Development and Regulation of the Extreme Biofilm Formation of Deinococcus radiodurans R1 under Extreme Environmental Conditions
Abstract
:1. Introduction
2. Results
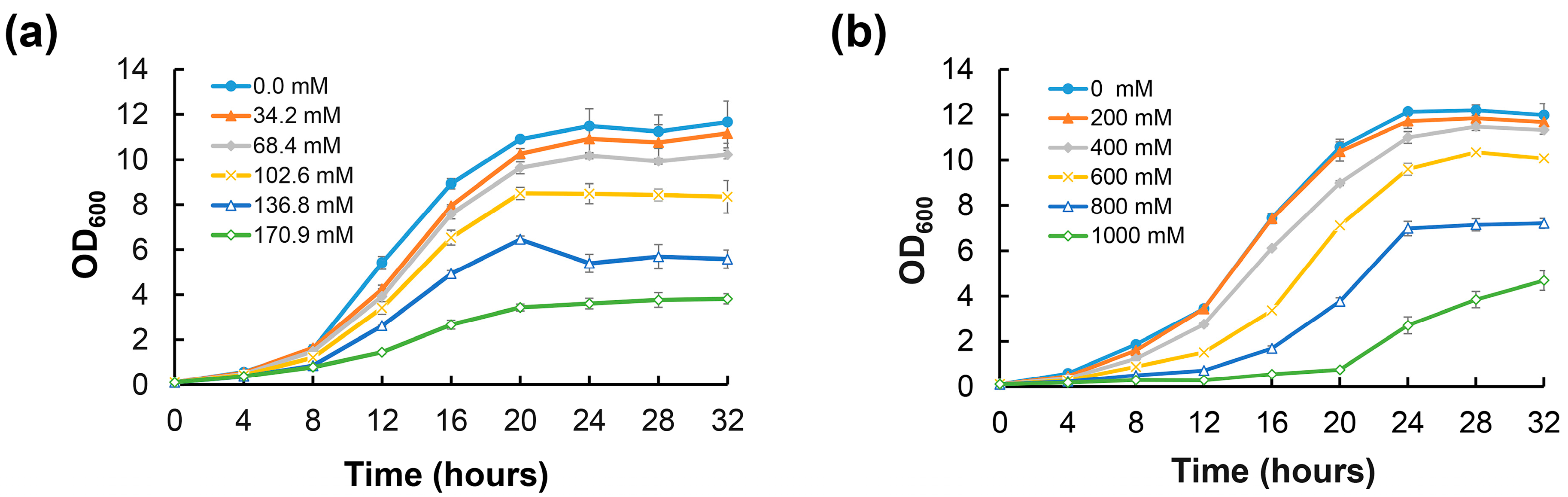
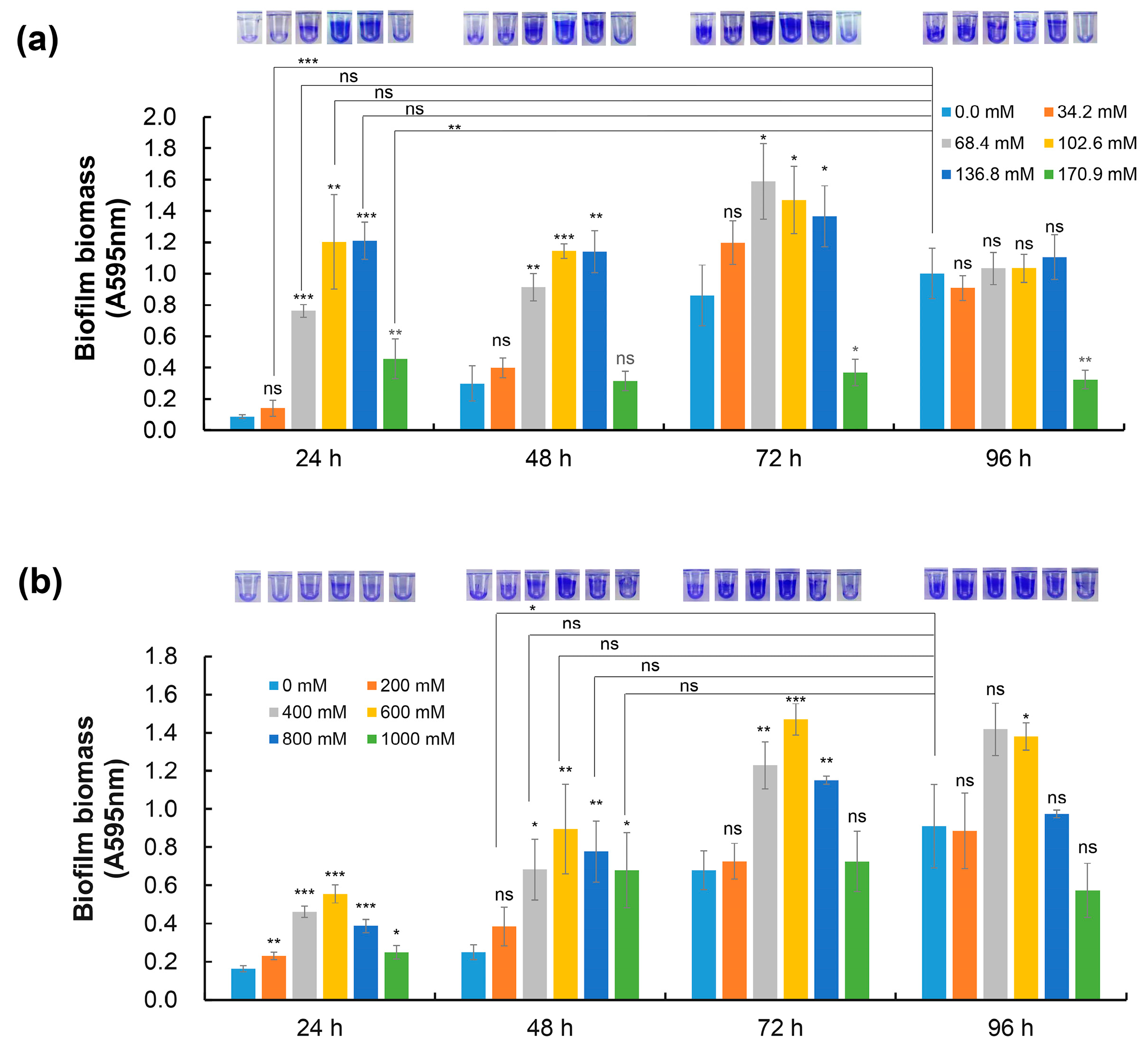
2.1. NaCl and Sorbitol Stress Induce Biofilm Formation of D. radiodurans
2.2. Biofilms Are Beneficial to the Adaptation of D. radiodurans to Extreme Conditions
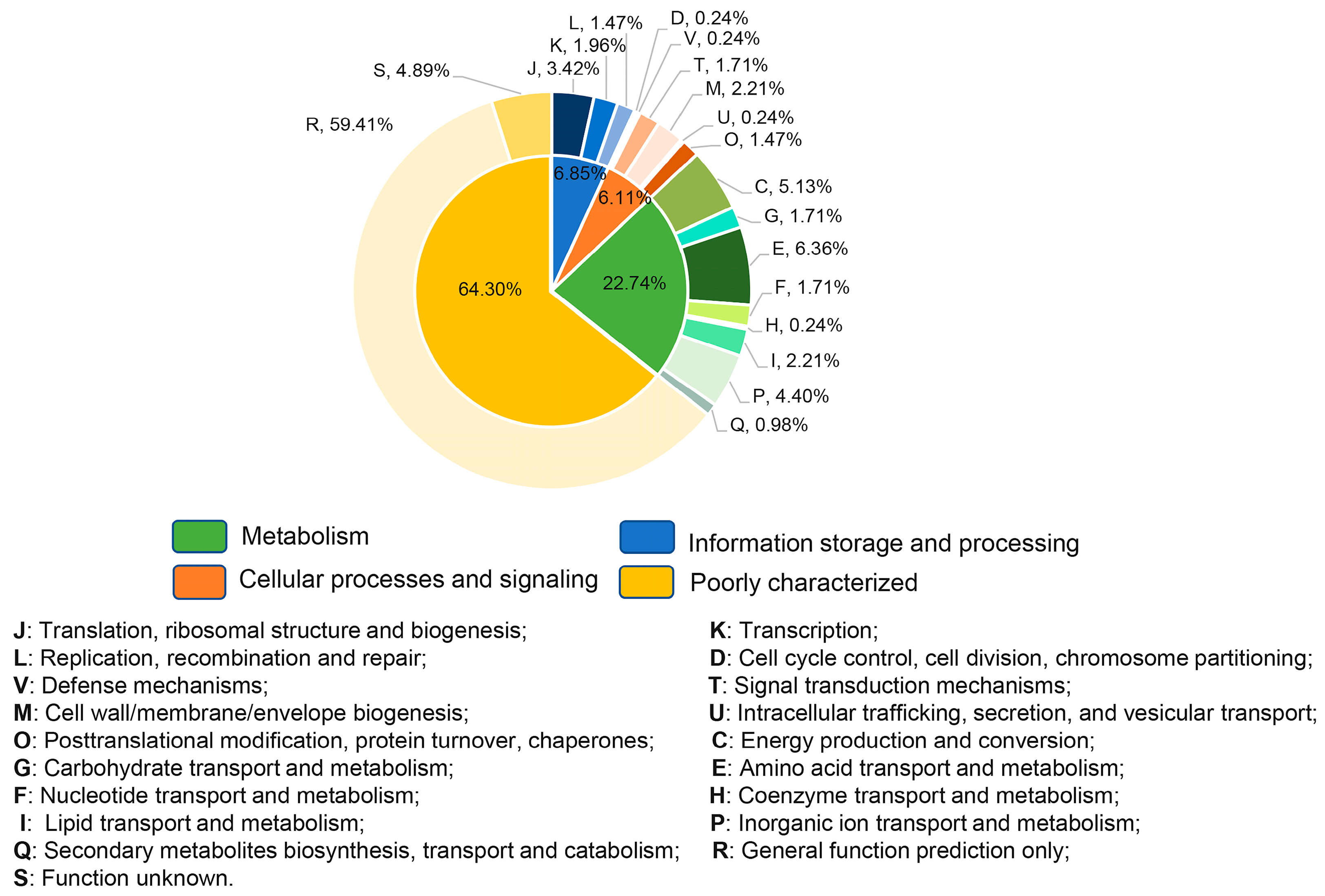
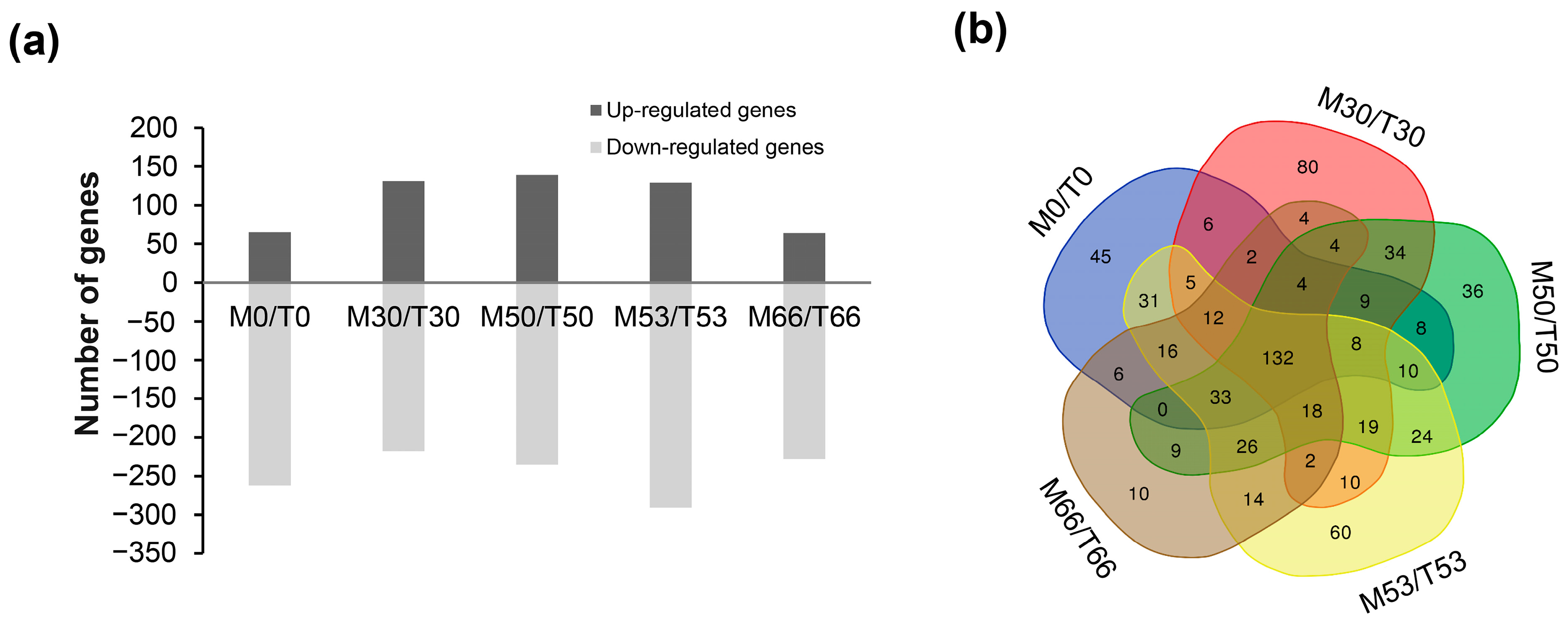
2.3. Genome-Wide Transcriptome Analyses of D. radiodurans Cells under Biofilm Formation Conditions
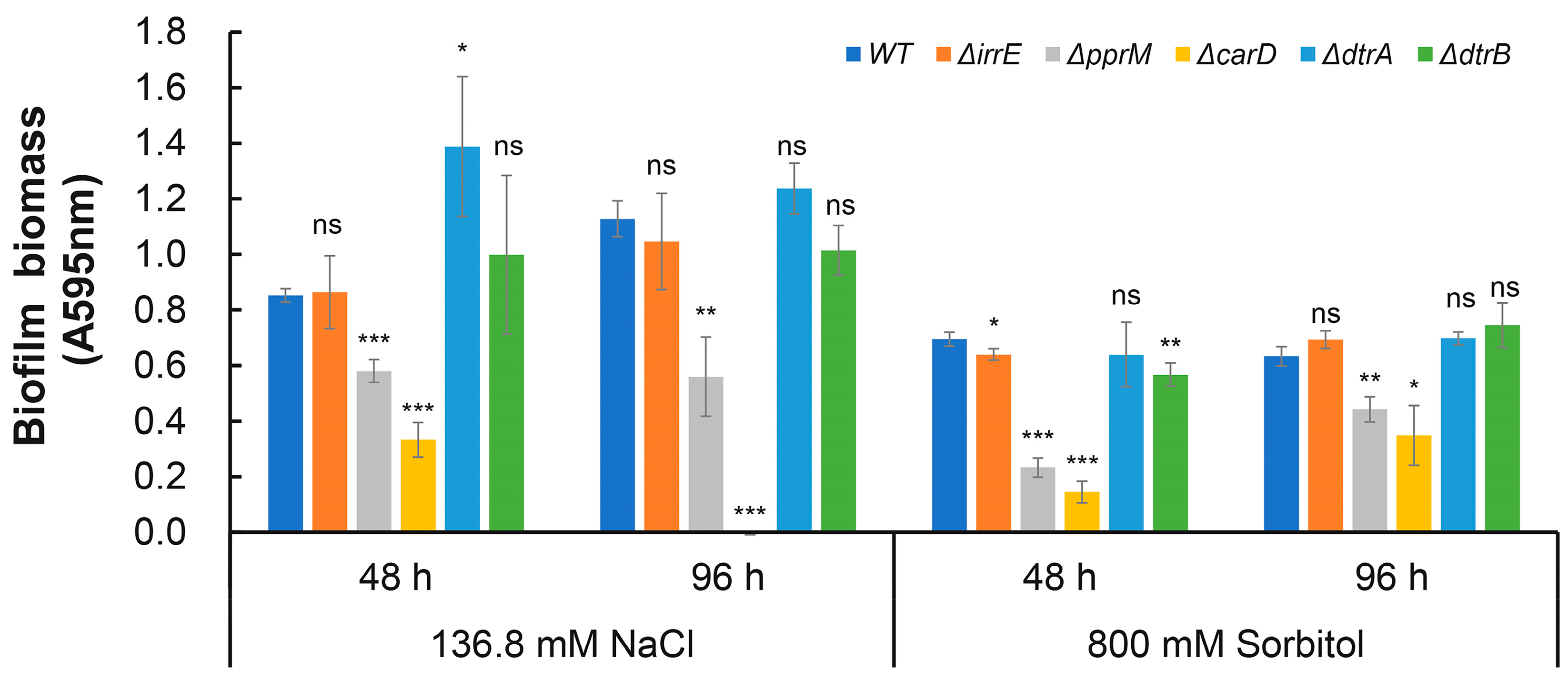
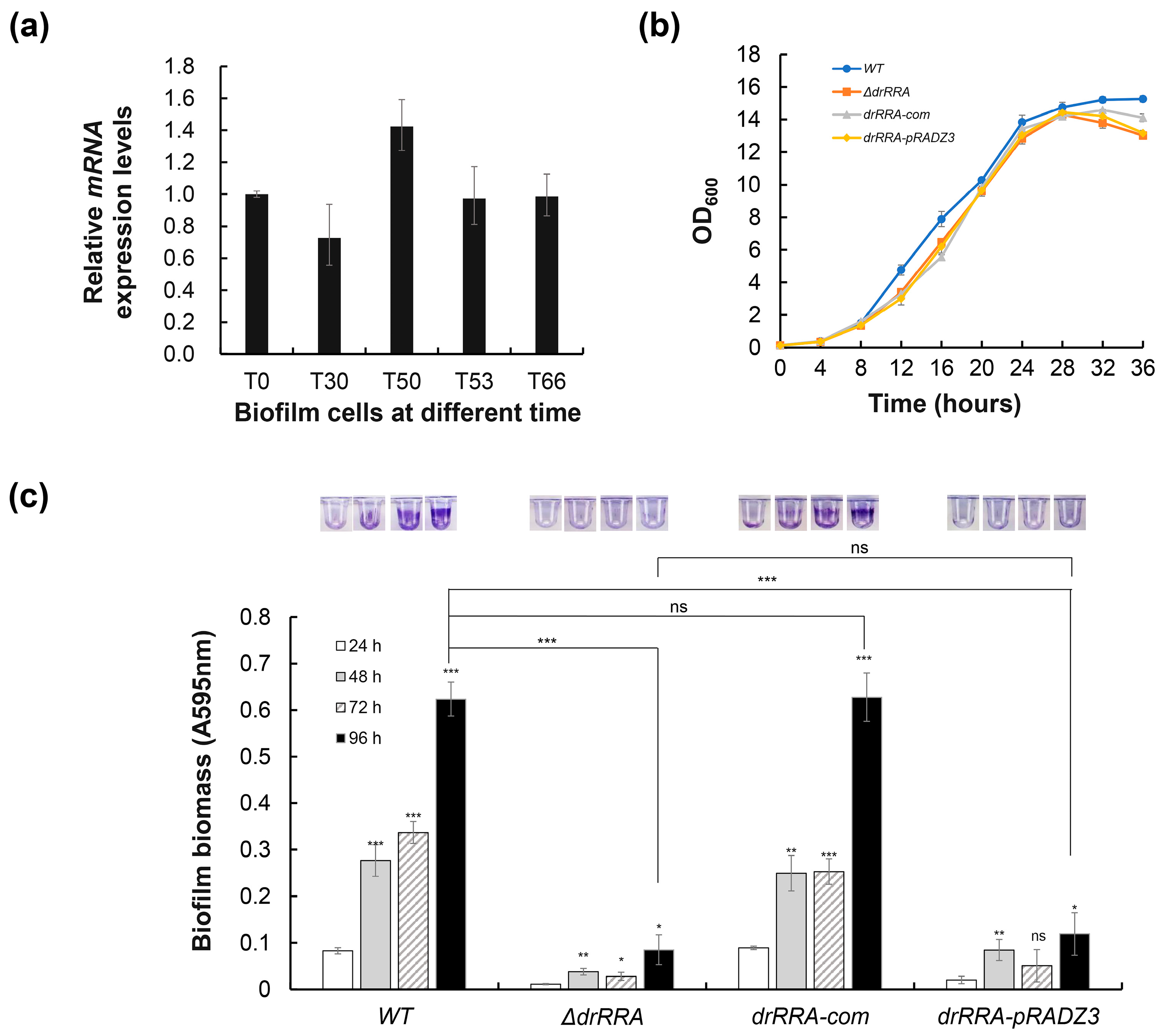
2.4. Identification of the Core drRRA Regulon Governing Biofilm Formation in D. radiodurans
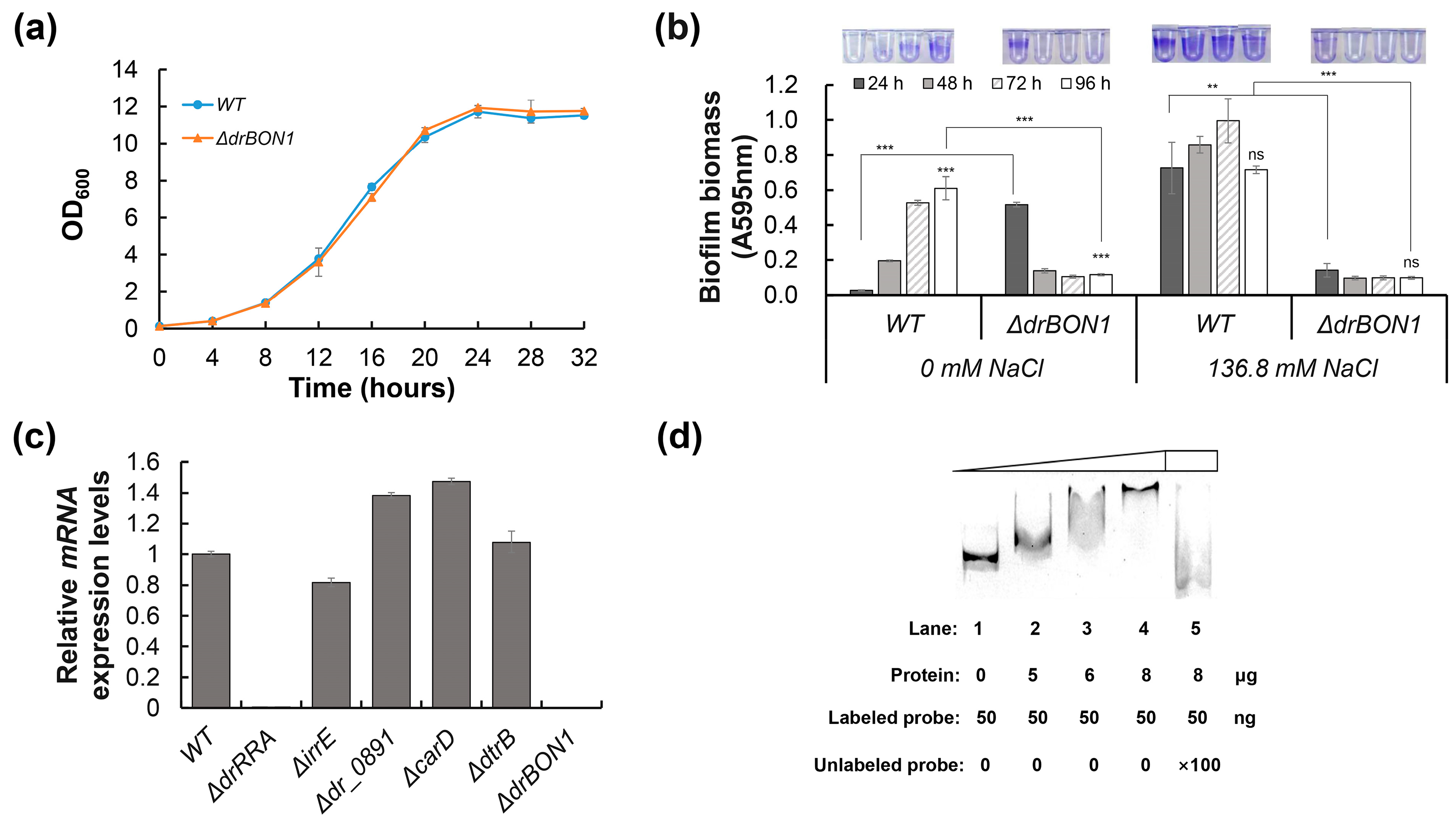
2.5. DrRRA Regulates Biofilm Formation via Transcriptional Activation of dr_0392
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmids, and Growth Conditions
4.2. Construction of the Mutants and Complementary Strains
4.3. Biofilm Formation Assays under Normal, NaCl, and Sorbitol Conditions
4.4. RNA Isolation and Quantitative Real-Time PCR (qRT–PCR)
4.5. RNA Sequencing and Analysis
4.6. Cell Survival under Oxidative Stress, Sorbitol Stress, and UV Radiation
4.7. Electrophoretic Mobility Shift Assay (EMSA)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Watnick, P.; Kolter, R. Biofilm, city of microbes. J. Bacteriol. 2000, 182, 2675–2679. [Google Scholar] [CrossRef] [PubMed]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.D.; Wirsen, C.O.; Gaill, F. Rapid microbial production of filamentous sulfur mats at hydrothermal vents. Appl. Environ. Microbiol. 1999, 65, 2253–2255. [Google Scholar] [CrossRef] [PubMed]
- Reysenbach, A.L.; Cady, S.L. Microbiology of ancient and modern hydrothermal systems. Trends Microbiol. 2001, 9, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Harrell, J.E.; Hahn, M.M.; D’Souza, S.J.; Vasicek, E.M.; Sandala, J.L.; Gunn, J.S.; McLachlan, J.B. Salmonella biofilm formation, chronic infection, and immunity within the intestine and hepatobiliary tract. Front. Cell. Infect. Microbiol. 2020, 10, 624622. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Wang, Y.; Liu, L.; He, J. Biofilms: The microbial “protective clothing” in extreme environments. Int. J. Mol. Sci. 2019, 20, 3423. [Google Scholar] [CrossRef] [PubMed]
- van Wolferen, M.; Orell, A.; Albers, S.V. Archaeal biofilm formation. Nat. Rev. Microbiol. 2018, 16, 699–713. [Google Scholar] [CrossRef]
- Koerdt, A.; Gödeke, J.; Berger, J.; Thormann, K.M.; Albers, S.V. Crenarchaeal biofilm formation under extreme conditions. PLoS ONE 2010, 5, e14104. [Google Scholar] [CrossRef]
- Rinaudi, L.V.; Giordano, W. An integrated view of biofilm formation in rhizobia. FEMS Microbiol. Lett. 2010, 304, 1–11. [Google Scholar] [CrossRef]
