Genome-Wide Identification of the CBF Gene Family and ICE Transcription Factors in Walnuts and Expression Profiles under Cold Conditions
Abstract
:1. Introduction
2. Results
2.1. Genome-Wide Identification and Phylogenetic Analysis of the CBF and ICE Gene Family in Three Walnut Genomes
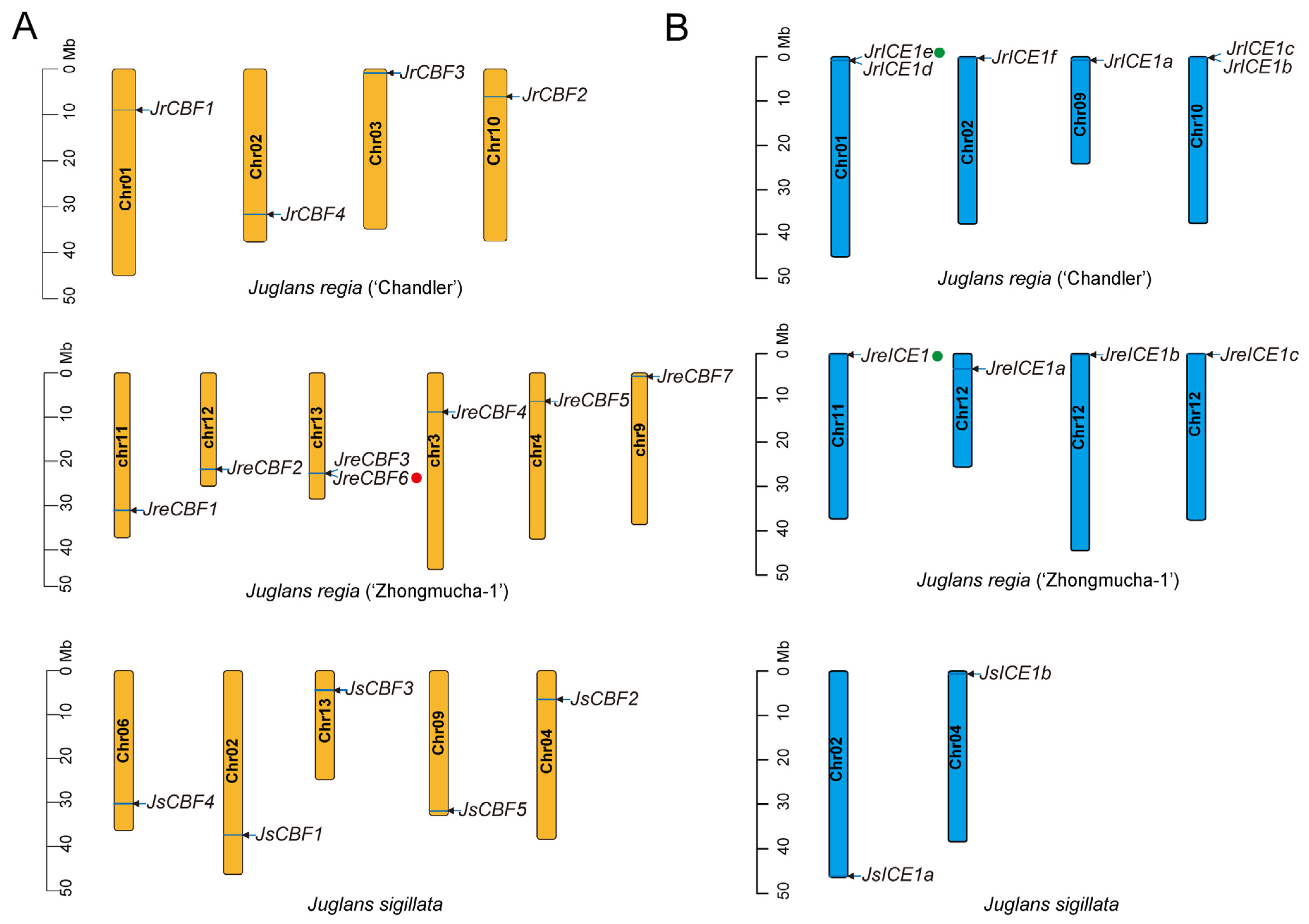
2.2. Chromosomal Distribution and Duplication Patterns of Juglans regia and Juglans sigillata
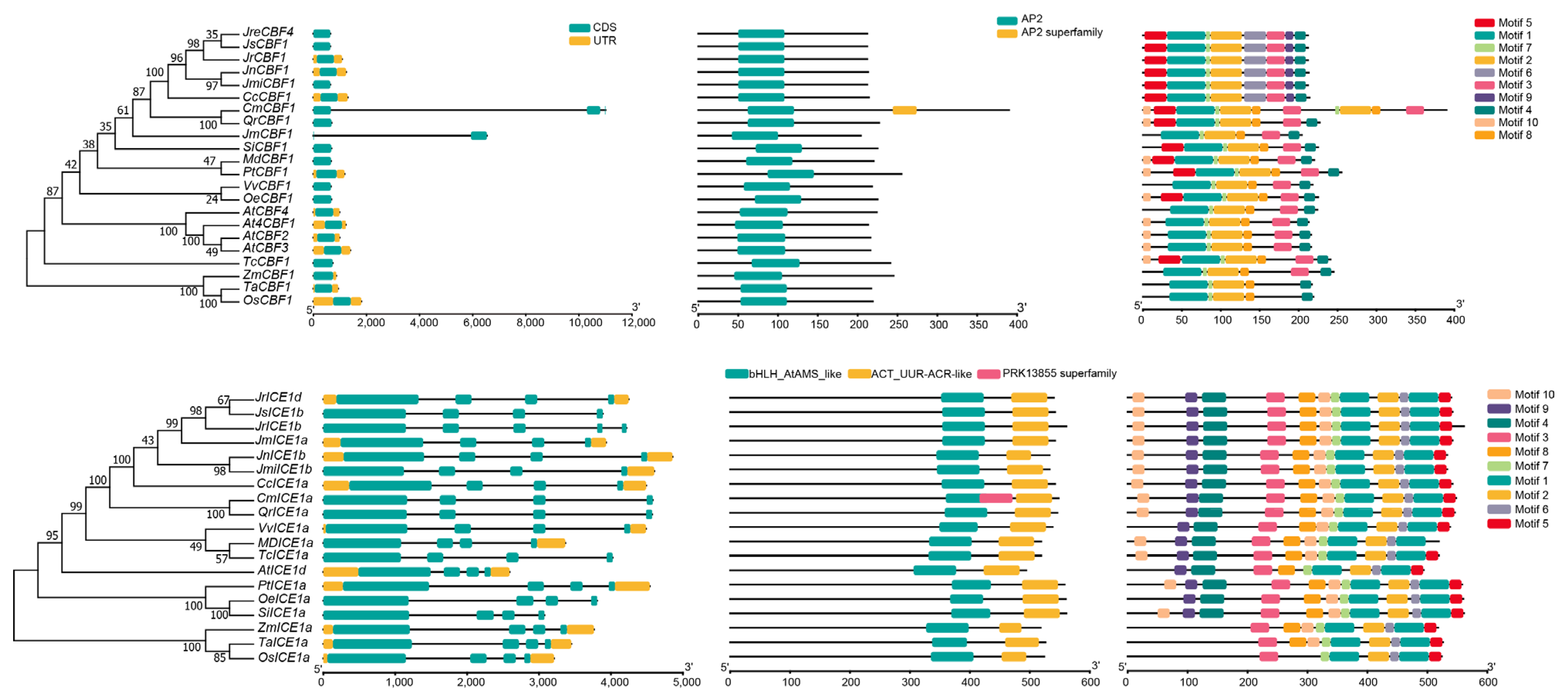
2.3. Protein Domain and Gene Structure Analysis
2.4. Collinearity Analysis and Selective Pressure
2.5. Cis-Acting Elements Analysis of Promoters
2.6. Gene Expression Patterns Analysis
2.7. Protein–Protein and Protein-microRNA Interaction Predictions
2.8. Evolution of CBF1 and ICE1 in Angiosperms
3. Discussion
3.1. Characteristics of CBF and ICE1 in Three Walnut Genomes
3.2. Conserved Domain and Cis-Acting Elements of CBF and ICE1 in Three Walnut Genomes
3.3. Gene Expression Profiles of CBF and ICE1 in Response to Cold Stress
3.4. Protein–Protein and Protein Interactions, and the Putative microRNA Interaction Predictions
4. Materials and Methods
4.1. Plant Materials and RNA Sequencing
4.2. Identification of the Gene Members in Three Juglans Regia Cultivars
4.3. Construction of Phylogenetic Tree and Analysis of Gene Structure, Conserved Motif
4.4. Chromosomal Location, Collinearity of Transcription Factors, and Selective Pressure Analysis in Juglans Regia
4.5. Analysis of Subcellular Localization and Protein Physicochemical Properties
4.6. Cis-Acting Elements Analysis of Promoter and Gene Expression
4.7. Protein–Protein Interactions and microRNA Targeting Analysis
4.8. Evolution of CBF1 and ICE1 Genes in Angiosperms
