Analysis of Lsm Protein-Mediated Regulation in the Haloarchaeon Haloferax mediterranei
Abstract
1. Introduction
2. Results
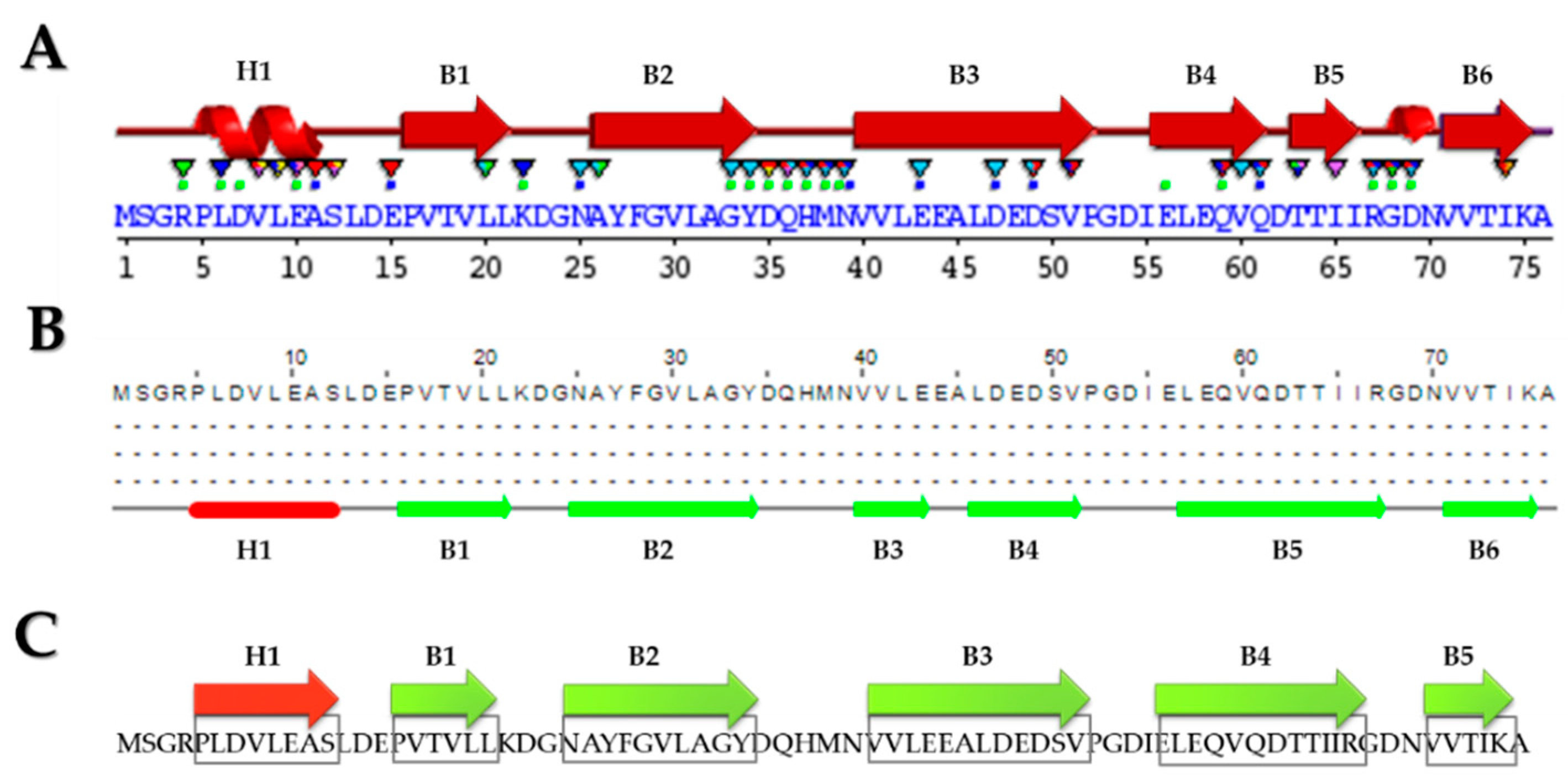
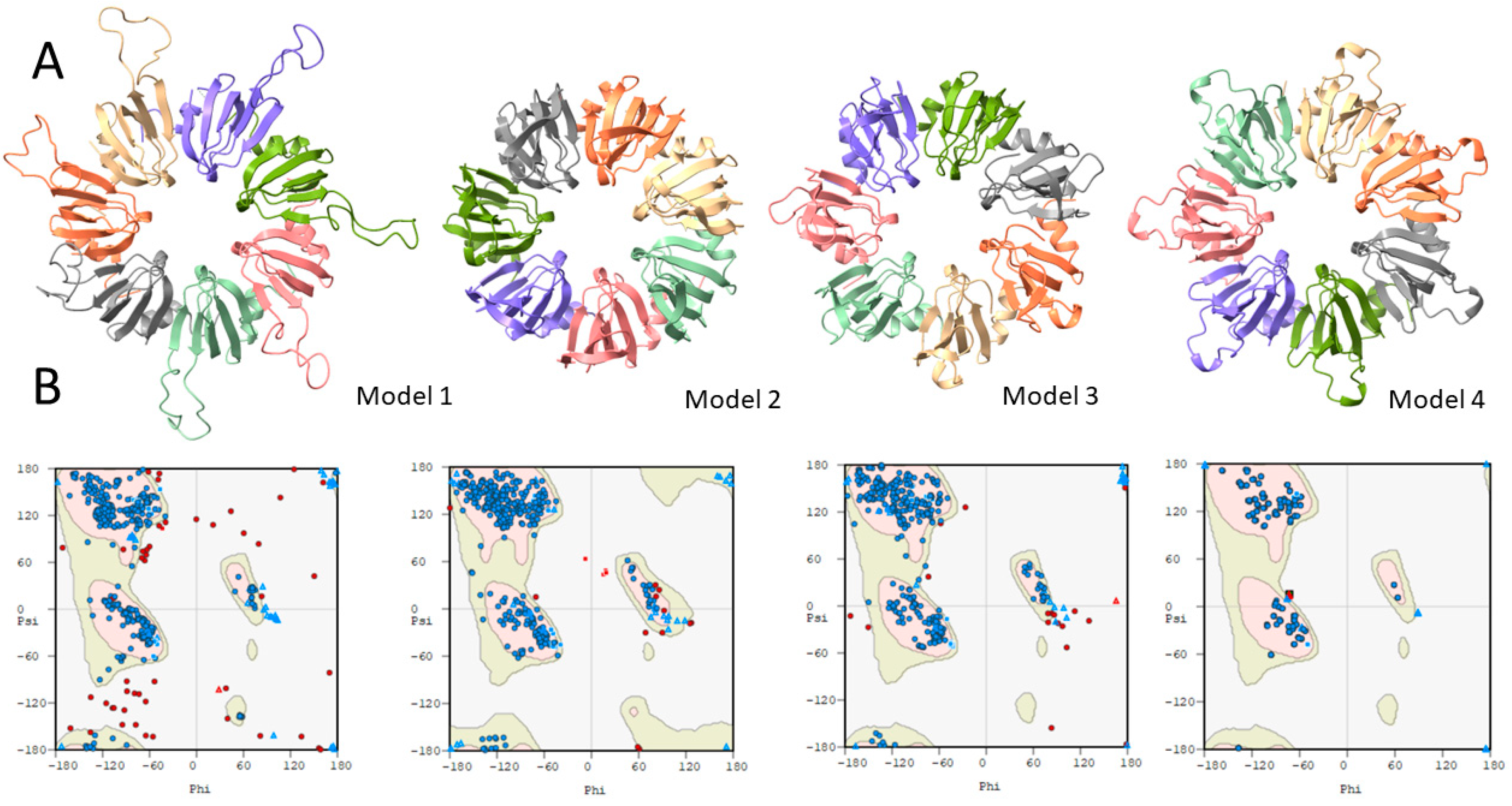
2.1. Bioinformatic Analysis of Haloferax mediterranei Lsm Protein
2.1.1. In Silico Analysis at the Primary and Secondary Structure Levels
2.1.2. Homology Modeling
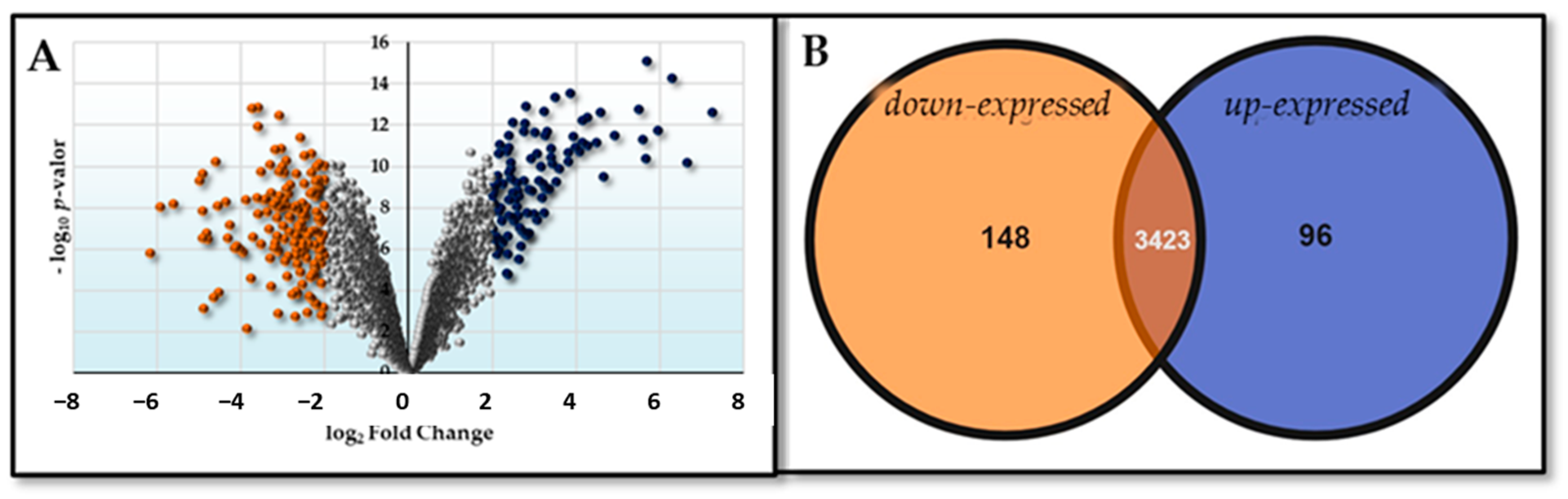
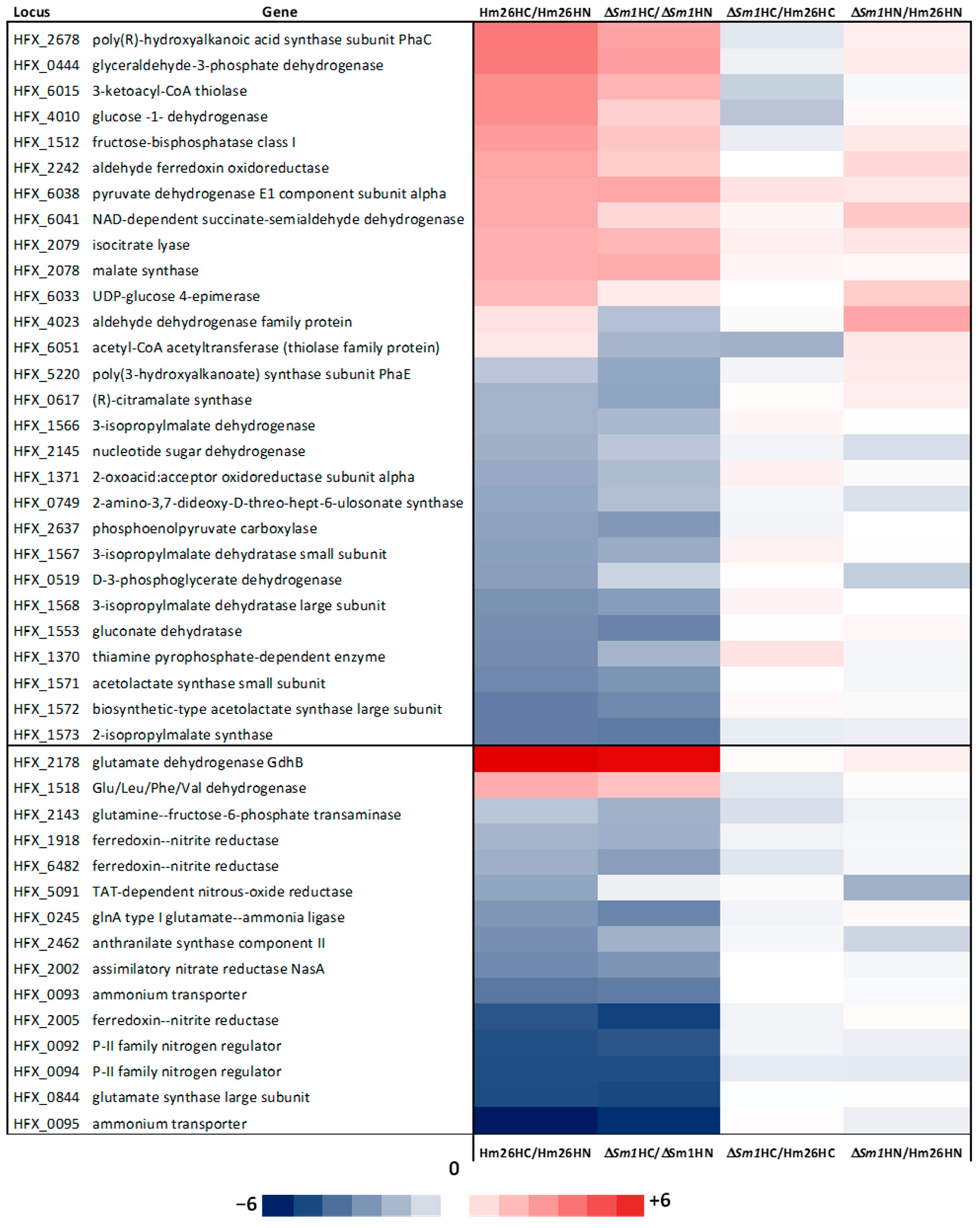
2.2. DNA Microarray Analysis
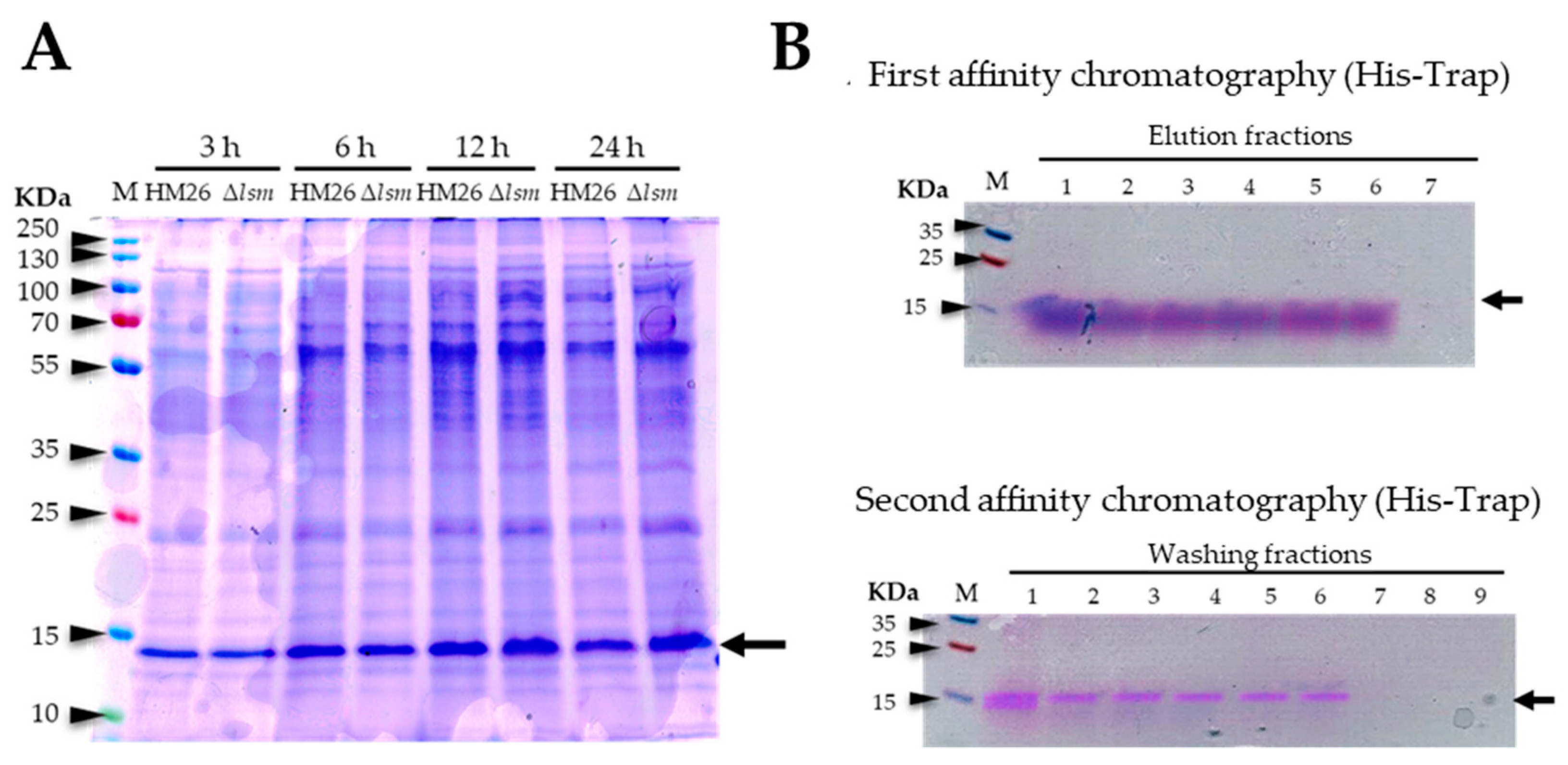
2.3. Overexpression and Purification
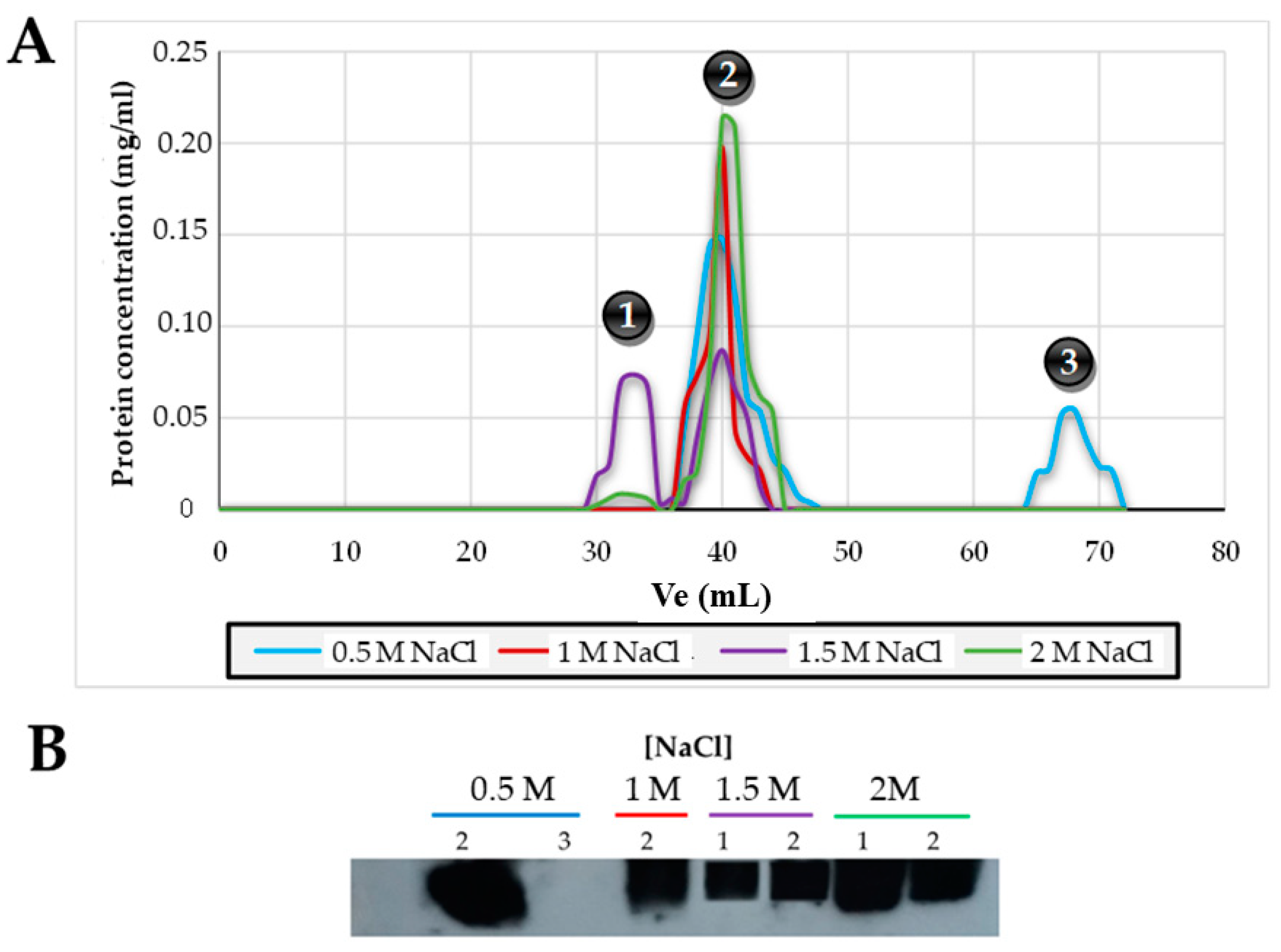
2.4. Molecular Mass Determination of Lsm Protein
3. Discussion
4. Materials and Methods
4.1. Bioinformatic Analysis
4.2. Strains, Plasmids, and Culture Conditions
4.3. DNA Microarray Analysis
4.4. Homologous Overexpression of lsm in Hfx. mediterranei HM26
4.5. Protein Purification
4.6. Determination of Molecular Mass
4.7. Western Blot
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Séraphin, B. Sm and Sm-like proteins belong to a large family: Identification of proteins of the U6 as well as the U1, U2, U4, and U5 snRNPs. EMBO J. 1995, 14, 2089–2098. [Google Scholar] [CrossRef] [PubMed]
- Scofield, D.G.; Lynch, M. Evolutionary diversification of the Sm family of RNA-associated proteins. Mol. Biol. Evol. 2008, 25, 2255–2267. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.; Luisi, B.F. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011, 9, 578–589. [Google Scholar] [CrossRef] [PubMed]
- Sauter, C.; Basquin, J.; Suck, D. Sm-like proteins in Eubacteria: The crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res. 2003, 31, 4091–4098. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.S.; Bøggild, A.; Andersen, C.B.; Nielsen, G.; Boysen, A.; Brodersen, D.E.; Valentin-Hansen, P. An Hfq-like protein in archaea: Crystal structure and functional characterization of the Sm protein from Methanococcus jannaschii. RNA 2007, 13, 2213–2223. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Valentin-Hansen, P.; Eriksen, M.; Udesen, C. The bacterial Sm-like protein Hfq: A key player in RNA transactions. Mol. Microbiol. 2004, 51, 1525–1533. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, C.; Wilusz, J. Eukaryotic Lsm proteins: Lessons from bacteria. Nat. Struct. Mol. Biol. 2005, 12, 1031–1036. [Google Scholar] [CrossRef]
- Törö, I.; Thore, S.; Mayer, C.; Basquin, J.; Séraphin, B.; Suck, D. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J. 2001, 20, 2293–2303. [Google Scholar] [CrossRef]
