Application of Long-Chained Auxin Conjugates Influenced Auxin Metabolism and Transcriptome Response in Brassica rapa L. ssp. pekinensis
Abstract
1. Introduction
2. Results
2.1. Root Growth Bioassay
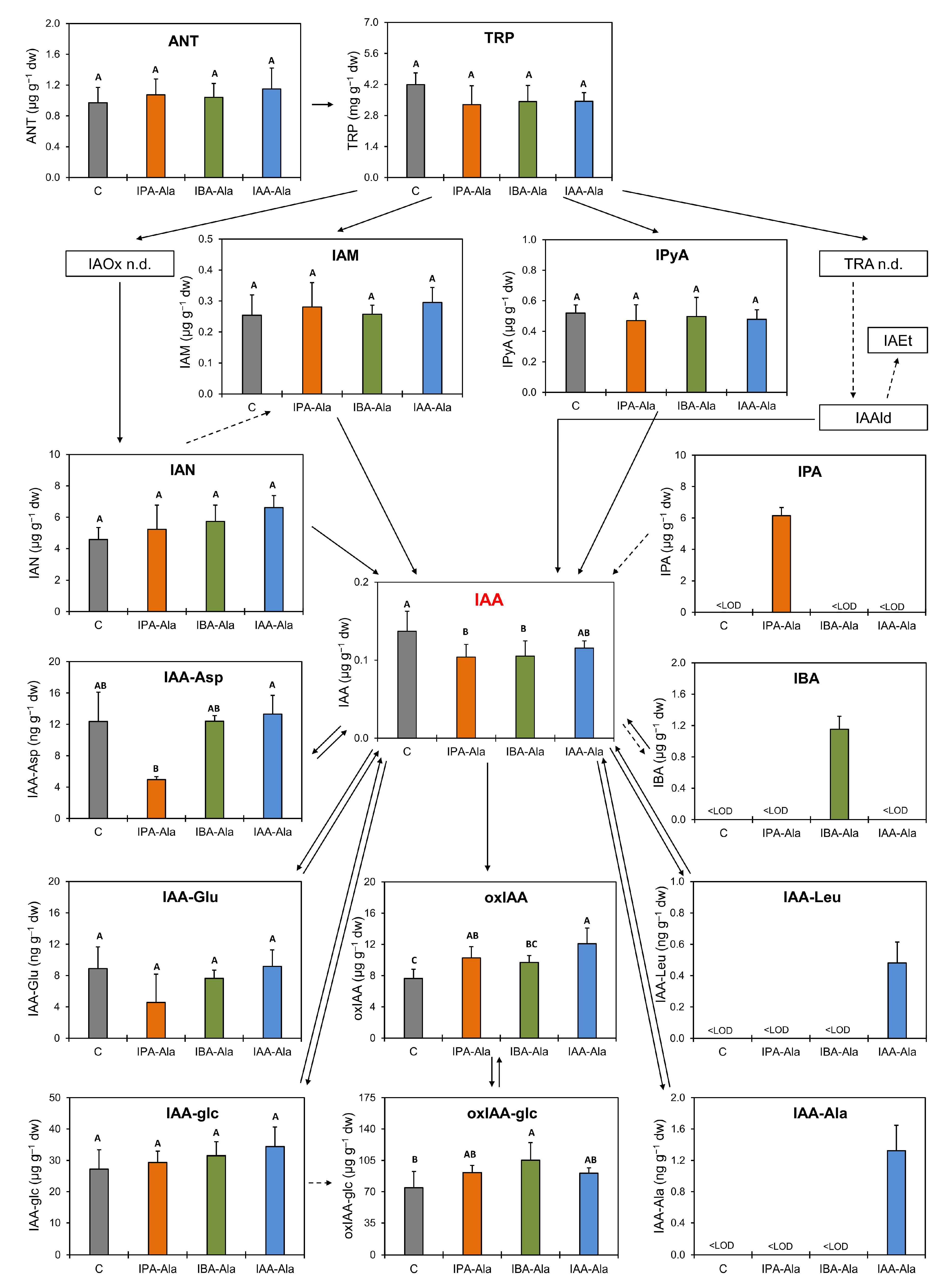
2.2. Analysis of Auxin, Auxin Precursors and Metabolites
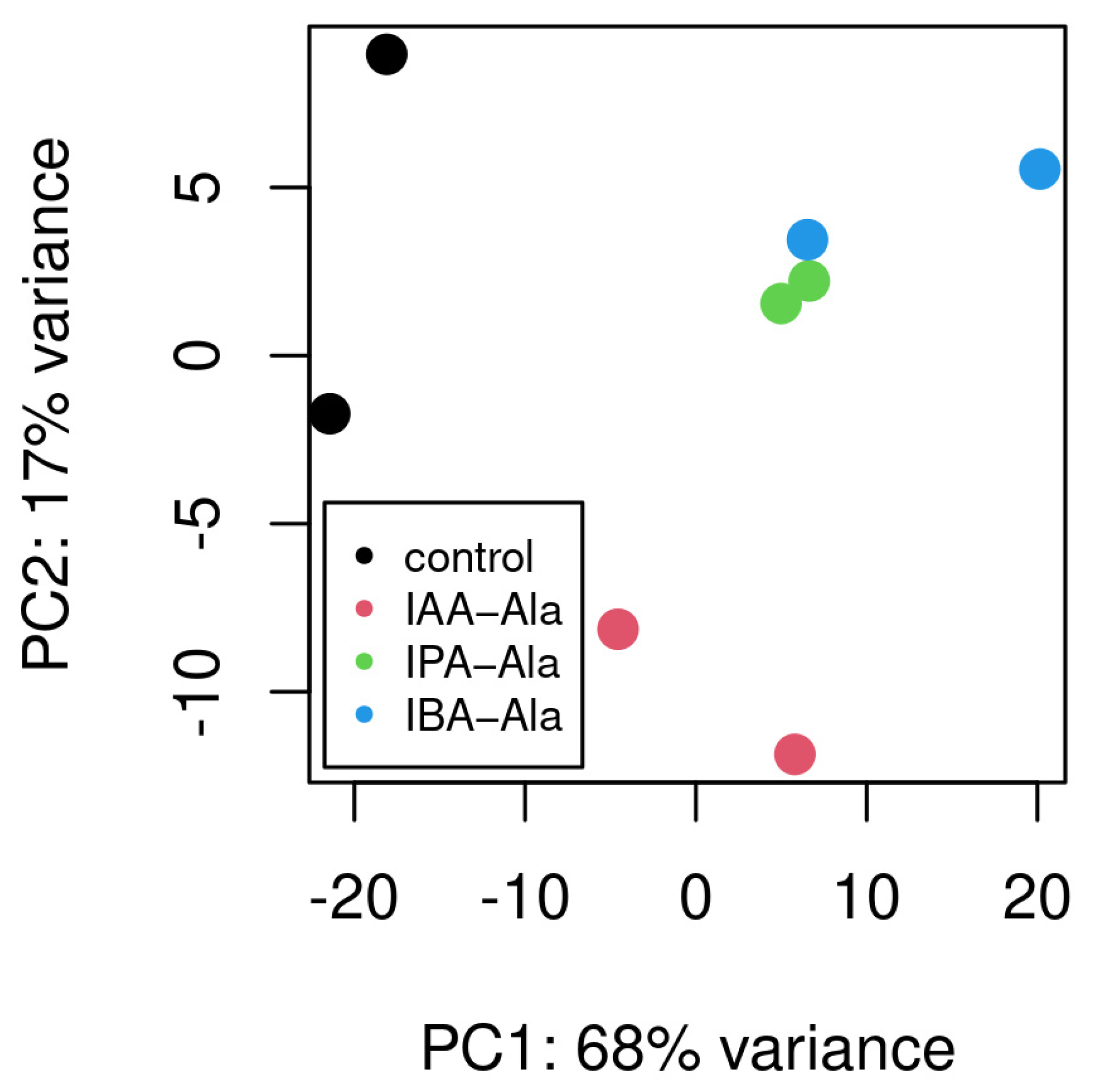
2.3. RNA-Seq Analysis
2.3.1. Overall Changes in Gene Expression upon Treatments
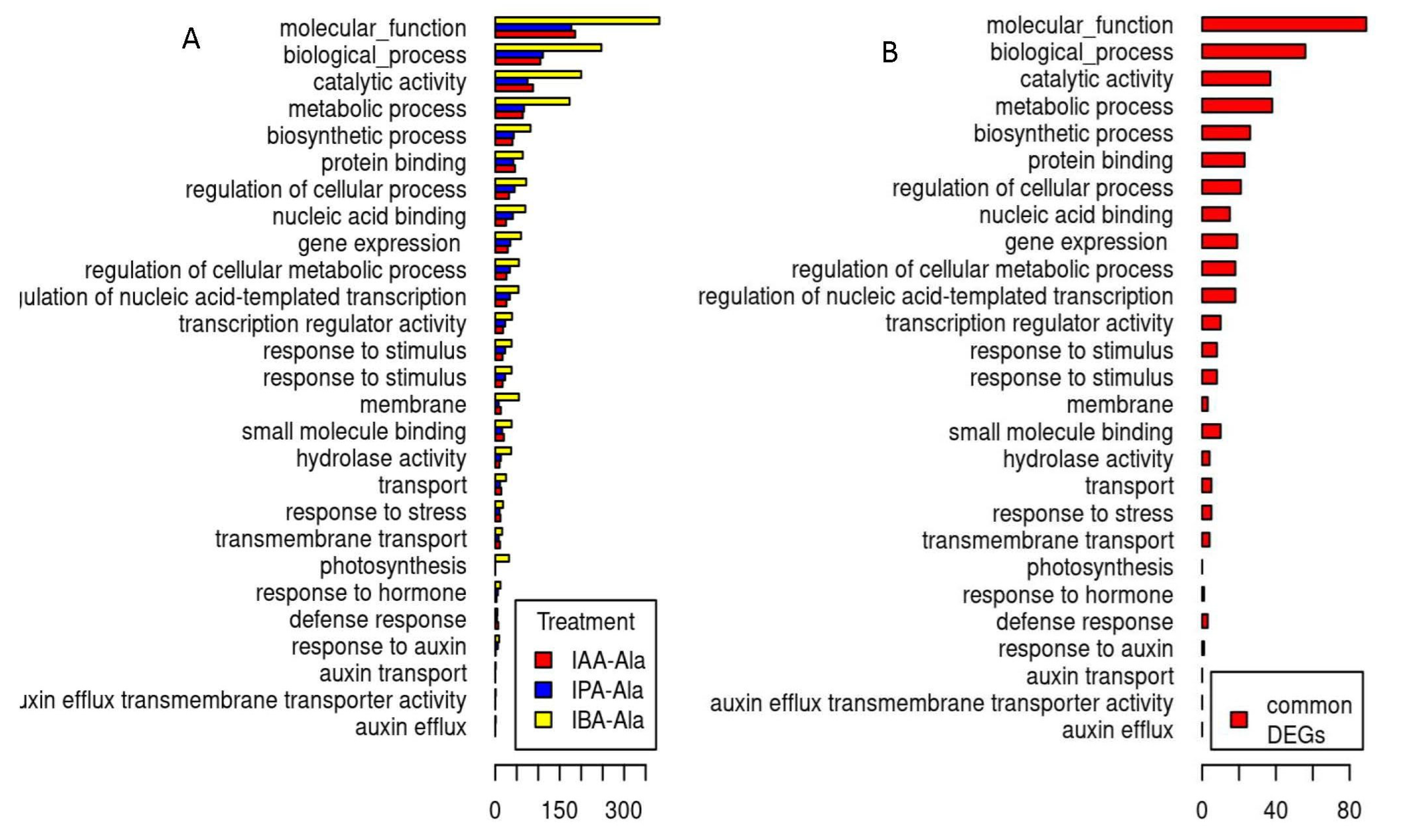
2.3.2. Differentially Expressed Genes (DEGs) across Gene Ontology (GO) Categories Linked to Treatments
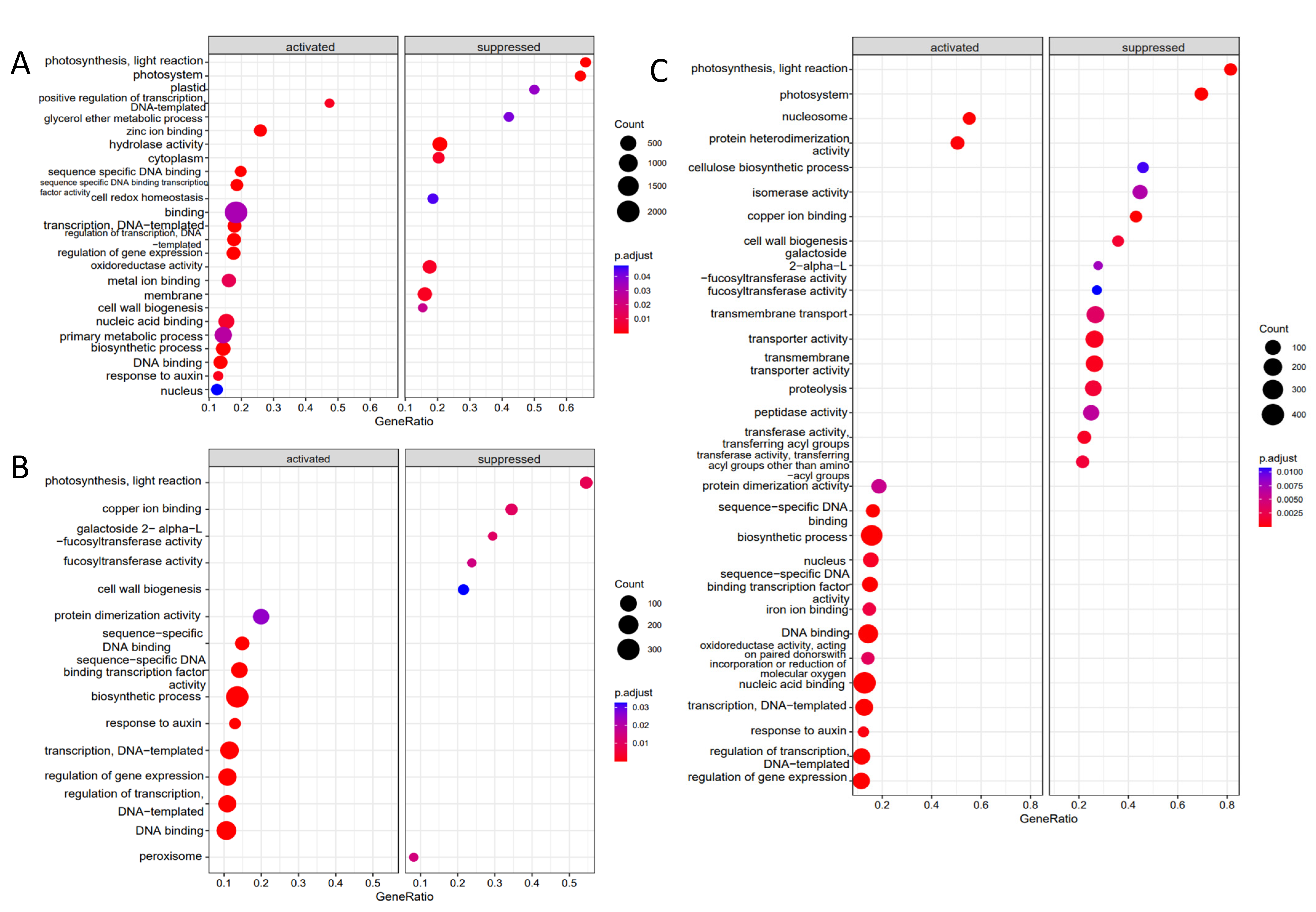
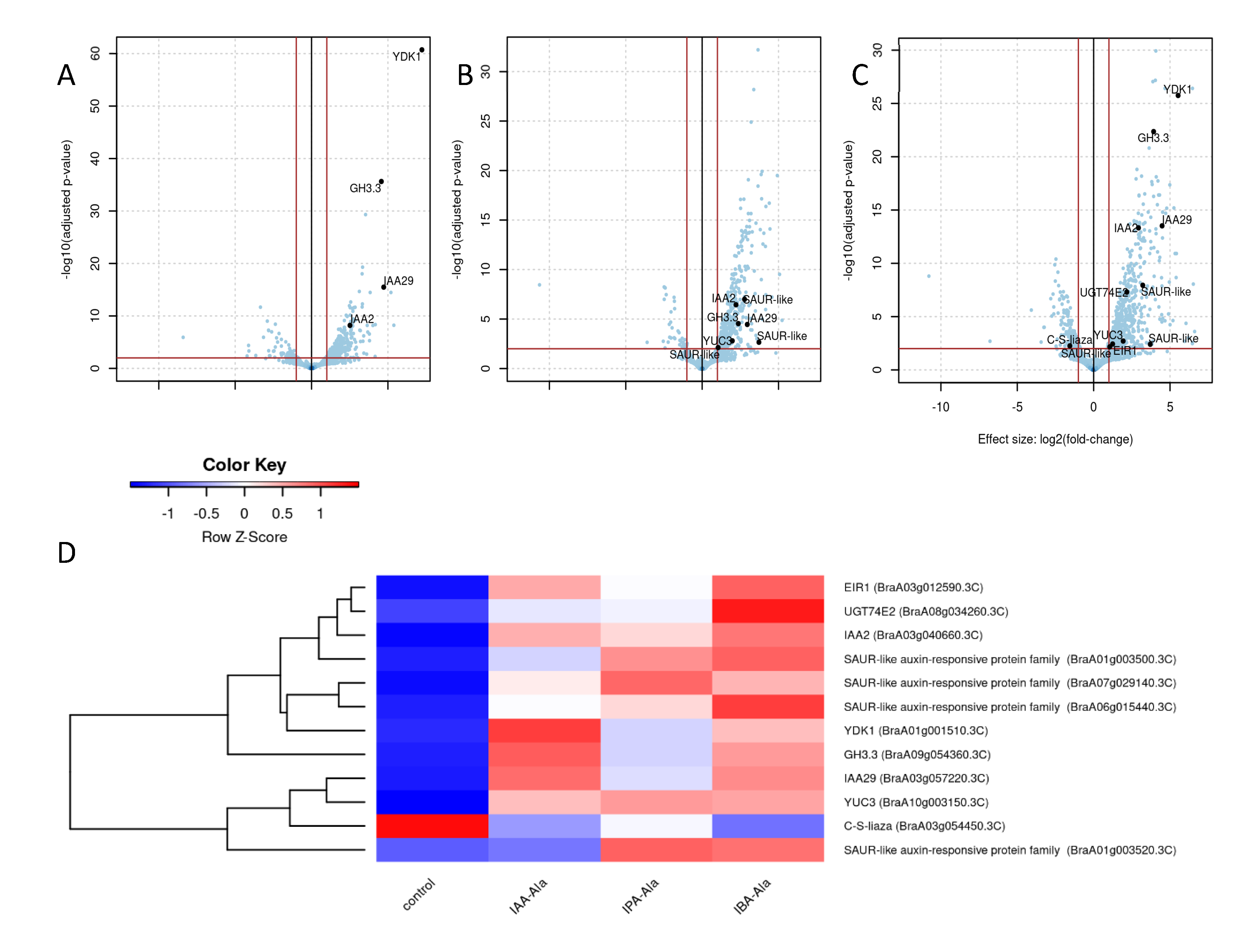
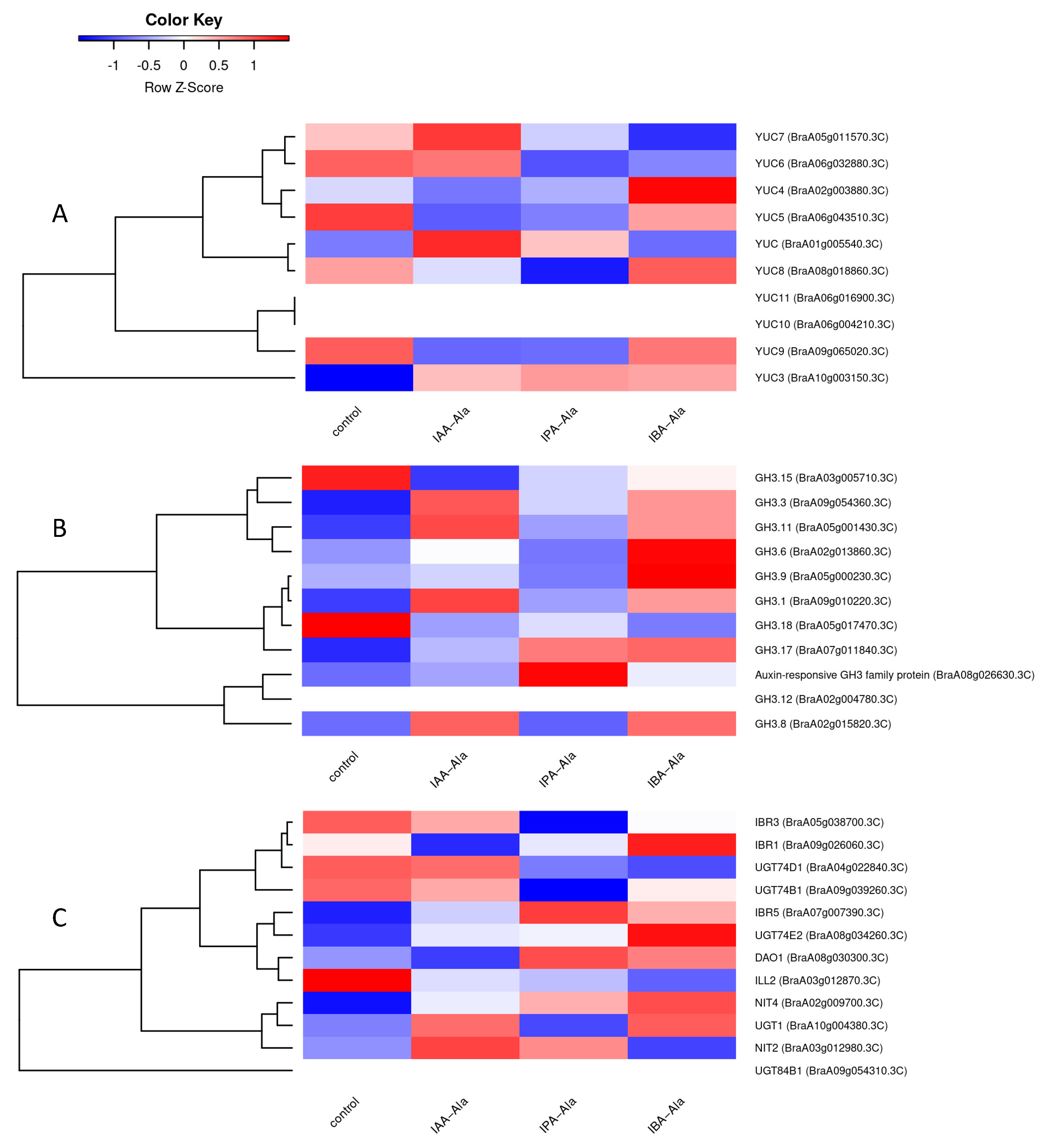
2.3.3. DEGs Linked to Auxin Metabolism
3. Discussion
3.1. Root Growth Inhibition by Auxin Amino Acid Conjugates
3.2. Auxin Metabolome
3.3. Transcriptome Analysis
4. Materials and Methods
4.1. Plant Materials
4.2. Auxin Quantification
4.3. Statistical Analysis
4.4. RNA-Seq Analysis
4.5. Mapping of B. rapa Genes to A. thaliana Genes and Gene Annotations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gomes, G.L.B.; Scortecci, K.C. Auxin and its role in plant development: Structure, signalling, regulation and response mechanisms. Plant Biol. 2021, 23, 894–904. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zhang, F.; Friml, J.; Ding, Z. Auxin signaling: Research advances over the past 30 years. J. Integr. Plant Biol. 2022, 64, 371–392. [Google Scholar] [CrossRef] [PubMed]
- Bartel, B.; LeClere, S.; Magidin, M.; Zolman, B.K. Inputs to the active indole-3-acetic acid pool: De novo synthesis, conjugate hydrolysis, and indole-3-butyric acid b-oxidation. J. Plant Growth Regul. 2001, 20, 198–216. [Google Scholar] [CrossRef]
