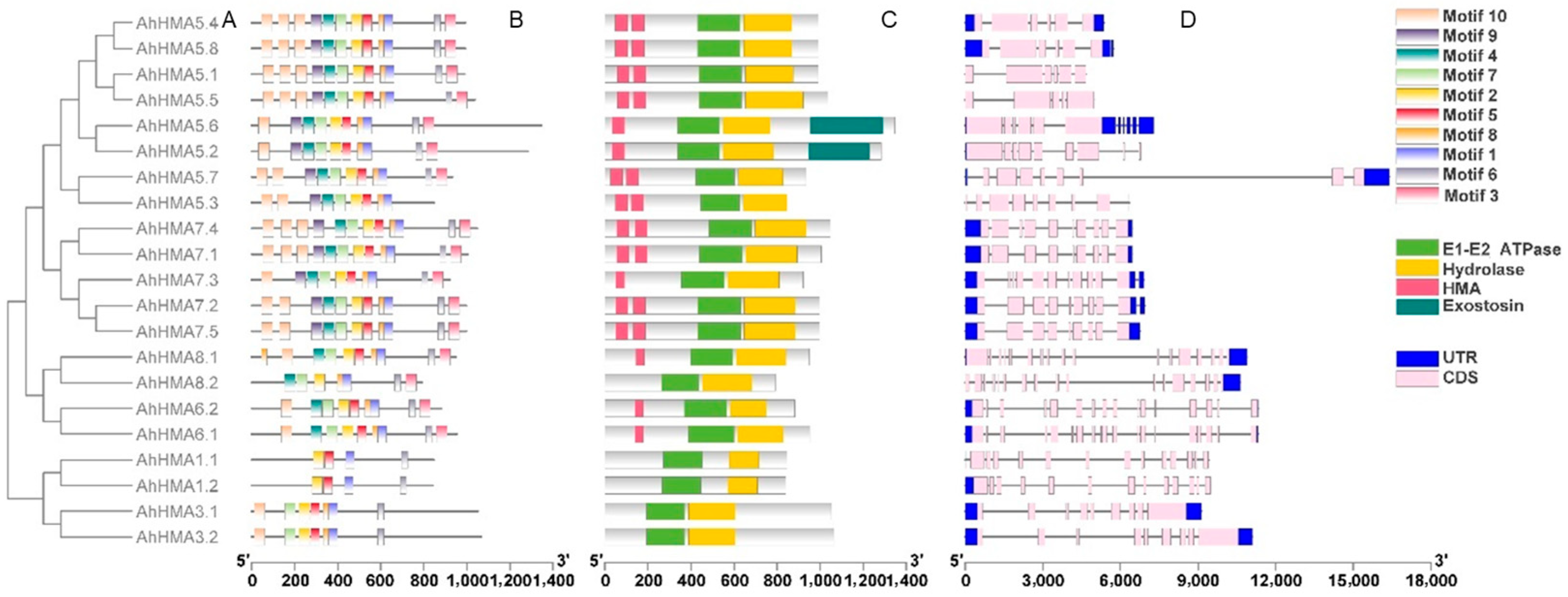
2.3. Conserved Motifs, Domain Architectures, and Models of AhHMA Proteins
2.4. Exon/Intron Organization and Duplication of AhHMA Genes
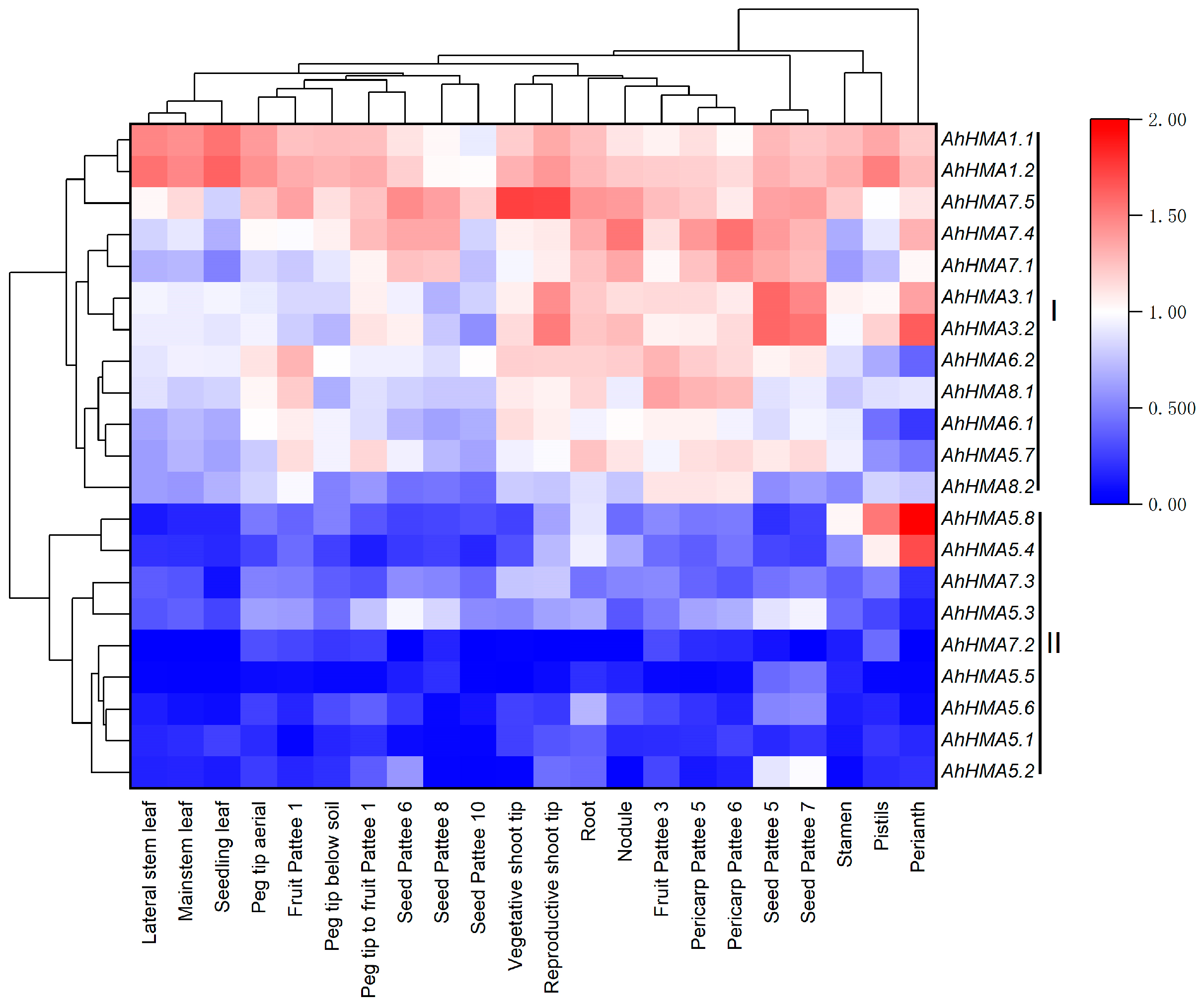
2.5. Expression Profiles of AhHMA Genes in Different Tissues of Peanut
2.6. Differential Expression of AhHMA Genes in the Root of Two Peanut Cultivars
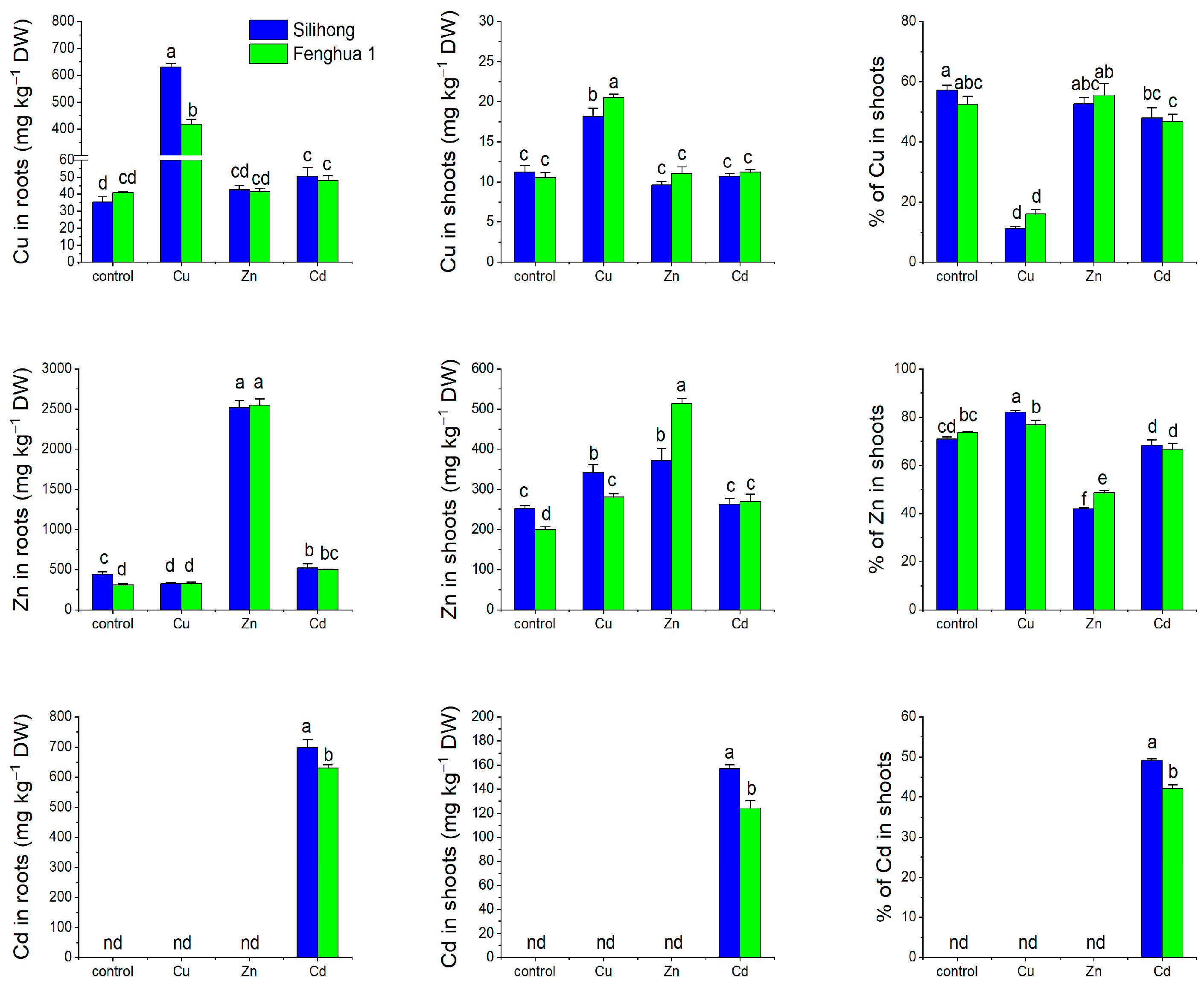
2.7. Metal Accumulation and Translocation in Two Peanut Cultivars
3. Discussion
3.1. WGD Expanded the Number of AhHMA Genes in Peanut
3.2. Conservation and Divergence of AhHMA Proteins in Peanut
3.3. Divergence of Duplicated AhHMA Genes in Peanut
3.4. Potential Role of AhHMA Genes in Metal Transport in Peanut
4. Materials and Methods
4.1. Plant Growth and Treatment
4.2. Metal Determination
4.3. Identification of HMA Genes in Peanut and Phylogenetic Analysis
4.4. Physicochemical and Structural Characteristics of AhHMA Proteins
4.5. Exon/intron Structure, Duplication and Ka/Ks of AhHMA Genes
4.6. Expression Profiles of AhHMA Genes in Different Tissues
4.7. RNA Extraction and qRT-PCR
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bhardwaj, E.; Shukla, R.; Das, S. Plant Roots and Mineral Nutrition: An Overview of Molecular Basis of Uptake and Regulation, and Strategies to Improve Nutrient Use Efficiency (NUE). In Plant Stress Biology: Strategies and Trends; Giri, B., Sharma, M.P., Eds.; Springer: Singapore, 2020; pp. 131–184. [Google Scholar]
- Su, Y.; Wang, X.; Liu, C.; Shi, G. Variation in cadmium accumulation and translocation among peanut cultivars as affected by iron deficiency. Plant Soil 2013, 363, 201–213. [Google Scholar] [CrossRef]
- Zhao, F.J.; Tang, Z.; Song, J.J.; Huang, X.Y.; Wang, P. Toxic metals and metalloids: Uptake, transport, detoxification, phytoremediation, and crop improvement for safer food. Mol. Plant 2022, 15, 27–44. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Wang, H.Q.; Chen, J.; Chang, J.D.; Zhao, F.J. Molecular mechanisms underlying the toxicity and detoxification of trace metals and metalloids in plants. J. Integr. Plant Biol. 2023, 65, 570–593. [Google Scholar] [CrossRef] [PubMed]
- Williams, L.E.; Mills, R.F. P1B-ATPases—An ancient family of transition metal pumps with diverse functions in plants. Trends Plant Sci. 2005, 10, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Cobbett, C.S.; Hussain, D.; Haydon, M.J. Structural and functional relationships between type 1B heavy metal-transporting P-type ATPases in Arabidopsis. New Phytol. 2003, 159, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Baxter, I.; Tchieu, J.; Sussman, M.R.; Boutry, M.; Palmgren, M.G.; Gribskov, M.; Harper, J.F.; Axelsen, K.B. Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol. 2003, 132, 618–628. [Google Scholar] [CrossRef] [PubMed]