- Frösler, J.; Panitz, C.; Wingender, J.; Flemming, H.-C.; Rettberg, P. Survival of Deinococcus geothermalis in biofilms under desiccation and simulated space and martian conditions. Astrobiology 2017, 17, 431–447. [Google Scholar] [CrossRef] [PubMed]
- Panitz, C.; Frösler, J.; Wingender, J.; Flemming, H.-C.; Rettberg, P. Tolerances of Deinococcus geothermalis biofilms and planktonic cells exposed to space and simulated martian conditions in low earth orbit for almost two years. Astrobiology 2019, 19, 979–994. [Google Scholar] [CrossRef]
- Sauer, K.; Stoodley, P.; Goeres, D.M.; Hall-Stoodley, L.; Burmølle, M.; Stewart, P.S.; Bjarnsholt, T. The biofilm life cycle: Expanding the conceptual model of biofilm formation. Nat. Rev. Microbiol. 2022, 20, 608–620. [Google Scholar] [CrossRef] [PubMed]
- Sauer, K.; Camper, A.K.; Ehrlich, G.D.; Costerton, J.W.; Davies, D.G. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 2002, 184, 1140–1154. [Google Scholar] [CrossRef] [PubMed]
- Moormeier, D.E.; Bayles, K.W. Staphylococcus aureus biofilm: A complex developmental organism. Mol. Microbiol. 2017, 104, 365–376. [Google Scholar] [CrossRef]
- Flemming, H.C.; van Hullebusch, E.D.; Neu, T.R.; Nielsen, P.H.; Seviour, T.; Stoodley, P.; Wingender, J.; Wuertz, S. The biofilm matrix: Multitasking in a shared space. Nat. Rev. Microbiol. 2023, 21, 70–86. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcal biofilms. Microbiol. Spectr. 2018, 6, 207–228. [Google Scholar] [CrossRef]
- Dubrac, S.; Boneca, I.G.; Poupel, O.; Msadek, T. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J. Bacteriol. 2007, 189, 8257–8269. [Google Scholar] [CrossRef]
- Mashruwala, A.A.; Gries, C.M.; Scherr, T.D.; Kielian, T.; Boyd, J.M. SaeRS is responsive to cellular respiratory status and regulates fermentative biofilm formation in Staphylococcus aureus. Infect. Immun. 2017, 85, e00157-17. [Google Scholar] [CrossRef]
- Ulrich, M.; Bastian, M.; Cramton, S.E.; Ziegler, K.; Pragman, A.A.; Bragonzi, A.; Memmi, G.; Wolz, C.; Schlievert, P.M.; Cheung, A.; et al. The staphylococcal respiratory response regulator SrrAB induces ica gene transcription and polysaccharide intercellular adhesin expression, protecting Staphylococcus aureus from neutrophil killing under anaerobic growth conditions. Mol. Microbiol. 2007, 65, 1276–1287. [Google Scholar] [CrossRef]
- Toledo-Arana, A.; Merino, N.; Vergara-Irigaray, M.; Débarbouillé, M.; Penadés, J.R.; Lasa, I. Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J. Bacteriol. 2005, 187, 5318–5329. [Google Scholar] [CrossRef] [PubMed]
- Sharma-Kuinkel, B.K.; Mann, E.E.; Ahn, J.S.; Kuechenmeister, L.J.; Dunman, P.M.; Bayles, K.W. The Staphylococcus aureus LytSR two-component regulatory system affects biofilm formation. J. Bacteriol. 2009, 191, 4767–4775. [Google Scholar] [CrossRef] [PubMed]
- Boles, B.R.; Thoendel, M.; Roth, A.J.; Horswill, A.R. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS ONE 2010, 5, e10146. [Google Scholar] [CrossRef] [PubMed]