- Schumacher, M.A.; Pearson, R.F.; Møller, T.; Valentin-Hansen, P.; Brennan, R.G. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: A bacterial Sm-like protein. EMBO J. 2002, 21, 3546–3556. [Google Scholar] [CrossRef]
- Törö, I.; Basquin, J.; Teo-Dreher, H.; Suck, D. Archaeal Sm proteins form heptameric and hexameric complexes: Crystal structures of the Sm1 and Sm2 proteins from the hyperthermophile Archaeoglobus fulgidus. J. Mol. Biol. 2002, 320, 129–142. [Google Scholar] [CrossRef]
- Achsel, T.; Stark, H.; Lührmann, R. The Sm domain is an ancient RNA-binding motif with oligo(U) specificity. Proc. Natl. Acad. Sci. USA 2001, 98, 3685–3689. [Google Scholar] [CrossRef] [PubMed]
- Collins, B.M.; Harrop, S.J.; Kornfeld, G.D.; Dawes, I.W.; Curmi, P.M.; Mabbutt, B.C. Crystal structure of a heptameric Sm-like protein complex from archaea: Implications for the structure and evolution of snRNPs. J. Mol. Biol. 2001, 309, 915–923. [Google Scholar] [CrossRef] [PubMed]
- Mura, C.; Cascio, D.; Sawaya, M.R.; Eisenberg, D.S. The crystal structure of a heptameric archaeal Sm protein: Implications for the eukaryotic snRNP core. Proc. Natl. Acad. Sci. USA 2001, 98, 5532–5537. [Google Scholar] [CrossRef] [PubMed]
- Fando, M.S.; Mikhaylina, A.O.; Lekontseva, N.V.; Tishchenko, S.V.; Nikulin, A.D. Structure and RNA-Binding Properties of Lsm Protein from Halobacterium salinarum. Biochemistry 2021, 86, 833–842. [Google Scholar] [CrossRef] [PubMed]
- Kilic, T.; Sanglier, S.; Van Dorsselaer, A.; Suck, D. Oligomerization behavior of the archaeal Sm2-type protein from Archaeoglobus fulgidus. Protein Sci. 2006, 15, 2310–2317. [Google Scholar] [CrossRef]
- Fischer, S.; Benz, J.; Späth, B.; Maier, L.K.; Straub, J.; Granzow, M.; Raabe, M.; Urlaub, H.; Hoffmann, J.; Brutschy, B.; et al. The archaeal Lsm protein binds to small RNAs. J. Biol. Chem. 2010, 285, 34429–34438. [Google Scholar] [CrossRef]
- He, W.; Parker, R. Functions of Lsm proteins in mRNA degradation and splicing. Curr. Opin. Cell Biol. 2003, 12, 346–350. [Google Scholar] [CrossRef]
- Kufel, J.; Allmang, C.; Petfalski, E.; Beggs, J.; Tollervey, D. Lsm Poteins are required for normal processing and stability of ribosomal RNAs. J. Biol. Chem. 2003, 278, 2147–2156. [Google Scholar] [CrossRef]
- Yong, J.; Wan, L.; Dreyfuss, G. Why do cells need an assembly machine for RNA-protein complexes? Trends Cell Biol. 2004, 14, 226–232. [Google Scholar] [CrossRef]