- Tabata, K.; Kashiwagi, S.; Mori, H.; Ueguchi, C.; Mizuno, T. Cloning of a cDNA encoding a putative metal-transporting P-type ATPase from Arabidopsis thaliana. Biochim. Biophy. Acta 1997, 1326, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Seigneurin-Berny, D.; Gravot, A.; Auroy, P.; Mazard, C.; Kraut, A.; Finazzi, G.; Grunwald, D.; Rappaport, F.; Vavasseur, A.; Joyard, J.; et al. HMA1, a new Cu-ATPase of the chsloroplast envelope, is essential for growth under adverse light conditions. J. Biol. Chem. 2006, 281, 2882–2892. [Google Scholar] [CrossRef]
- Boutigny, S.; Sautron, E.; Finazzi, G.; Rivasseau, C.; Frelet-Barrand, A.; Pilon, M.; Rolland, N.; Seigneurin-Berny, D. HMA1 and PAA1, two chloroplast-envelope PIB-ATPases, play distinct roles in chloroplast copper homeostasis. J. Exp. Bot. 2014, 65, 1529–1540. [Google Scholar] [CrossRef]
- Abdel-Ghany, S.E.; Müller-Moulé, P.; Niyogi, K.K.; Pilon, M.; Shikanai, T. Two P-type ATPases are required for copper delivery in Arabidopsis thaliana chloroplasts. Plant Cell 2005, 17, 1233–1251. [Google Scholar] [CrossRef]
- Kim, Y.-Y.; Choi, H.; Segami, S.; Cho, H.-T.; Martinoia, E.; Maeshima, M.; Lee, Y. AtHMA1 contributes to the detoxification of excess Zn(II) in Arabidopsis. Plant J. 2009, 58, 737–753. [Google Scholar] [CrossRef] [PubMed]
- Andrés-Colás, N.; Sancenón, V.; Rodríguez-Navarro, S.; Mayo, S.; Thiele, D.J.; Ecker, J.R.; Puig, S.; Peñarrubia, L. The Arabidopsis heavy metal P-type ATPase HMA5 interacts with metallochaperones and functions in copper detoxification of roots. Plant J. 2006, 45, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Kuroda, K.; Kimura, K.; Southron-Francis, J.L.; Furuzawa, A.; Kimura, K.; Iuchi, S.; Kobayashi, M.; Taylor, G.J.; Koyama, H. Amino acid polymorphisms in strictly conserved domains of a P-Type ATPase HMA5 are involved in the mechanism of copper tolerance variation in Arabidopsis. Plant Physiol. 2008, 148, 969–980. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, T.; Kieber, J.J.; Hirayama, N.; Kogan, M.; Guzman, P.; Nourizadeh, S.; Alonso, J.M.; Dailey, W.P.; Dancis, A.; Ecker, J.R. RESPONSIVE-TO-ANTAGONIST1, a Menkes/Wilson disease–related copper transporter, is required for ethylene signaling in Arabidopsis. Cell 1999, 97, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Woeste, K.E.; Kieber, J.J. A Strong loss-of-function mutation in RAN1 results in constitutive activation of the ethylene response pathway as well as a rosette-lethal phenotype. Plant Cell 2000, 12, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.K.E.; Cobbett, C.S. HMA P-type ATPases are the major mechanism for root-to-shoot Cd translocation in Arabidopsis thaliana. New Phytol. 2009, 181, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Hussain, D.; Haydon, M.J.; Wang, Y.; Wong, E.; Sherson, S.M.; Young, J.; Camakaris, J.; Harper, J.F.; Cobbett, C.S. P-Type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell 2004, 16, 1327–1339. [Google Scholar] [CrossRef]