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Waadt, R.; Seller, C.A.; Hsu, P.K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ding, Y.; Yang, Y.; Song, C.; Wang, B.; Yang, S.; Guo, Y.; Gong, Z. Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 2021, 63, 53–78. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Shi, Y.; Peng, Y.; Jia, Y.; Yan, Y.; Dong, X.; Li, H.; Dong, J.; Li, J.; Gong, Z.; et al. Cold-induced CBF-PIF3 interaction enhances freezing tolerance by stabilizing the phyB thermosensor in Arabidopsis. Mol. Plant 2020, 13, 894–906. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhang, Z.; Xie, S.; Si, T.; Li, Y.; Zhu, J.K. Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 2016, 171, 2744–2759. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.M.; Thomashow, M.F. Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2012, 109, 15054–15059. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Ding, Y.; Yang, S. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Han, R.; Cai, K.; Yan, H.; Li, Y.; Qu, G.; Liu, L.; Zhao, X. Identification and analysis of the CBF gene family in three species of Acer under cold stress. Int. J. Mol. Sci. 2023, 24, 2088. [Google Scholar] [CrossRef]
- Maruyama, K.; Mizoi, J.; Myint, P.S.H.N.; Fujita, Y.; Sekita, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Soybean DREB1/CBF-type transcription factors function in heat and drought as well as cold stress-responsive gene expression. Plant J. 2015, 81, 505–518. [Google Scholar]
- Ma, L.F.; Zhang, J.M.; Huang, G.Q.; Li, Y.; Li, X.B.; Zheng, Y. Molecular characterization of cotton C-repeat/dehydration-responsive element binding factor genes that are involved in response to cold stress. Mol. Biol. Rep. 2014, 41, 4369–4379. [Google Scholar] [CrossRef]
- Liang, X.; Luo, G.; Li, W.; Yao, A.; Liu, W.; Xie, L.; Han, M.; Li, X.; Han, D. Overexpression of a Malus baccata CBF transcription factor gene, MbCBF1, increases cold and salinity tolerance in Arabidopsis thaliana. Plant Physiol. Biochem. 2022, 192, 230–242. [Google Scholar] [CrossRef]