- Ludwig-Müller, J. Auxin conjugates: Their role for plant development and in the evolution of land plants. J. Exp. Bot. 2011, 62, 1757–1773. [Google Scholar] [CrossRef] [PubMed]
- Ljung, K. Auxin metabolism and homeostasis during plant development. Development 2013, 140, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Strader, L.C.; Bartel, B. Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol. Plant 2011, 4, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Mihaljević, S.; Salopek-Sondi, B. Amide conjugate of Indole-3-butyric acid improves rooting of highbush blueberry. Plant Soil Env. 2012, 58, 236–241. [Google Scholar] [CrossRef]
- Savić, B.; Tomić, S.; Magnus, V.; Gruden, K.; Barle, K.; Grenković, R.; Ludwig-Müller, J.; Salopek-Sondi, B. Auxin amidohydrolases from Brassica rapa cleave the alanine conjugate of indolepropionic acid as a preferable substrate: A biochemical and modeling approach. Plant Cell Physiol. 2009, 50, 1577–1589. [Google Scholar] [CrossRef][Green Version]
- Ludwig-Müller, J. Indole-3-butyric acid in plant growth and development. Plant Growth Regul. 2000, 32, 219–230. [Google Scholar] [CrossRef]
- Novák, O.; Hényková, E.; Sairanen, I.; Kowalczyk, M.; Pospíšil, T.; Ljung, K. Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J. 2012, 72, 523–536. [Google Scholar] [CrossRef]
- Zolman, B.K.; Yoder, A.; Bartel, B. Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 2000, 156, 1323–1337. [Google Scholar] [CrossRef] [PubMed]
- Kreiser, M.; Giblin, C.; Murphy, R.; Fiesel, P.; Braun, L.; Johnson, G.; Wyse, D.; Cohen, J.D. Conversion of indole-3-butyric acid to indole-3-acetic acid in shoot tissue of hazelnut (Corylus) and elm (Ulmus). J. Plant Growth Regul. 2016, 35, 710–721. [Google Scholar] [CrossRef]