- Kolar, S.L.; Nagarajan, V.; Oszmiana, A.; Rivera, F.E.; Miller, H.K.; Davenport, J.E.; Riordan, J.T.; Potempa, J.; Barber, D.S.; Koziel, J.; et al. NsaRS is a cell-envelope-stress-sensing two-component system of Staphylococcus aureus. Microbiology 2011, 157 Pt 8, 2206–2219. [Google Scholar] [CrossRef]
- Liu, C.; Sun, D.; Zhu, J.; Liu, W. Two-component signal transduction systems: A major strategy for connecting input stimuli to biofilm formation. Front. Microbiol. 2018, 9, 3279. [Google Scholar] [CrossRef] [PubMed]
- Petrova, O.E.; Sauer, K. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 2009, 5, e1000668. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Shin, B.; Kim, W.; Cheong, H.; Park, S.; Park, W. Comparative genomics of wild-type and laboratory-evolved biofilm-overproducing Deinococcus metallilatus strains. Microb. Genom. 2020, 6, mgen000464. [Google Scholar] [CrossRef]
- Battista, J.R. Against all odds: The survival strategies of Deinococcus radiodurans. Annu. Rev. Microbiol. 1997, 51, 203–224. [Google Scholar] [CrossRef]
- Slade, D.; Radman, M. Oxidative stress resistance in Deinococcus radiodurans. Microbiol. Mol. Biol. Rev. 2011, 75, 133–191. [Google Scholar] [CrossRef]
- Wang, L.; Xu, G.; Chen, H.; Zhao, Y.; Xu, N.; Tian, B.; Hua, Y. DrRRA: A novel response regulator essential for the extreme radioresistance of Deinococcus radiodurans. Mol. Microbiol. 2008, 67, 1211–1222. [Google Scholar] [CrossRef]
- Earl, A.M.; Mohundro, M.M.; Mian, I.S.; Battista, J.R. The IrrE protein of Deinococcus radiodurans R1 is a novel regulator of recA expression. J. Bacteriol. 2002, 184, 6216–6224. [Google Scholar] [CrossRef] [PubMed]
- Ohba, H.; Satoh, K.; Sghaier, H.; Yanagisawa, T.; Narumi, I. Identification of PprM: A modulator of the PprI-dependent DNA damage response in Deinococcus radiodurans. Extremophiles 2009, 13, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.W.; Seo, H.S.; Kim, M.K.; Choi, J.I.; Lim, H.M.; Lim, S. PprM is necessary for up-regulation of katE1, encoding the major catalase of Deinococcus radiodurans, under unstressed culture conditions. J. Microbiol. 2016, 54, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Ma, Y.; Xiao, F.; Wang, W.; He, S. Knockout of pprM decreases resistance to desiccation and oxidation in Deinococcus radiodurans. Indian J. Microbiol. 2017, 57, 316–321. [Google Scholar] [CrossRef] [PubMed]
- Manobala, T.; Shukla, S.K.; Rao, T.S.; Kumar, M.D. A new uranium bioremediation approach using radio-tolerant Deinococcus radiodurans biofilm. J. Biosci. 2019, 44, 122. [Google Scholar] [CrossRef]
- Manobala, T.; Shukla, S.K.; Rao, T.S.; Kumar, M.D. Kinetic modelling of the uranium biosorption by Deinococcus radiodurans biofilm. Chemosphere 2021, 269, 128722. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Oh, S.; Kim, S.H. Released exopolysaccharide (r-EPS) produced from probiotic bacteria reduce biofilm formation of enterohemorrhagic Escherichia coli O157:H7. Biochem. Biophys. Res. Commun. 2009, 379, 324–329. [Google Scholar] [CrossRef]