- Zhang, X.; Fowler, S.G.; Cheng, H.; Lou, Y.; Rhee, S.Y.; Stockinger, E.J.; Thomashow, M.F. Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis. Plant J. 2004, 39, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yu, J.; Wang, R.; Liu, W.; Chen, S.; Wang, Y.; Yu, Y.; Qu, G.; Chen, S. Genome-wide identification and expression profiles of C-repeat binding factor transcription factors in Betula platyphylla under abiotic stress. Int. J. Mol. Sci. 2023, 24, 10573. [Google Scholar] [CrossRef]
- Yamasaki, Y.; Randall, S.K. Functionality of soybean CBF/DREB1 transcription factors. Plant Sci. 2016, 246, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, T.; Wang, X.; Mildenhall, F.; Ibrahim, I.M.; Puthiyaveetil, S.; Varala, K. Chilling stress drives organ-specific transcriptional cascades and dampens diurnal oscillation in tomato. Hortic. Res. 2023, 10, uhad137. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Lee, M.; Lee, J.H.; Lee, H.J.; Park, C.M. The unified ICE-CBF pathway provides a transcriptional feedback control of freezing tolerance during cold acclimation in Arabidopsis. Plant Mol. Biol. 2015, 89, 187–201. [Google Scholar] [CrossRef]
- Skinner, J.S.; von Zitzewitz, J.; Szucs, P.; Marquez-Cedillo, L.; Filichkin, T.; Amundsen, K.; Stockinger, E.J.; Thomashow, M.F.; Chen, T.H.; Hayes, P.M. Structural, functional, and phylogenetic characterization of a large CBF gene family in barley. Plant Mol. Biol. 2005, 59, 533–551. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Jawad, U.M.; Yang, M.; Hou, Y.; Gereziher, M.T.; Zheng, J.; Wang, H.; Liu, J.; Dong, W.; Xu, Y.; et al. Genome-wide identification and functional analysis of ICE genes reveal that Gossypium thurberi “GthICE2” is responsible for cold and drought stress tolerance. Plant Physiol. Biochem. 2023, 199, 107708. [Google Scholar] [CrossRef]
- Thomashow, M.F.; Torii, K.U. SCREAMing Twist on the role of ICE1 in freezing tolerance. Plant Cell 2020, 32, 816–819. [Google Scholar] [CrossRef]
- Marrano, A.; Britton, M.; Zaini, P.A.; Zimin, A.V.; Workman, R.E.; Puiu, D.; Bianco, L.; Pierro, E.A.D.; Allen, B.J.; Chakraborty, S.; et al. High-quality chromosome-scale assembly of the walnut (Juglans regia L.) reference genome. Gigascience 2020, 9, giaa050. [Google Scholar] [CrossRef]