- Aryal, B.; Huynh, J.; Schneuwly, J.; Siffert, A.; Liu, J.; Alejandro, S.; Ludwig-Müller, J.; Martinoia, E.; Geisler, M. ABCG36/PEN3/PDR8 is an exporter of the auxin precursor, indole-3-butyric acid, and involved in auxin-controlled development. Front. Plant Sci. 2019, 10, 899. [Google Scholar] [CrossRef] [PubMed]
- Segal, L.M.; Wightman, F. Gas chromatographic and GC–MS evidence for the occurrence of 3-indolylpropionic acid and 3-indolylacetic acid in seedlings of Cucurbita pepo. Physiol. Plant. 1982, 56, 367–370. [Google Scholar] [CrossRef]
- Schneider, E.A.; Kazakoff, C.W.; Wightman, F. Gas chromatography–mass spectrometry evidence for several endogenous auxins in pea seedling organs. Planta 1985, 165, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Walker, T.S.; Bais, H.P.; Halligan, K.M.; Stermitz, F.R.; Vivanco, J.M. Metabolic profiling of root exudates of Arabidopsis thaliana. J. Agric. Food Chem. 2003, 51, 2548–2554. [Google Scholar] [CrossRef] [PubMed]
- Elsden, S.R.; Hilton, M.G.; Waller, J.M. The end products of the metabolism of aromatic amino acids by Clostridia. Arch. Microbiol. 1976, 107, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, N.; Onodera, R.; Or-Rashid, M.M. Degradation of tryptophan and related indolic compounds by ruminal bacteria, protozoa and their mixture in vitro. Amino Acids 2003, 24, 73–80. [Google Scholar] [CrossRef]
- Barkawi, L.S.; Yuen-Yee Tam, Y.-Y.; Tillman, J.A.; Pederson, B.; Calio, J.; Al-Amier, H.; Emerick, M.; Normanly, J.; Cohen, J.D. A high-throughput method for the quantitative analysis of indole-3-acetic acid and other auxins from plant tissue. Anal. Biochem. 2008, 372, 177–188. [Google Scholar] [CrossRef]