- Dudin, O.; Geiselmann, J.; Ogasawara, H.; Ishihama, A.; Lacour, S. Repression of flagellar genes in exponential phase by CsgD and CpxR, two crucial modulators of Escherichia coli biofilm formation. J. Bacteriol. 2014, 196, 707–715. [Google Scholar] [CrossRef]
- Ouyang, J.; Feng, W.; Lai, X.; Chen, Y.; Zhang, X.; Rong, L.; Sun, F.; Chen, Y. Quercetin inhibits Pseudomonas aeruginosa biofilm formation via the vfr-mediated lasIR system. Microb. Pathog. 2020, 149, 104291. [Google Scholar] [CrossRef]
- Kang, H.; Gan, J.; Zhao, J.; Kong, W.; Zhang, J.; Zhu, M.; Li, F.; Song, Y.; Qin, J.; Liang, H. Crystal structure of Pseudomonas aeruginosa RsaL bound to promoter DNA reaffirms its role as a global regulator involved in quorum-sensing. Nucleic Acids Res. 2017, 45, 699–710. [Google Scholar] [CrossRef]
- Neidig, A.; Yeung, A.T.Y.; Rosay, T.; Tettmann, B.; Strempel, N.; Rueger, M.; Lesouhaitier, O.; Overhage, J. TypA is involved in virulence, antimicrobial resistance and biofilm formation in Pseudomonas aeruginosa. BMC Microbiol. 2013, 13, 77. [Google Scholar] [CrossRef] [PubMed]
- Petrova, O.E.; Schurr, J.R.; Schurr, M.J.; Sauer, K. Microcolony formation by the opportunistic pathogen Pseudomonas aeruginosa requires pyruvate and pyruvate fermentation. Mol. Microbiol. 2012, 86, 819–835. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Murphy, K.; Lo, R.Y.; Khursigara, C.M.; Lam, J.S.; O’Toole, G.A. Single-nucleotide polymorphisms found in the migA and wbpX glycosyltransferase genes account for the intrinsic lipopolysaccharide defects exhibited by Pseudomonas aeruginosa PA14. J. Bacteriol. 2015, 197, 2780–2791. [Google Scholar] [CrossRef] [PubMed]
- Grande, R.; Nistico, L.; Sambanthamoorthy, K.; Longwell, M.; Iannitelli, A.; Cellini, L.; Di Stefano, A.; Hall Stoodley, L.; Stoodley, P. Temporal expression of agrB, cidA, and alsS in the early development of Staphylococcus aureus UAMS-1 biofilm formation and the structural role of extracellular DNA and carbohydrates. Pathog. Dis. 2014, 70, 414–422. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.-H.; Choi, Y.-S.; Cho, Y.-H. Unusual properties of catalase A (KatA) of Pseudomonas aeruginosa PA 14 are associated with its biofilm peroxide resistance. J. Bacteriol. 2008, 190, 2663–2670. [Google Scholar] [CrossRef] [PubMed]
- Merino, N.; Toledo-Arana, A.; Vergara-Irigaray, M.; Valle, J.; Solano, C.; Calvo, E.; Lopez, J.A.; Foster, T.J.; Penadés, J.R.; Lasa, I.I. Protein A-mediated multicellular behavior in Staphylococcus aureus. J. Bacteriol. 2009, 191, 832–843. [Google Scholar] [CrossRef]
- Kato, F.; Yabuno, Y.; Yamaguchi, Y.; Sugai, M.; Inouye, M. Deletion of mazF increases Staphylococcus aureus biofilm formation in an ica-dependent manner. Pathog. Dis. 2017, 75, ftx026. [Google Scholar] [CrossRef] [PubMed]
- Sung, B.H.; Lee, C.H.; Yu, B.J.; Lee, J.H.; Lee, J.Y.; Kim, M.S.; Blattner, F.R.; Kim, S.C. Development of a biofilm production-deficient Escherichia coli strain as a host for biotechnological applications. Appl. Environ. Microbiol. 2006, 72, 3336–3342. [Google Scholar] [CrossRef]