- Salgado-Garrido, J.; Bragado-Nilsson, E.; Kandels-Lewis, S.; Séraphin, B. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 1999, 18, 3451–3462. [Google Scholar] [CrossRef]
- Mura, C.; Randolph, P.S.; Patterson, J.; Cozen, A.E. Archaeal and eukaryotic homologs of Hfq: A structural and evolutionary perspective on Sm function. RNA Biol. 2013, 10, 636–651. [Google Scholar] [CrossRef] [PubMed]
- Wassarman, K.M.; Repoila, F.; Rosenow, C.; Storz, G.; Gottesman, S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001, 15, 1637–1651. [Google Scholar] [CrossRef] [PubMed]
- Märtens, B.; Bezerra, G.A.; Kreuter, M.J.; Grishkovskaya, I.; Manica, A.; Arkhipova, V.; Djinovic-Carugo, K.; Bläsi, U. The Heptameric SmAP1 and SmAP2 Proteins of the Crenarchaeon Sulfolobus solfataricus Bind to Common and Distinct RNA Targets. Life 2015, 5, 1264–1281. [Google Scholar] [CrossRef] [PubMed]
- Märtens, B.; Hou, L.; Amman, F.; Wolfinger, M.T.; Evguenieva-Hackenberg, E.; Bläsi, U. The SmAP1/2 proteins of the crenarchaeon Sulfolobus solfataricus interact with the exosome and stimulate A-rich tailing of transcripts. Nucleic Acids Res. 2017, 45, 7938–7949. [Google Scholar] [CrossRef] [PubMed]
- Tsui, H.C.; Leung, H.C.; Winkler, M.E. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 1994, 13, 35–49. [Google Scholar] [CrossRef]
- Chambers, J.R.; Bender, K.S. The RNA chaperone Hfq is important for growth and stress tolerance in Francisella novicida. PLoS ONE 2011, 6, e19797. [Google Scholar] [CrossRef]
- Chiang, M.K.; Lu, M.C.; Liu, L.C.; Lin, C.T.; Lai, Y.C. Impact of Hfq on global gene expression and virulence in Klebsiella pneumoniae. PLoS ONE 2011, 6, e22248. [Google Scholar] [CrossRef]
- Brennan, C.M.; Keane, M.L.; Hunt, T.M.; Goulet, M.T.; Mazzucca, N.Q.; Sexton, Z.; Mezoian, T.; Douglas, K.E.; Osborn, J.M.; Pellock, B.J. Shewanella oneidensis Hfq promotes exponential phase growth, stationary phase culture density, and cell survival. BMC Microbiol. 2013, 13, 33. [Google Scholar] [CrossRef]
- Lai, J.L.; Tang, D.J.; Liang, Y.W.; Zhang, R.; Chen, Q.; Qin, Z.P.; Ming, Z.H.; Tang, J.L. The RNA chaperone Hfq is important for the virulence, motility and stress tolerance in the phytopathogen Xanthomonas campestris. Environ. Microbiol. Rep. 2018, 10, 542–554. [Google Scholar] [CrossRef]
- Yao, H.; Kang, M.; Wang, Y.; Feng, Y.; Kong, S.; Cai, X.; Ling, Z.; Chen, S.; Jiao, X.; Yin, Y. An essential role for hfq involved in biofilm formation and virulence in serotype 4b Listeria monocytogenes. Microbiol. Res. 2018, 215, 148–154. [Google Scholar] [CrossRef]