- Verret, F.; Gravot, A.; Auroy, P.; Leonhardt, N.; David, P.; Nussaume, L.; Vavasseur, A.; Richaud, P. Overexpression of AtHMA4 enhances root-to-shoot translocation of zinc and cadmium and plant metal tolerance. FEBS Lett. 2004, 576, 306–312. [Google Scholar] [CrossRef]
- Morel, M.l.; Crouzet, J.r.m.; Gravot, A.; Auroy, P.; Leonhardt, N.; Vavasseur, A.; Richaud, P. AtHMA3, a P1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiol. 2008, 149, 894–904. [Google Scholar] [CrossRef]
- Takahashi, R.; Ishimaru, Y.; Shimo, H.; Ogo, Y.; Senoura, T.; Nishizawa, N.K.; Nakanishi, H. The OsHMA2 transporter is involved in root-to-shoot translocation of Zn and Cd in rice. Plant Cell Environ. 2012, 35, 1948–1957. [Google Scholar] [CrossRef]
- Satoh-Nagasawa, N.; Mori, M.; Nakazawa, N.; Kawamoto, T.; Nagato, Y.; Sakurai, K.; Takahashi, H.; Watanabe, A.; Akagi, H. Mutations in rice (Oryza sativa) heavy metal ATPase 2 (OsHMA2) restrict the translocation of zinc and cadmium. Plant Cell Physiol. 2012, 53, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, A.; Yamaji, N.; Ma, J.F. Overexpression of OsHMA3 enhances Cd tolerance and expression of Zn transporter genes in rice. J. Exp. Bot. 2014, 65, 6013–6021. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Huang, S.; Che, J.; Yamaji, N.; Ma, J.F. The tonoplast-localized transporter OsHMA3 plays an important role in maintaining Zn homeostasis in rice. J. Exp. Bot. 2019, 70, 2717–2725. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.Y.; Deng, F.; Yamaji, N.; Pinson, S.R.; Fujii-Kashino, M.; Danku, J.; Douglas, A.; Guerinot, M.L.; Salt, D.E.; Ma, J.F. A heavy metal P-type ATPase OsHMA4 prevents copper accumulation in rice grain. Nat. Commun. 2016, 7, 12138. [Google Scholar] [CrossRef] [PubMed]
- Deng, F.; Yamaji, N.; Xia, J.; Ma, J.F. A member of the heavy metal P-type ATPase OsHMA5 is involved in xylem loading of copper in rice. Plant Physiol. 2013, 163, 1353–1362. [Google Scholar] [CrossRef]
- Lee, S.; Kim, Y.Y.; Lee, Y.; An, G. Rice P1B-type heavy-metal ATPase, OsHMA9, is a metal efflux protein. Plant Physiol. 2007, 145, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Kappara, S.; Neelamraju, S.; Ramanan, R. Down regulation of a heavy metal transporter gene influences several domestication traits and grain Fe-Zn content in rice. Plant Sci. 2018, 276, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Su, G.; Li, F.; Lin, J.; Liu, C.; Shi, G. Peanut as a potential crop for bioenergy production via Cd-phytoextraction: A life-cycle pot experiment. Plant Soil 2013, 365, 337–345. [Google Scholar] [CrossRef]
- Wang, X.; Wang, C.; Zhang, Z.; Shi, G. Genome-wide identification of metal tolerance protein genes in peanut: Differential expression in the root of two contrasting cultivars under metal stresses. Front. Plant Sci. 2022, 13, 791200. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, N.; Zhang, Z.; Shi, G. Genome-wide identification and expression profile reveal potential roles of peanut ZIP family genes in zinc/iron-deficiency tolerance. Plants 2022, 11, 786. [Google Scholar] [CrossRef]