- Nepper, J.F.; Lin, Y.C.; Weibel, D.B.; Mullineaux, C.W. Rcs phosphorelay activation in cardiolipin-deficient Escherichia coli reduces biofilm formation. J. Bacteriol. 2019, 201, e00804-18. [Google Scholar] [CrossRef]
- Im, S.; Joe, M.; Kim, D.; Park, D.-H.; Lim, S. Transcriptome analysis of salt-stressed Deinococcus radiodurans and characterization of salt-sensitive mutants. Res. Microbiol. 2013, 164, 923–932. [Google Scholar] [CrossRef]
- Stallings, C.L.; Stephanou, N.C.; Chu, L.; Hochschild, A.; Nickels, B.E.; Glickman, M.S. CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell 2009, 138, 146–159. [Google Scholar] [CrossRef] [PubMed]
- Potrykus, K.; Cashel, M. (p)ppGpp: Still magical? Annu. Rev. Microbiol. 2008, 62, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhu, J.; Xu, J.; Shao, H.; Zhou, R. Regulation of (p)ppGpp and its homologs on environmental adaptation, survival, and pathogenicity of Streptococci. Front. Microbiol. 2020, 11, 1842. [Google Scholar] [CrossRef] [PubMed]
- Steinchen, W.; Ahmad, S.; Valentini, M.; Eilers, K.; Majkini, M.; Altegoer, F.; Lechner, M.; Filloux, A.; Whitney, J.C.; Bange, G. Dual role of a (p)ppGpp- and (p)ppApp-degrading enzyme in biofilm formation and interbacterial antagonism. Mol. Microbiol. 2021, 115, 1339–1356. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xiao, Y.; Nie, H.; Huang, Q.; Chen, W. Influence of (p)ppGpp on biofilm regulation in Pseudomonas putida KT2440. Microbiol. Res. 2017, 204, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.R.; Kasetty, S.; Chatterji, D.; Kivisaar, M. Novel functions of (p)ppGpp and cyclic di-GMP in Mycobacterial physiology revealed by phenotype microarray analysis of wild-type and isogenic strains of Mycobacterium smegmatis. Appl. Environ. Microbiol. 2015, 81, 2571–2578. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tian, Y.; Zhou, Z.; Zhang, L.; Zhang, W.; Lin, M.; Chen, M. Identification and functional analysis of RelA/SpoT homolog (RSH) genes in Deinococcus radiodurans. J. Microbiol. Biotechnol. 2016, 26, 2106–2115. [Google Scholar] [CrossRef]
- Im, S.; Song, D.; Joe, M.; Kim, D.; Park, D.H.; Lim, S. Comparative survival analysis of 12 histidine kinase mutants of Deinococcus radiodurans after exposure to DNA-damaging agents. Bioprocess. Biosyst. Eng. 2013, 36, 781–789. [Google Scholar] [CrossRef]
- Makarova, K.S.; Aravind, L.; Wolf, Y.I.; Tatusov, R.L.; Minton, K.W.; Koonin, E.V.; Daly, M.J. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 2001, 65, 44–79. [Google Scholar] [CrossRef]
- Lim, S.; Jung, J.H.; Blanchard, L.; de Groot, A. Conservation and diversity of radiation and oxidative stress resistance mechanisms in Deinococcus species. FEMS Microbiol. Rev. 2019, 43, 19–52. [Google Scholar] [CrossRef]