- Chao, Y.; Vogel, J. The role of Hfq in bacterial pathogens. Curr. Opin. Microbiol. 2010, 13, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Maier, L.K.; Benz, J.; Fischer, S.; Alstetter, M.; Jaschinski, K.; Hilker, R.; Becker, A.; Allers, T.; Soppa, J.; Marchfelder, A. Deletion of the Sm1 encoding motif in the lsm gene results in distinct changes in the transcriptome and enhanced swarming activity of Haloferax cells. Biochimie 2015, 117, 129–137. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Payá, G.; Bautista, V.; Camacho, M.; Bonete, M.J.; Esclapez, J. Functional analysis of Lsm protein under multiple stress conditions in the extreme haloarchaeon Haloferax mediterranei. Biochimie 2021, 187, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Payá, G.; Bautista, V.; Camacho, M.; Esclapez, J.; Bonete, M.J. Comprehensive Bioinformatics Analysis of the Biodiversity of Lsm Proteins in the Archaea Domain. Microorganisms 2023, 11, 1196. [Google Scholar] [CrossRef] [PubMed]
- Lorenzetti, A.P.R.; Kusebauch, U.; Zaramela, L.S.J.; Wu, W.J.; de Almeida, J.P.P.; Turkarslan, S.; de Lomana, A.L.G.; Gomes-Filho, J.V.; Vêncio, R.Z.N.; Moritz, R.L.; et al. A Genome-Scale Atlas Reveals Complex Interplay of Transcription and Translation in an Archaeon. mSystems 2023, 8, e0081622. [Google Scholar] [CrossRef] [PubMed]
- Danson, M.D.; Hough, D.W. The Structural Basis of Protein Halophilicity. Comp. Biochem. Physiol. Part A Physiol. 1997, 117, 307–312. [Google Scholar] [CrossRef]
- Mevarech, M.; Frolow, F.; Gloss, L.M. Halophilic enzymes: Proteins with a grain of salt. Biophys. Chem. 2000, 86, 155–164. [Google Scholar] [CrossRef]
- Britton, K.L.; Baker, P.J.; Fisher, M.; Ruzheinikov, S.; Gilmour, D.J.; Bonete, M.J.; Ferrer, J.; Pire, C.; Esclapez, J.; Rice, D.W. Analysis of protein solvent interactions in glucose dehydrogenase from the extreme halophile Haloferax mediterranei. Proc. Natl. Acad. Sci. USA 2006, 103, 4846–4851. [Google Scholar] [CrossRef]
- Jolley, K.A.; Rapaport, E.; Hough, D.W.; Danson, M.J.; Woods, W.G.; Dyall-Smith, M.L. Dihydrolipoamide dehydrogenase from the halophilic archaeon Haloferax volcanii: Homologous overexpression of the cloned gene. J. Bacteriol. 1996, 178, 3044–3048. [Google Scholar] [CrossRef][Green Version]
- Fine, A.; Irihimovitch, V.; Dahan, I.; Konrad, Z.; Eichler, J. Cloning, expression, and purification of functional Sec11a and Sec11b, type I signal peptidases of the archaeon Haloferax volcanii. J. Bacteriol. 2006, 188, 1911–1919. [Google Scholar] [CrossRef]