- Tan, Z.; Li, J.; Guan, J.; Wang, C.; Zhang, Z.; Shi, G. Genome-wide identification and expression analysis reveals roles of the NRAMP gene family in iron/cadmium interactions in peanut. Int. J. Mol. Sci. 2023, 24, 1713. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, X.; Li, J.; Guan, J.; Tan, Z.; Zhang, Z.; Shi, G. Genome-wide identification and transcript analysis reveal potential roles of oligopeptide transporter genes in iron deficiency induced cadmium accumulation in peanut. Front. Plant Sci. 2022, 13, 894848. [Google Scholar] [CrossRef] [PubMed]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Bertioli, D.J.; Jenkins, J.; Clevenger, J.; Dudchenko, O.; Gao, D.; Seijo, G.; Leal-Bertioli, S.C.M.; Ren, L.; Farmer, A.D.; Pandey, M.K.; et al. The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 2019, 51, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yu, R.; Shi, G. Effects of drought on the accumulation and redistribution of cadmium in peanuts at different developmental stages. Arch. Agron. Soil Sci. 2017, 63, 1049–1057. [Google Scholar] [CrossRef]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef] [PubMed]
- Clevenger, J.; Chu, Y.; Scheffler, B.; Ozias-Akins, P. A developmental transcriptome map for allotetraploid Arachis hypogaea. Front. Plant Sci. 2016, 7, 1446. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Wang, L.; Deng, X.; Wang, P.; Ma, Q.; Nian, H.; Wang, Y.; Yang, C. Genome-wide characterization of soybean P1B-ATPases gene family provides functional implications in cadmium responses. BMC Genom. 2016, 17, 376. [Google Scholar] [CrossRef]
- Ma, Y.; Wei, N.; Wang, Q.; Liu, Z.; Liu, W. Genome-wide identification and characterization of the heavy metal ATPase (HMA) gene family in Medicago truncatula under copper stress. Int. J. Biol. Macromol. 2021, 193, 893–902. [Google Scholar] [CrossRef]
- Li, N.; Xiao, H.; Sun, J.; Wang, S.; Wang, J.; Chang, P.; Zhou, X.; Lei, B.; Lu, K.; Luo, F.; et al. Genome-wide analysis and expression profiling of the HMA gene family in Brassica napus under Cd stress. Plant Soil 2018, 426, 365–381. [Google Scholar] [CrossRef]
- Khan, N.; You, F.M.; Datla, R.; Ravichandran, S.; Jia, B.; Cloutier, S. Genome-wide identification of ATP binding cassette (ABC) transporter and heavy metal associated (HMA) gene families in flax (Linum usitatissimum L.). BMC Genom. 2020, 21, 722. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Xu, X.; Hu, X.; Liu, Q.; Wang, Z.; Zhang, H.; Wang, H.; Wei, M.; Wang, H.; Liu, H.; et al. Genome-wide analysis and heavy metal-induced expression profiling of the HMA gene family in Populus trichocarpa. Front. Plant Sci. 2015, 6, 1149. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lu, Q.; Liu, H.; Zhang, J.; Hong, Y.; Lan, H.; Li, H.; Wang, J.; Liu, H.; Li, S.; et al. Sequencing of cultivated peanut, Arachis hypogaea, yields insights into genome evolution and oil improvement. Mol. Plant 2019, 12, 920–934. [Google Scholar] [CrossRef]