- Lin, L.; Dai, S.; Tian, B.; Li, T.; Yu, J.; Liu, C.; Wang, L.; Xu, H.; Zhao, Y.; Hua, Y. DqsIR quorum sensing-mediated gene regulation of the extremophilic bacterium Deinococcus radiodurans in response to oxidative stress. Mol. Microbiol. 2016, 100, 527–541. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Yang, N.; Zheng, S.; Yan, F.; Jiang, C.; Yu, Y.; Guo, J.; Chai, Y.; Chen, Y. The spo0A-sinI-sinR regulatory circuit plays an essential role in biofilm formation, nematicidal activities, and plant protection in Bacillus cereus AR156. Mol. Plant-Microbe Interact. 2017, 30, 603–619. [Google Scholar] [CrossRef] [PubMed]
- Boles, B.R.; Horswill, A.R. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008, 4, e1000052. [Google Scholar] [CrossRef] [PubMed]
- Alshalchi, S.A.; Anderson, G.G. Involvement of stress-related genes polB and PA14_46880 in biofilm formation of Pseudomonas aeruginosa. Infect. Immun. 2014, 82, 4746–4757. [Google Scholar] [PubMed]
- Amores, G.R.; de Las Heras, A.; Sanches-Medeiros, A.; Elfick, A.; Silva-Rocha, R. Systematic identification of novel regulatory interactions controlling biofilm formation in the bacterium Escherichia coli. Sci. Rep. 2017, 7, 16768. [Google Scholar] [CrossRef] [PubMed]
- Rasamiravaka, T.; Labtani, Q.; Duez, P.; El Jaziri, M. The formation of biofilms by Pseudomonas aeruginosa: A review of the natural and synthetic compounds interfering with control mechanisms. BioMed Res. Int. 2015, 2015, 759348. [Google Scholar] [CrossRef] [PubMed]
- Chambers, J.R.; Sauer, K. Small RNAs and their role in biofilm formation. Trends Microbiol. 2013, 21, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.H.; Liao, R.; Chou, B.; Contreras, L.M. Transcriptional analysis of Deinococcus radiodurans reveals novel small RNAs that are differentially expressed under ionizing radiation. Appl. Environ. Microbiol. 2015, 81, 1754–1764. [Google Scholar] [CrossRef]
- Powell, L.C.; Pritchard, M.F.; Ferguson, E.L.; Powell, K.A.; Patel, S.U.; Rye, P.D.; Sakellakou, S.M.; Buurma, N.J.; Brilliant, C.D.; Copping, J.M.; et al. Targeted disruption of the extracellular polymeric network of Pseudomonas aeruginosa biofilms by alginate oligosaccharides. NPJ Biofilms Microbiomes 2018, 4, 13. [Google Scholar] [CrossRef]
- Remminghorst, U.; Rehm, B.H. Alg44, a unique protein required for alginate biosynthesis in Pseudomonas aeruginosa. FEBS Lett. 2006, 580, 3883–3888. [Google Scholar] [CrossRef]
- Lin, S.M.; Baek, C.Y.; Jung, J.-H.; Kim, W.S.; Song, H.-Y.; Lee, J.H.; Ji, H.J.; Zhi, Y.; Kang, B.S.; Bahn, Y.-S.; et al. Antioxidant activities of an exopolysaccharide (deinoPol) produced by the extreme radiation-resistant bacterium Deinococcus radiodurans. Sci. Rep. 2020, 10, 55. [Google Scholar] [CrossRef] [PubMed]