- Timpson, L.M.; Liliensiek, A.K.; Alsafadi, D.; Cassidy, J.; Sharkey, M.A.; Liddell, S.; Allers, T.; Paradisi, F. A comparison of two novel alcohol dehydrogenase enzymes (ADH1 and ADH2) from the extreme halophile Haloferax volcanii. Appl. Microbiol. Biotechnol. 2013, 97, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Esclapez, J.; Zafrilla, B.; Martínez-Espinosa, R.M.; Bonete, M.J. Cu-NirK from Haloferax mediterranei as an example of metalloprotein maturation and exportation via Tat system. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2013, 1834, 1003–1009. [Google Scholar] [CrossRef] [PubMed]
- Matarredona, L.; Camacho, M.; García-Bonete, M.J.; Esquerra, B.; Zafrilla, B.; Esclapez, J.; Bonete, M.J. Analysis of Haloferax mediterranei Lrp Transcriptional Regulator. Genes 2021, 12, 802. [Google Scholar] [CrossRef] [PubMed]
- Thore, S.; Mayer, C.; Sauter, C.; Weeks, S.; Suck, D. Crystal structures of the Pyrococcus abyssi Sm core and its complex with RNA. Common features of RNA binding in archaea and eukarya. J. Biol. Chem. 2003, 278, 1239–1247. [Google Scholar] [CrossRef]
- Milburn, D.; Laskowski, R.A.; Thornton, J.M. Sequences annotated by structure: A tool to facilitate the use of structural information in sequence analysis. Protein Eng. 1998, 11, 855–859. [Google Scholar] [CrossRef] [PubMed]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 2015, 43, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.; Henrick, K.; Nakamura, H. Announcing the worldwide Protein Data Bank. Nat. Struct. Mol. Biol. 2003, 10, 980. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Ignoul, S.; Eggermont, J. CBS domains: Structure, function, and pathology in human proteins. Am. J. Physiol. Cell Physiol. 2005, 289, C1369–C1378. [Google Scholar] [CrossRef]
- Baykov, A.A.; Tuominen, H.K.; Lahti, R. The CBS domain: A protein module with an emerging prominent role in regulation. ACS Chem. Biol. 2011, 6, 1156–1163. [Google Scholar] [CrossRef] [PubMed]
- Oren, A. Microbial life at high salt concentrations: Phylogenetic and metabolic diversity. Saline Syst. 2008, 15, 4. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.N.; Sharma, D.; Gulati, S.; Singh, S.; Dey, R.; Pal, K.K.; Kaushik, R.; Saxena, A.K. Haloarchaea Endowed with Phosphorus Solubilization Attribute Implicated in Phosphorus Cycle. Sci. Rep. 2015, 5, 12293. [Google Scholar] [CrossRef] [PubMed]
- Soppa, J.; Baumann, A.; Brenneis, M.; Dambeck, M.; Hering, O.; Lange, C. Genomics and functional genomics with haloarchaea. Arch. Microbiol. 2008, 190, 197–215. [Google Scholar] [CrossRef] [PubMed]