- Ostrowski, M.; Ciarkowska, A.; Jakubowska, A. The auxin conjugate indole-3-acetyl-aspartate affects responses to cadmium and salt stress in Pisum sativum L. J. Plant Physiol. 2016, 191, 63–72. [Google Scholar] [CrossRef]
- Fu, X.; Shi, Z.; Jiang, Y.; Jiang, L.; Qi, M.; Xu, T.; Li, T. A family of auxin conjugate hydrolases from Solanum lycopersicum and analysis of their roles in flower pedicel abscission. BMC Plant Biol. 2019, 19, 233. [Google Scholar] [CrossRef] [PubMed]
- LeClere, S.; Tellez, R.; Rampey, R.A.; Seiichi, P.T.; Matsuda, S.P.T.; Bartel, B. Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 2002, 277, 20446–20452. [Google Scholar] [CrossRef] [PubMed]
- Campanella, J.J.; Olajide, A.F.; Magnus, V.; Ludwig-Müller, J. A novel auxin conjugate hydrolase from wheat with substrate specificity for longer side-chain auxin amide conjugates. Plant Physiol. 2004, 135, 2230–2240. [Google Scholar] [CrossRef] [PubMed]
- Campanella, J.J.; Smith, S.M.; Leibu, D.; Wexler, S.; Ludwig-Müller, J. The auxin conjugate hydrolase family of Medicago truncatula and their expression during the interaction with two symbionts. J. Plant Growth Regul. 2008, 27, 26–38. [Google Scholar] [CrossRef]
- Smolko, A.; Šupljika, F.; Martinčić, J.; Jajčanin-Jozić, N.; Grabar-Branilović, M.; Tomić, S.; Ludwig-Müller, J.; Piantanida, I.; Salopek-Sondi, B. The role of conserved Cys residues in Brassica rapa auxin amidohydrolase: The Cys139 is crucial for the enzyme activity and the Cys320 regulates enzyme stability. Phys. Chem. Chem. Phys. 2016, 18, 8890–8900. [Google Scholar] [CrossRef]
- Smolko, A.; Ludwig-Müller, J.; Salopek-Sondi, B. Auxin amidohydrolases—From structure to function: Revisited. Croat. Chem. Acta 2018, 91, 233–239. [Google Scholar] [CrossRef]