- Gravot, A.; Lieutaud, A.; Verret, F.; Auroy, P.; Vavasseur, A.; Richaud, P. AtHMA3, a plant P1B-ATPase, functions as a Cd/Pb transporter in yeast. FEBS Lett. 2004, 561, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Verret, F.; Gravot, A.; Auroy, P.; Preveral, S.; Forestier, C.; Vavasseur, A.; Richaud, P. Heavy metal transport by AtHMA4 involves the N-terminal degenerated metal binding domain and the C-terminal His11 stretch. FEBS Lett. 2005, 579, 1515–1522. [Google Scholar] [CrossRef] [PubMed]
- Mills, R.F.; Francini, A.; Ferreira da Rocha, P.S.C.; Baccarini, P.J.; Aylett, M.; Krijger, G.C.; Williams, L.E. The plant P1B-type ATPase AtHMA4 transports Zn and Cd and plays a role in detoxification of transition metals supplied at elevated levels. FEBS Lett. 2005, 579, 783–791. [Google Scholar] [CrossRef]
- Toyoshima, C.; Nomura, H.; Sugita, Y. Structural basis of ion pumping by Ca2+-ATPase of sarcoplasmic reticulum. FEBS Lett. 2003, 555, 106–110. [Google Scholar] [CrossRef]
- Alberts, P.; Daumke, O.; Deverson, E.V.; Howard, J.C.; Knittler, M.R. Distinct functional properties of the TAP subunits coordinate the nucleotide-dependent transport cycle. Curr. Biol. 2001, 11, 242–251. [Google Scholar] [CrossRef]
- Jorgensen, P.L.; Håkansson, K.O.; Karlish, S.J.D. Structure and mechanism of Na,K-ATPase: Functional sites and their interactions. Annu. Rev. Physiol. 2003, 65, 817–849. [Google Scholar] [CrossRef]
- Grønberg, C.; Hu, Q.; Mahato, D.R.; Longhin, E.; Salustros, N.; Duelli, A.; Lyu, P.; Bågenholm, V.; Eriksson, J.; Rao, K.U.; et al. Structure and ion-release mechanism of P(IB-4)-type ATPases. eLife 2021, 10, e73124. [Google Scholar] [CrossRef]
- Yang, G.M.; Xu, L.; Wang, R.M.; Tao, X.; Zheng, Z.W.; Chang, S.; Ma, D.; Zhao, C.; Dong, Y.; Wu, S.; et al. Structures of the human Wilson disease copper transporter ATP7B. Cell Rep. 2023, 42, 112417. [Google Scholar] [CrossRef] [PubMed]
- Bitter, R.M.; Oh, S.; Deng, Z.; Rahman, S.; Hite, R.K.; Yuan, P. Structure of the Wilson disease copper transporter ATP7B. Sci. Adv. 2022, 8, eabl5508. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.; Mattle, D.; Sitsel, O.; Klymchuk, T.; Nielsen, A.M.; Møller, L.B.; White, S.H.; Nissen, P.; Gourdon, P. Copper-transporting P-type ATPases use a unique ion-release pathway. Nat. Struct. Mol. Biol. 2014, 21, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.-H. Evolution of gene duplication in plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Liao, B.-Y.; Chang, A.Y.F.; Zhang, J. Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet. 2010, 26, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. Genetic redundancies and their evolutionary maintenance. In Evolutionary Systems Biology; Soyer, O.S., Ed.; Springer: New York, NY, USA, 2012; pp. 279–300. [Google Scholar]