- Sauer, E. Structure and RNA-binding properties of the bacterial LSm protein Hfq. RNA Biol. 2013, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Zaigler, A.; Schuster, S.C.; Soppa, J. Construction and usage of a onefold-coverage shotgun DNA microarray to characterize the metabolism of the archaeon Haloferax volcanii. Mol. Microbiol. 2003, 48, 4. [Google Scholar] [CrossRef]
- Pire, C.; Martínez-Espinosa, R.M.; Pérez-Pomares, F.; Esclapez, J.; Bonete, M.J. Ferredoxin-dependent glutamate synthase: Involvement in ammonium assimilation in Haloferax mediterranei. Extremophiles 2014, 18, 147–159. [Google Scholar] [CrossRef]
- Esclapez, J.; Pire, C.; Camacho, M.; Bautista, V.; Martínez-Espinosa, R.M.; Zafrilla, B.; Vegara, A.; Alcaraz, L.A.; Bonete, M.J. Transcriptional profiles of Haloferax mediterranei based on nitrogen availability. J. Biotechnol. 2015, 193, 100–107. [Google Scholar] [CrossRef]
- Nyström, T.; Neidhardt, F.C. Cloning, mapping and nucleotide sequencing of a gene encoding a universal stress protein in Escherichia coli. Mol. Microbiol. 1992, 6, 3187–3198. [Google Scholar] [CrossRef]
- Nyström, T.; Neidhardt, F.C. Expression and role of the universal stress protein, UspA, of Escherichia coli during growth arrest. Mol. Microbiol. 1994, 11, 537–544. [Google Scholar] [CrossRef]
- Gustavsson, N.; Diez, A.; Nyström, T. The universal stress protein paralogues of Escherichia coli are co-ordinately regulated and co-operate in the defence against DNA damage. Mol. Microbiol. 2002, 43, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Kvint, K.; Nachin, L.; Diez, A.; Nyström, T. The bacterial universal stress protein: Function and regulation. Curr. Opin. Microbiol. 2003, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Tkaczuk, K.L.; Shumilin, I.A.; Chruszcz, M.; Evdokimova, E.; Savchenko, A.; Minor, W. Structural and functional insight into the universal stress protein family. Evol. Appl. 2013, 6, 434–449. [Google Scholar] [CrossRef] [PubMed]
- Arluison, V.; Mura, C.; Guzman, M.R.; Liquier, J.; Pellegrini, O.; Gingery, M.; Régnier, P.; Marco, S. Three-dimensional structures of fibrillar Sm proteins: Hfq and other Sm-like proteins. J. Mol. Biol. 2006, 356, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Watts, N.R.; Misra, M.; Wingfield, P.T.; Stahl, S.J.; Cheng, N.; Trus, B.L.; Steven, A.C.; Williams, R.W. Three-dimensional structure of HIV-1 Rev protein filaments. J. Struct. Biol. 1998, 121, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Blanco, F.J.; Hess, S.; Pannell, L.K.; Rizzo, N.W.; Tycko, R. Solid-state NMR data support a helix-loop-helix structural model for the N-terminal half of HIV-1 Rev in fibrillar form. J. Mol. Biol. 2001, 313, 845–859. [Google Scholar] [CrossRef] [PubMed]