- Korasick, D.A.; Enders, T.A.; Strader, L.C. Auxin biosynthesis and storage forms. J. Exp. Bot. 2013, 64, 2541–2555. [Google Scholar] [CrossRef]
- Tanaka, K.; Hayashi, K.; Natsume, M.; Kamiya, Y.; Sakakibara, H.; Kawaide, H.; Kasahara, H. UGT74D1 catalyzes the glucosylation of 2-oxindole-3-acetic acid in the auxin metabolic pathway in Arabidopsis. Plant Cell Physiol. 2014, 55, 218–228. [Google Scholar] [CrossRef]
- Porco, S.; Pěnčík, A.; Rashe, A.; Voß, U.; Casanova-Sáez, R.; Bishopp, A.; Golebiowska, A.; Bhosale, R.; Swarup, R.; Swarup, K.; et al. Dioxygenase-encoding AtDAO1 gene controls IAA oxidation and homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 11016–11021. [Google Scholar] [CrossRef]
- Hayashi, K.-I.; Arai, K.; Aoi, Y.; Tanaka, Y.; Hira, H.; Guo, R.; Hu, Y.; Ge, C.; Zhao, Y.; Kasahara, H.; et al. The main oxidative inactivation pathway of the plant hormone auxin. Nat. Commun. 2021, 12, 6752. [Google Scholar] [CrossRef]
- Brunoni, F.; Pěnčík, A.; Žukauskaitė, A.; Ament, A.; Kopečná, M.; Collani, S.; Kopečný, D.; Novák, O. Amino acid conjugation of oxIAA is a secondary metabolic regulation involved in auxin homeostasis. New Phytol. 2023, 238, 2264–2270. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, M.D.; Naur, P.; Halkier, B.A. Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J. 2004, 37, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Rampey, R.A.; LeClere, S.; Kowalczyk, M.; Ljung, K.; Sandberg, G.; Bartel, B. A Family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol. 2004, 135, 978–988. [Google Scholar] [CrossRef] [PubMed]