- Yeats, C.; Bateman, A. The BON domain: A putative membrane-binding domain. Trends Biochem. Sci. 2003, 28, 352–355. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhao, M.; Lv, M.; Lin, M.; Wang, J.; Zuo, K. A Novel Small RNA, DsrO, in Deinococcus radiodurans Promotes Methionine Sulfoxide Reductase (msrA) Expression for Oxidative Stress Adaptation. Appl. Environ. Microbiol. 2022, 88, e0003822. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Wang, L.; Chen, H.; Lu, H.; Ying, N.; Tian, B.; Hua, Y. RecO is essential for DNA damage repair in Deinococcus radiodurans. J. Bacteriol. 2008, 190, 2624–2628. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]

| Locus Tag | Annotation | Homology Protein | Identity (%) | Coverage (%) | Log2(Fold Change) | |||
|---|---|---|---|---|---|---|---|---|
| T30/T0 | T50/T0 | T53/T0 | T66/T0 | |||||
| DR_0408 | response regulator | CheY (NP_416396.1; E. coli) [37] | 32.74 | 69 | −0.24 | −1.40 | −1.09 | −1.28 |
| CpxR (NP_418348.1; E. coli) [38] | 30.47 | 49 | ||||||
| DR_A0350 | response regulator PilH | CpxR (NP_418348.1; E. coli) [38] | 30.25 | 49 | 2.16 | 2.16 | 1.10 | 1.01 |
| DR_0997 | Crp/Fnr family transcriptional regulator | Vfr (NP_249343.1; P. aeruginosa) [39] | 20.92 | 91 | −2.15 | -2.15 | −0.93 | −0.82 |
| DR_B0067 | salicylate monooxygenase-related protein | PqsH (NP_251277.1; P. aeruginosa) [40] | 34.41 | 23 | −0.70 | −4.43 | −4.88 | −2.26 |
| DR_B0072 | salicylate monooxygenase-related protein | PqsH (NP_251277.1; P. aeruginosa) [40] | 40.79 | 18 | −1.00 | −4.43 | −4.78 | −2.20 |
| DR_1198 | GTP-binding elongation factor family protein TypA/BipA | TypA (NP_253804.1; P. aeruginosa) [41] | 56.83 | 98 | −0.61 | −1.72 | −0.77 | −0.84 |
| DR_1291 | D-3-phosphoglycerate dehydrogenase | LdhA (NP_249618.1; P. aeruginosa) [42] | 30.88 | 85 | 1.54 | 1.79 | 1.72 | 2.08 |
| DR_0713 | lipopolysaccharide glycosyltransferase, putative | WbpX (NP_254136.1; P. aeruginosa) [43] | 16.38 | 61 | −0.68 | 1.37 | 2.03 | 2.30 |
| DR_1516 | acetolactate synthase | AlsS (WP_000130155.1; S. aureus) [44] | 29.4 | 96 | 1.47 | 1.58 | 2.28 | 2.17 |
| DR_1998 | catalase | KatA (NP_252926.1; P. aeruginosa) [45] | 42.8 | 99 | 2.38 | 2.69 | 2.30 | 2.17 |
| DR_0888 | hypothetical protein | Spa (AFD54305.1; S. aureus) [46] | 35.29 | 10 | −0.66 | −1.45 | −0.55 | −0.83 |
| DR_0662 | hypothetical protein | MazF (BBJ19047.1; S. aureus) [47] | 26.79 | 93 | −0.31 | −1.38 | −1.17 | −0.79 |
| DR_0687 | hypothetical protein | Wza (NP_416566.1; E. coli) [48,49] | 26.17 | 27 | −2.96 | −0.34 | −0.22 | 0.03 |
| Locus Tag | Functional Description | Log2(Fold Change) | |||
|---|---|---|---|---|---|
| T30/T0 | T50/T0 | T53/T0 | T66/T0 | ||
| Energy metabolism | |||||
| DR_1501 | NADH-quinone oxidoreductase | 2.58 | 2.89 | 2.73 | 2.28 |
| DR_1498 | NADH-quinone oxidoreductase subunit H | 2.92 | 2.59 | 3.62 | 2.89 |
| DR_1497 | NADH-quinone oxidoreductase subunit I | 3.03 | 2.67 | 3.65 | 2.85 |
| DR_1496 | NADH dehydrogenase subunit J | 2.33 | 2.46 | 3.30 | 2.64 |
| DR_1495 | NADH-quinone oxidoreductase subunit K | 2.76 | 2.77 | 3.82 | 3.20 |
| DR_1493 | NADH dehydrogenase subunit M | 2.76 | 2.76 | 3.72 | 3.02 |
| DR_1492 | NADH-quinone oxidoreductase subunit N | 2.31 | 2.35 | 3.19 | 2.67 |