- Jain, C.; Belasco, J.G. Structural model for the cooperative assembly of HIV-1 Rev multimers on the RRE as deduced from analysis of assembly-defective mutants. Mol. Cell 2001, 7, 603–614. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Chenn, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. All-atom structure validation for macromolecular crystallography. Acta Cryst. 2010, 66, 16–21. [Google Scholar] [CrossRef]
- Rodriguez-Valera, F.; Juez, G.; Kushner, D. Halobacterium mediterranei spec, nov., a New Carbohydrate-Utilizing Extreme Halophile. Syst. Appl. Microbiol. 1983, 4, 369–381. [Google Scholar] [CrossRef]
- Pedro-Roig, L.; Lange, C.; Bonete, M.J.; Soppa, J.; Maupin-Furlow, J. Nitrogen regulation of protein-protein interactions and transcript levels of GlnK PII regulator and AmtB ammonium transporter homologs in Archaea. MicrobiologyOpen 2013, 2, 826–840. [Google Scholar] [CrossRef] [PubMed]
- Haque, R.U.; Paradisi, F.; Allers, T. Haloferax volcanii for biotechnology applications: Challenges, current state and perspectives. Appl. Microbiol. Biotechnol. 2020, 104, 1371–1382. [Google Scholar] [CrossRef] [PubMed]
- Haque, R.U.; Paradisi, F.; Allers, T. Haloferax volcanii as immobilised whole cell biocatalyst: New applications for halophilic systems. Appl. Microbiol. Biotechnol. 2019, 103, 3807–3817. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
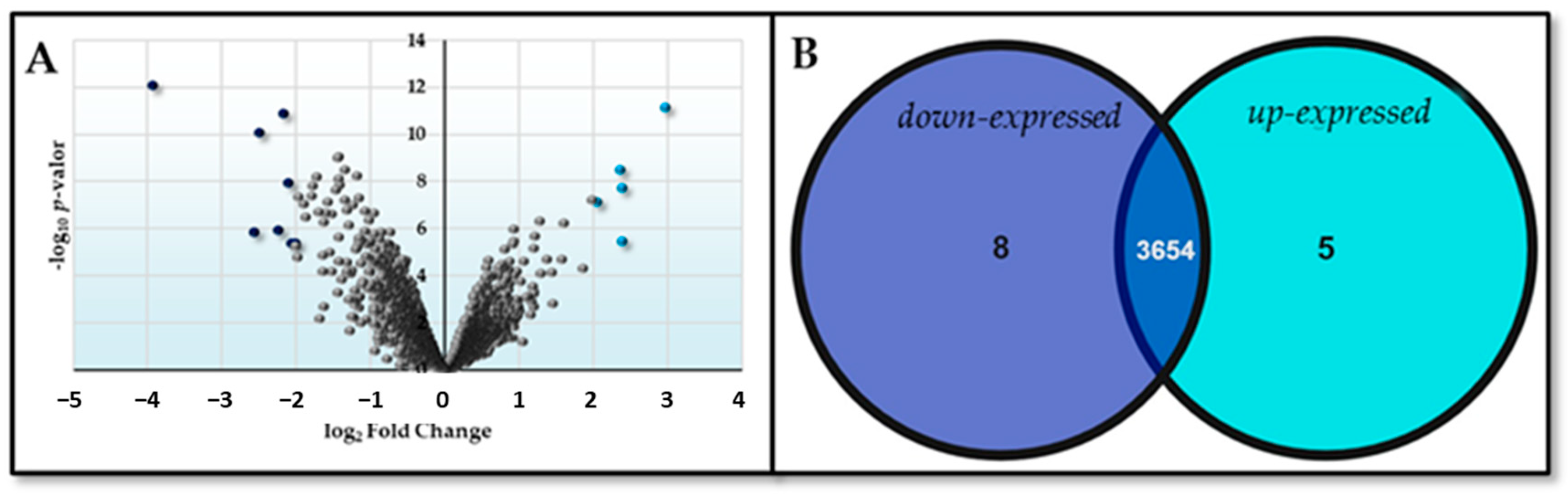
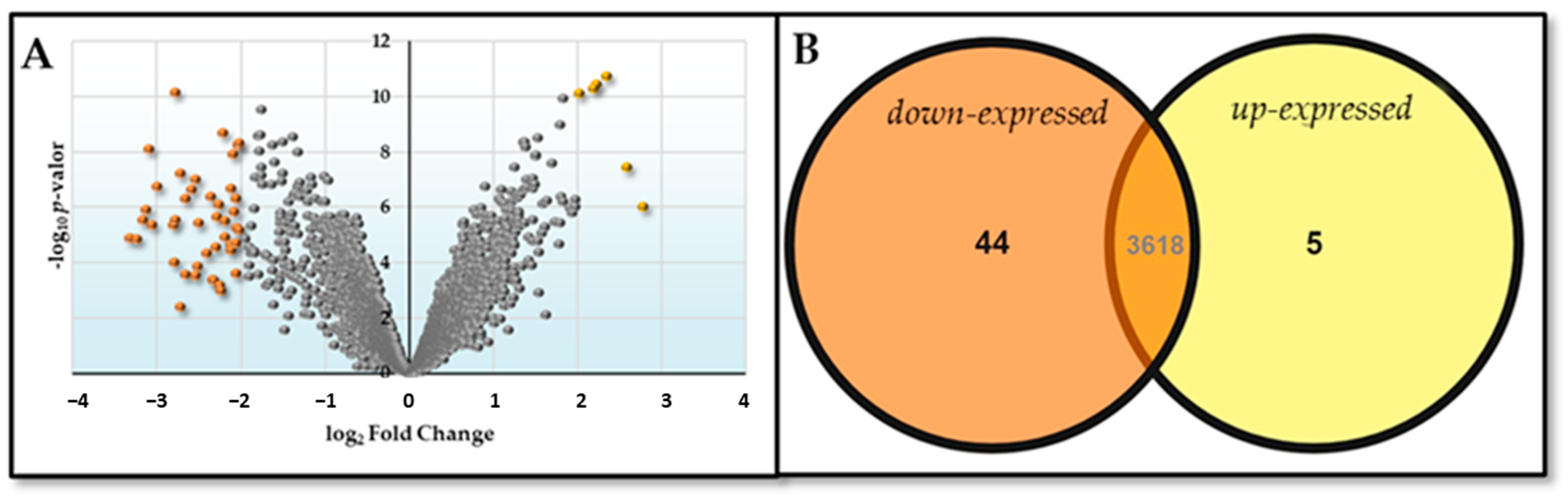
| C Deficit | N Deficit | ||
| Energy metabolism | |||
| HFX_0944 | b(o/a)3-type cytochrome-c oxidase subunit 1 | −0.41 | −2.07 |
| HFX_0429 | cytochrome ubiquinol oxidase subunit I | −0.33 | −2.34 |
| HFX_1925 | FAD-dependent oxidoreductase | −0.34 | −2.55 |
| HFX_0428 | cytochrome d ubiquinol oxidase subunit II | −0.77 | −2.72 |
| HFX_0943 | cytochrome c oxidase subunit II | −0.89 | −2.82 |
| HFX_1927 | electron transfer flavoprotein subunit beta/FixA family protein | −0.49 | −3.14 |
| HFX_1926 | electron transfer flavoprotein subunit alpha/FixB family protein | −0.53 | −3.33 |
| Genes that encode stress proteins | |||
| HFX_0946 | universal stress protein | 0.22 | −2.05 |
| HFX_1094 | universal stress protein | −0.94 | −2.09 |