- Birchler, J.A.; Yang, H. The multiple fates of gene duplications: Deletion, hypofunctionalization, subfunctionalization, neofunctionalization, dosage balance constraints, and neutral variation. Plant Cell 2022, 34, 2466–2474. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon–intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef] [PubMed]
- Ueno, D.; Milner, M.J.; Yamaji, N.; Yokosho, K.; Koyama, E.; Clemencia Zambrano, M.; Kaskie, M.; Ebbs, S.; Kochian, L.V.; Ma, J.F. Elevated expression of TcHMA3 plays a key role in the extreme Cd tolerance in a Cd-hyperaccumulating ecotype of Thlaspi caerulescens. Plant J. 2011, 66, 852–862. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, C.; Liu, Y.; Yu, K.; Zhou, Y. GmHMA3 sequesters Cd to the root endoplasmic reticulum to limit translocation to the stems in soybean. Plant Sci. 2018, 270, 23–29. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef]
- Tsirigos, K.D.; Peters, C.; Shu, N.; Käll, L.; Elofsson, A. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 2015, 43, W401–W407. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2020, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]


| Gene Name | Gene ID | Gene Length (bp) | CDS (bp) | Aa a | MW b (kDa) | Instability | Aliphatic Index | GRAVY c | pI d | No. of TMD e | Location |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AhHMA1.1 | Arahy.VMCJ1G | 9517 | 2541 | 91.92 | 846 | 37.86 | 102.86 | 0.121 | 7.86 | 8 | Chl. f |
| AhHMA1.2 | Arahy.FTH7SR | 9482 | 2523 | 91.23 | 840 | 37.43 | 102.32 | 0.093 | 7.5 | 8 | Chl. |
| AhHMA3.1 | Arahy.NH81W7 | 9130 | 3159 | 114.12 | 1052 | 37.96 | 88.34 | −0.211 | 6.25 | 8 | PM g |
| AhHMA3.2 | Arahy.6QFK0R | 11,108 | 3198 | 115.79 | 1065 | 35.17 | 87.62 | −0.215 | 6.26 | 8 | PM |
| AhHMA5.1 | Arahy.C06U18 | 4665 | 2973 | 107.41 | 990 | 36.52 | 100.37 | 0.138 | 6.16 | 9 | PM |
| AhHMA5.2 | Arahy.1HUL85 | 6784 | 3855 | 142.51 | 1284 | 38.21 | 96.36 | −0.09 | 6.19 | 7 | PM |
| AhHMA5.3 | Arahy.694CK1 | 11,328 | 2541 | 91.90 | 846 | 36.14 | 102.87 | 0.155 | 5.73 | 6 | PM |
| AhHMA5.4 | Arahy.9DR5PQ | 5362 | 2979 | 107.31 | 992 | 34.58 | 100.77 | 0.146 | 5.95 | 9 | PM |
| AhHMA5.5 | Arahy.NXU65W | 4972 | 3111 | 113.03 | 1036 | 35.08 | 100.89 | 0.137 | 7.25 | 9 | PM |
| AhHMA5.6 | Arahy.NB9ZER | 7292 | 4041 | 149.65 | 1346 | 37.40 | 97.86 | −0.073 | 6.94 | 8 | PM |
| AhHMA5.7 | Arahy.3M9T3M | 16,396 | 2805 | 100.97 | 934 | 37.39 | 105.61 | 0.245 | 5.53 | 8 | PM |
| AhHMA5.8 | Arahy.LQF816 | 5780 | 2979 | 107.45 | 992 | 35.35 | 100.18 | 0.128 | 5.96 | 9 | PM |
| AhHMA6.1 | Arahy.8T0CRQ | 11,318 | 2862 | 101.18 | 953 | 36.11 | 101.19 | 0.192 | 8.48 | 8 | Chl. |
| AhHMA6.2 | Arahy.KT5IG2 | 11,336 | 2649 | 93.68 | 882 | 36.93 | 98.84 | 0.133 | 8.88 | 8 | Chl. |
| AhHMA7.1 | Arahy.H8X8VQ | 6457 | 3018 | 107.24 | 1005 | 30.82 | 104.75 | 0.314 | 5.14 | 8 | PM |