| DR_B0106 | cytochrome C oxidase subunit II | 4.59 | 3.60 | 2.22 | 2.18 |
| DR_1440 | ATPase | 3.06 | 3.55 | 4.24 | 3.63 |
| Carbohydrate metabolism | |||||
| DR_0828 | isocitrate lyase | 4.74 | 4.57 | 6.13 | 4.72 |
| DR_1778 | 3-isopropylmalate dehydratase large subunit 2 | 1.92 | 1.94 | 2.13 | 2.00 |
| Environmental information processing | |||||
| DR_B0073 | PTS fructose transporter subunit IIBC | −1.41 | −3.92 | −5.02 | −2.06 |
| DR_B0074 | PTS fructose transporter subunit IIA | −1.39 | −3.59 | −4.38 | −1.63 |
| DR_B0075 | 1-phosphofructokinase | −1.69 | −4.22 | −4.57 | −1.77 |
| DR_1220 | iron transporter | −2.67 | −2.65 | −1.72 | −1.57 |
| Nucleotide metabolism | |||||
| DR_B0107 | ribonucleotide reductase, NrdI family | 2.85 | 2.20 | 2.04 | 2.04 |
| DR_B0108 | ribonucleotide-diphosphate reductase subunit alpha | 2.83 | 2.16 | 1.80 | 1.85 |
| DR_B0109 | ribonucleotide-diphosphate reductase subunit beta | 2.86 | 1.93 | 1.45 | 1.55 |
| DR_B0121 | Iron ABC transporter, ATP-binding protein | −1.83 | −3.24 | −2.73 | −2.79 |
| DR_B0122 | iron ABC transporter permease | −2.03 | −3.30 | −2.70 | −2.80 |
| DR_B0123 | iron ABC transporter permease | −2.34 | −3.10 | −2.73 | −2.61 |
| Stress response | |||||
| DR_1998 | catalase | 2.38 | 2.69 | 2.29 | 2.17 |
| Unknown function | |||||
| DR_1710 | hypothetical protein | 2.69 | 3.36 | 2.97 | 2.34 |
| DR_1082 | hypothetical protein | 2.34 | 2.42 | 1.67 | 1.91 |
| rna4 | tRNA-Phe | ND * | ND * | ND * | ND * |
| rna8 | tRNA-Glu | ND * | 2.18 | ND * | 1.71 |
| rna26 | tRNA-Ser | ND * | 1.62 | ND * | ND * |
| rna54 | tRNA-Ala | ND * | 2.07 | 2.37 | 2.55 |
| DR_2028 | DNA-binding protein | ND * | −2.16 | −3.07 | −1.65 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Q.; Zhan, Y.; Zhang, W.; Wang, J.; Yan, Y.; Wang, W.; Lin, M. Development and Regulation of the Extreme Biofilm Formation of Deinococcus radiodurans R1 under Extreme Environmental Conditions. Int. J. Mol. Sci. 2024, 25, 421. https://doi.org/10.3390/ijms25010421
Guo Q, Zhan Y, Zhang W, Wang J, Yan Y, Wang W, Lin M. Development and Regulation of the Extreme Biofilm Formation of Deinococcus radiodurans R1 under Extreme Environmental Conditions. International Journal of Molecular Sciences. 2024; 25(1):421. https://doi.org/10.3390/ijms25010421
Chicago/Turabian StyleGuo, Qiannan, Yuhua Zhan, Wei Zhang, Jin Wang, Yongliang Yan, Wenxiu Wang, and Min Lin. 2024. "Development and Regulation of the Extreme Biofilm Formation of Deinococcus radiodurans R1 under Extreme Environmental Conditions" International Journal of Molecular Sciences 25, no. 1: 421. https://doi.org/10.3390/ijms25010421
APA StyleGuo, Q., Zhan, Y., Zhang, W., Wang, J., Yan, Y., Wang, W., & Lin, M. (2024). Development and Regulation of the Extreme Biofilm Formation of Deinococcus radiodurans R1 under Extreme Environmental Conditions. International Journal of Molecular Sciences, 25(1), 421. https://doi.org/10.3390/ijms25010421