| HFX_2288 | universal stress protein | −1.55 | −2.13 |
| HFX_1885 | universal stress protein | −0.40 | −3.10 |
| HFX_1928 | universal stress protein | −0.11 | −3.24 |
| DNA metabolism and processing | |||
| HFX_0688 | PadR family transcriptional regulator | 2.37 | 0.29 |
| HFX_4107 | C2H2-type zinc finger protein | 2.40 | 0.18 |
| HFX_1341 | helix-turn-helix domain-containing protein | 0.14 | −2.06 |
| HFX_0165 | winged helix-turn-helix transcriptional regulator | 0.15 | −2.42 |
| Carbon metabolism | |||
| HFX_4023 | aldehyde dehydrogenase family protein | −0.09 | 2.58 |
| HFX_6051 | thiolase family protein | −2.16 | 0.66 |
| Metabolism of cofactors and vitamins | |||
| HFX_5076 | dethiobiotin synthase | −0.52 | −2.22 |
| HFX_5079 | biotin synthase BioB | −1.98 | −2.66 |
| HFX_5077 | 8-amino-7-oxononanoate synthase | −1.39 | −2.73 |
| Nitrogen metabolism | |||
| HFX_5091 | TAT-dependent nitrous-oxide reductase | −0.09 | −2.19 |
| Lipid metabolism | |||
| HFX_1281 | beta-ketoacyl-ACP reductase | −0.47 | 2.22 |
| Cell motility proteins | |||
| HFX_6257 | type IV pilin N-terminal domain-containing protein | 2.98 | −1.80 |
| Transporters | |||
| HFX_1359 | substrate-binding domain-containing protein | −1.68 | −2.26 |
| Environmental signal processing | |||
| HFX_5231 | CBS domain-containing protein | −1.63 | −3.07 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Payá, G.; Bautista, V.; Pastor-Soler, S.; Camacho, M.; Esclapez, J.; Bonete, M.-J. Analysis of Lsm Protein-Mediated Regulation in the Haloarchaeon Haloferax mediterranei. Int. J. Mol. Sci. 2024, 25, 580. https://doi.org/10.3390/ijms25010580
Payá G, Bautista V, Pastor-Soler S, Camacho M, Esclapez J, Bonete M-J. Analysis of Lsm Protein-Mediated Regulation in the Haloarchaeon Haloferax mediterranei. International Journal of Molecular Sciences. 2024; 25(1):580. https://doi.org/10.3390/ijms25010580
Chicago/Turabian StylePayá, Gloria, Vanesa Bautista, Sandra Pastor-Soler, Mónica Camacho, Julia Esclapez, and María-José Bonete. 2024. "Analysis of Lsm Protein-Mediated Regulation in the Haloarchaeon Haloferax mediterranei" International Journal of Molecular Sciences 25, no. 1: 580. https://doi.org/10.3390/ijms25010580
APA StylePayá, G., Bautista, V., Pastor-Soler, S., Camacho, M., Esclapez, J., & Bonete, M.-J. (2024). Analysis of Lsm Protein-Mediated Regulation in the Haloarchaeon Haloferax mediterranei. International Journal of Molecular Sciences, 25(1), 580. https://doi.org/10.3390/ijms25010580