| AhHMA7.2 | Arahy.XIB6CP | 6910 | 2766 | 98.77 | 921 | 34.05 | 105.1 | 0.28 | 5.53 | 8 | PM |
| AhHMA7.3 | Arahy.GR9FC3 | 6947 | 2994 | 106.71 | 997 | 34.58 | 105.79 | 0.299 | 5.12 | 8 | PM |
| AhHMA7.4 | Arahy.PPR7D4 | 6457 | 3147 | 112.04 | 1048 | 30.85 | 106.68 | 0.313 | 5.43 | 8 | PM |
| AhHMA7.5 | Arahy.8Q2P9V | 6759 | 2994 | 106.65 | 997 | 34.49 | 105.4 | 0.296 | 5.09 | 8 | PM |
| AhHMA8.1 | Arahy.ALH963 | 10,892 | 2853 | 101.55 | 950 | 39.05 | 106.11 | 0.162 | 5.95 | 8 | Chl. |
| AhHMA8.2 | Arahy.39IB73 | 10,646 | 2379 | 84.17 | 792 | 32.40 | 106.87 | 0.224 | 5.15 | 8 | Chl. |
| Protein Name | Template | Sequence Identity (%) | Coverage | GMQE | QMEANDisCo Global | Description |
|---|---|---|---|---|---|---|
| AhHMA1.1 | 7qc0.1 | 0.3214 | A169-812 | 0.48 | 0.62 ± 0.05 | Cadmium translocating P-type ATPase |
| AhHMA1.2 | 7qc0.1 | 0.3214 | A163-806 | 0.48 | 0.62 ± 0.05 | Cadmium translocating P-type ATPase |
| AhHMA3.1 | 7si3.1 | 0.3006 | A11-702 | 0.42 | 0.62 ± 0.05 | P-type Cu(+) transporter |
| AhHMA3.2 | 7si3.1 | 0.3006 | A11-702 | 0.41 | 0.62 ± 0.05 | P-type Cu(+) transporter |
| AhHMA5.1 | 7si3.1 | 0.4293 | A128-976 | 0.62 | 0.65 ± 0.05 | P-type Cu(+) transporter |
| AhHMA5.2 | 7si3.1 | 0.4272 | A31-874 | 0.45 | 0.65 ± 0.05 | P-type Cu(+) transporter |
| AhHMA5.3 | 7si3.1 | 0.3942 | A117-845 | 0.64 | 0.67 ± 0.05 | P-type Cu(+) transporter |
| AhHMA5.4 | 7si3.1 | 0.4264 | A121-970 | 0.65 | 0.70 ± 0.05 | P-type Cu(+) transporter |
| AhHMA5.5 | 7si3.1 | 0.4292 | A128-1021 | 0.6 | 0.65 ± 0.05 | P-type Cu(+) transporter |
| AhHMA5.6 | 7si3.1 | 0.4486 | A28-869 | 0.42 | 0.63 ± 0.05 | P-type Cu(+) transporter |
| AhHMA5.7 | 7si3.1 | 0.4192 | A94-927 | 0.68 | 0.69 ± 0.05 | P-type Cu(+) transporter |
| AhHMA5.8 | 7si3.1 | 0.4247 | A121-970 | 0.64 | 0.69 ± 0.05 | P-type Cu(+) transporter |
| AhHMA6.1 | 7xuk.1 | 0.3249 | A136-934 | 0.46 | 0.56 ± 0.05 | Copper-transporting ATPase 2 |
| AhHMA6.2 | 7xuk.1 | 0.3276 | A136-855 | 0.51 | 0.61 ± 0.05 | Copper-transporting ATPase 2 |
| AhHMA7.1 | 7si3.1 | 0.3885 | A134-993 | 0.62 | 0.66 ± 0.05 | P-type Cu(+) transporter |
| AhHMA7.2 | 7si3.1 | 0.4012 | A44-910 | 0.6 | 0.62 ± 0.05 | P-type Cu(+) transporter |
| AhHMA7.3 | 7si3.1 | 0.3811 | A126-986 | 0.62 | 0.66 ± 0.05 | P-type Cu(+) transporter |
| AhHMA7.4 | 7si3.1 | 0.3862 | A134-1037 | 0.58 | 0.63 ± 0.05 | P-type Cu(+) transporter |
| AhHMA7.5 | 7si3.1 | 0.384 | A126-985 | 0.61 | 0.65 ± 0.05 | P-type Cu(+) transporter |
| AhHMA8.1 | 4bbj.1 | 0.3687 | A285-942 | 0.46 | 0.66 ± 0.05 | Copper efflux ATPase |
| AhHMA8.2 | 4bbj.1 | 0.3672 | A149-784 | 0.52 | 0.65 ± 0.05 | Copper efflux ATPase |
| Gene Pairs | Duplicate Type | Ka a | Ks b | Ka/Ks c | Positive Selection | Divergence Time (Mya) |
|---|---|---|---|---|---|---|
| AhHMA5.1/5.2 | Segmental | 0.150 | 1.338 | 0.112 | No | 82.41 |
| AhHMA5.5/5.6 | Segmental | 0.154 | 1.293 | 0.119 | No | 79.60 |
| AhHMA7.1/7.2 | Segmental | 0.099 | 0.631 | 0.157 | No | 38.87 |