- Pěnčík, A.; Casanova-Sáez, R.; Pilařová, V.; Žukauskaite, A.; Pinto, R.; Micol, J.L.; Ljung, K.; Novák, O. Ultra-rapid auxin metabolite profiling for high-throughput mutant screening in Arabidopsis. J. Exp. Bot. 2018, 69, 2569–2579. [Google Scholar] [CrossRef] [PubMed]
- Östin, A.; Kowalyczk, M.; Bhalerao, R.P.; Sandberg, G. Metabolism of indole-3-acetic acid in Arabidopsis. Plant Physiol. 1998, 118, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Barratt, N.; Dong, W.; Gage, D.; Magnus, V.; Town, C. Metabolism of exogenous auxin by Arabidopsis thaliana: Identification of the conjugate Na-(indol-3-ylacetyl)-glutamine and initiation of a mutant screen. Physiol Plant. 1999, 105, 207–217. [Google Scholar] [CrossRef]
- Pěnčík, A.; Simonovik, B.; Petersson, S.V.; Henyková, E.; Simon, S.; Greenham, K.; Zhang, Y.; Kowalczyk, M.; Estelle, M.; Zažímalová, E.; et al. Regulation of auxin homeostasis and gradients in Arabidopsis roots through the formation of the IAA catabolite oxIAA. Plant Cell 2013, 25, 3858–3870. [Google Scholar] [CrossRef]
- Ayala, P.G.; Acevedo, R.M.; Luna, C.V.; Rivarola, M.; Acuña, C.; Marcucci Poltri, S.; González, A.M.; Sansberro, P.A. Transcriptome dynamics of rooting zone and leaves during in vitro adventitious root formation in Eucalyptus nitens. Plants 2022, 11, 3301. [Google Scholar] [CrossRef]
- Paponova, I.A.; Paponov, M.; Teale, W.; Menges, M.; Chakrabortee, S.; Murray, J.A.H.; Palme, K. Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol. Plant 2008, 1, 321–337. [Google Scholar] [CrossRef]
- Wei, L.; Yang, B.; Jian, H.; Zhang, A.; Liu, R.; Zhu, Y.; Ma, J.; Shi, X.; Wang, R.; Li, J.-N.; et al. Genome-wide identification and characterization of Gretchen Hagen3 (GH3) family genes in Brassica napus. Genome 2019, 62, 597–608. [Google Scholar] [CrossRef]