| AhHMA7.4/7.5 | Segmental | 0.093 | 0.716 | 0.130 | No | 44.10 |
| AhHMA7.2/7.3 | Tandem | 0.007 | 0.009 | 0.755 | No | 0.58 |
| AhHMA1.1/1.2 | Whole-genome | 0.002 | 0.024 | 0.089 | No | 1.45 |
| AhHMA3.1/3.2 | Whole-genome | 0.010 | 0.043 | 0.228 | No | 2.68 |
| AhHMA5.1/5.5 | Whole-genome | 0.006 | 0.030 | 0.208 | No | 1.85 |
| AhHMA5.2/5.6 | Whole-genome | 0.014 | 0.040 | 0.343 | No | 2.46 |
| AhHMA5.3/5.7 | Whole-genome | 0.024 | 0.064 | 0.384 | No | 3.92 |
| AhHMA5.4/5.8 | Whole-genome | 0.011 | 0.020 | 0.565 | No | 1.25 |
| AhHMA6.1/6.2 | Whole-genome | 0.002 | 0.012 | 0.122 | No | 0.77 |
| AhHMA7.1/7.4 | Whole-genome | 0.005 | 0.019 | 0.256 | No | 1.17 |
| AhHMA7.2/7.5 | Whole-genome | 0.016 | 0.039 | 0.399 | No | 2.41 |
| AhHMA8.1/8.2 | Whole-genome | 0.048 | 0.098 | 0.489 | No | 6.03 |
| Gene Expression | [Cu]root | [Cu]shoot | % of Cu in Shoots | [Zn]root | [Zn]shoot | % of Zn in Shoots |
|---|---|---|---|---|---|---|
| AhHMA1.1 | 0.283 | 0.061 | −0.087 | 0.534 ** | 0.798 ** | −0.292 |
| AhHMA1.2 | −0.155 | −0.278 | 0.313 | 0.721 ** | 0.848 ** | −0.560 ** |
| AhHMA3.1 | −0.471 * | −0.589 ** | 0.546 ** | 0.785 ** | 0.593 ** | −0.776 ** |
| AhHMA3.2 | −0.291 | −0.504 * | 0.331 | 0.514 * | 0.245 | −0.579 ** |
| AhHMA5.7 | 0.091 | 0.037 | −0.110 | −0.188 | −0.120 | 0.179 |
| AhHMA6.1 | 0.097 | −0.206 | 0.032 | −0.248 | −0.248 | 0.222 |
| AhHMA6.2 | 0.050 | −0.209 | 0.156 | 0.189 | 0.325 | −0.082 |
| AhHMA7.1 | −0.382 | −0.523 ** | 0.430 * | 0.851 ** | 0.586 ** | −0.845 ** |
| AhHMA7.4 | −0.276 | −0.336 | 0.340 | 0.895 ** | 0.890 ** | −0.762 ** |
| AhHMA7.5 | 0.066 | −0.221 | −0.011 | 0.089 | 0.012 | −0.135 |
| AhHMA8.1 | −0.295 | −0.277 | 0.325 | 0.728 ** | 0.683 ** | −0.611 ** |
| AhHMA8.2 | 0.285 | −0.022 | −0.145 | 0.137 | 0.037 | −0.061 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Zhang, Z.; Shi, G. Genome-Wide Identification and Expression Profiling of Heavy Metal ATPase (HMA) Genes in Peanut: Potential Roles in Heavy Metal Transport. Int. J. Mol. Sci. 2024, 25, 613. https://doi.org/10.3390/ijms25010613
Li J, Zhang Z, Shi G. Genome-Wide Identification and Expression Profiling of Heavy Metal ATPase (HMA) Genes in Peanut: Potential Roles in Heavy Metal Transport. International Journal of Molecular Sciences. 2024; 25(1):613. https://doi.org/10.3390/ijms25010613
Chicago/Turabian StyleLi, Jinxiu, Zheng Zhang, and Gangrong Shi. 2024. "Genome-Wide Identification and Expression Profiling of Heavy Metal ATPase (HMA) Genes in Peanut: Potential Roles in Heavy Metal Transport" International Journal of Molecular Sciences 25, no. 1: 613. https://doi.org/10.3390/ijms25010613
APA StyleLi, J., Zhang, Z., & Shi, G. (2024). Genome-Wide Identification and Expression Profiling of Heavy Metal ATPase (HMA) Genes in Peanut: Potential Roles in Heavy Metal Transport. International Journal of Molecular Sciences, 25(1), 613. https://doi.org/10.3390/ijms25010613