- Toufighi, K.; Brady, S.M.; Austin, R.; Ly, E.; Provart, N.J. The botany array resource: E-northerns, expression angling, and promoter analyses. Plant J. 2005, 43, 153–163. [Google Scholar] [CrossRef]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provart, N.J. An “Electronic fluorescent pictograph” browser for exploring and analyzing large scale biological data sets. PLoS ONE 2007, 2, e718. [Google Scholar] [CrossRef] [PubMed]
- Goda, H.; Sasaki, E.; Akiyama, K.; Maruyama-Nakashita, A.; Nakabayashi, K.; Li, W.; Ogawa, M.; Yamauchi, Y.; Preston, J.; Aoki, K.; et al. The AtGenExpress hormone and chemical treatment data set: Experimental design, data evaluation, model data analysis and data access. Plant J. 2008, 55, 526–542. [Google Scholar] [CrossRef] [PubMed]
- Staswick, P.E.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 2005, 17, 616–627. [Google Scholar] [CrossRef] [PubMed]
- Rekhter, D.; Lüdke, D.; Ding, Y.; Feussner, K.; Zienkiewicz, K.; Lipka, V.; Wiermer, M.; Zhang, Y.; Feussner, I. Isochorismate-derived biosynthesis of the plant stress hormone salicylic acid. Science 2019, 365, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.J.; Park, J.W.; Lee, H.W.; Kim, J. Genome-wide analysis of the auxin-responsive transcriptome downstream of iaa1 and its expression analysis reveal the diversity and complexity of auxin-regulated gene expression. J. Exp. Bot. 2009, 60, 3935–3957. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Chen, H.; Wang, T.; He, X.; Cai, X.; Lin, R.; Liang, J.; Wu, J.; King, G.; Wang, X. BRAD V3.0: An upgraded Brassicaceae database. Nucleic Acids Res. 2022, 50, D1432–D1441. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. ClusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. MC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Poole, R.L. The TAIR Database. In Plant Bioinformatics. Methods in Molecular Biology™; Edwards, D., Ed.; Humana Press: Totowa, NJ, USA, 2005; Volume 406. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smolko, A.; Repar, J.; Matković, M.; Pavlović, I.; Pěnčík, A.; Novák, O.; Ludwig-Müller, J.; Salopek-Sondi, B. Application of Long-Chained Auxin Conjugates Influenced Auxin Metabolism and Transcriptome Response in Brassica rapa L. ssp. pekinensis. Int. J. Mol. Sci. 2024, 25, 447. https://doi.org/10.3390/ijms25010447
Smolko A, Repar J, Matković M, Pavlović I, Pěnčík A, Novák O, Ludwig-Müller J, Salopek-Sondi B. Application of Long-Chained Auxin Conjugates Influenced Auxin Metabolism and Transcriptome Response in Brassica rapa L. ssp. pekinensis. International Journal of Molecular Sciences. 2024; 25(1):447. https://doi.org/10.3390/ijms25010447
Chicago/Turabian StyleSmolko, Ana, Jelena Repar, Marija Matković, Iva Pavlović, Aleš Pěnčík, Ondřej Novák, Jutta Ludwig-Müller, and Branka Salopek-Sondi. 2024. "Application of Long-Chained Auxin Conjugates Influenced Auxin Metabolism and Transcriptome Response in Brassica rapa L. ssp. pekinensis" International Journal of Molecular Sciences 25, no. 1: 447. https://doi.org/10.3390/ijms25010447
APA StyleSmolko, A., Repar, J., Matković, M., Pavlović, I., Pěnčík, A., Novák, O., Ludwig-Müller, J., & Salopek-Sondi, B. (2024). Application of Long-Chained Auxin Conjugates Influenced Auxin Metabolism and Transcriptome Response in Brassica rapa L. ssp. pekinensis. International Journal of Molecular Sciences, 25(1), 447. https://doi.org/10.3390/ijms25010447