- Luo, X.; Zhou, H.; Cao, D.; Yan, F.; Chen, P.; Wang, J.; Woeste, K.; Chen, X.; Fei, Z.; An, H.; et al. Domestication and selection footprints in Persian walnuts (Juglans regia). PLoS Genet. 2022, 18, e1010513. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Zhou, H.; Zulfiqar, S.; Luo, X.; Hu, Y.; Feng, L.; Malvolti, M.E.; Woeste, K.; Zhao, P. The Phytogeographic history of common walnut in China. Front. Plant Sci. 2018, 9, 1399. [Google Scholar] [CrossRef] [PubMed]
- Ning, D.L.; Wu, T.; Xiao, L.J.; Ma, T.; Fang, W.L.; Dong, R.Q.; Cao, F.L. Chromosomal-level assembly of Juglans sigillata genome using Nanopore, BioNano, and Hi-C analysis. Gigascience 2020, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.W.; Hou, N.; Woeste, K.; Zhang, C.; Yue, M.; Yuan, X.Y.; Zhao, P. Population genetic structure and adaptive differentiation of iron walnut Juglans regia subsp. sigillata in southwestern China. Ecol. Evol. 2019, 9, 14154–14166. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.; Zou, H.; Li, P.; Yao, X.; Zhou, Z.; Gu, X.; Sun, R.; Liu, A. Genomic characterization of the NAC transcription factors, directed at understanding their functions involved in endocarp lignification of iron walnut (Juglans sigillata Dode). Front. Genet. 2023, 14, 1168142. [Google Scholar] [CrossRef] [PubMed]
- Bükücü, Ş.B.; Sütyemez, M.; Kefayati, S.; Paizila, A.; Jighly, A.; Kafkas, S. Major QTL with pleiotropic effects controlling time of leaf budburst and flowering-related traits in walnut (Juglans regia L.). Sci. Rep. 2020, 10, 15207. [Google Scholar] [CrossRef] [PubMed]
- Hassankhah, A.; Rahemi, M.; Ramshini, H.; Sarikhani, S.; Vahdati, K. Flowering in Persian walnut: Patterns of gene expression during flower development. BMC Plant Biol. 2020, 20, 136. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Li, Y.; Zhang, X.; You, X.; Yu, H.; Guo, R.; Zhao, X. Genome-wide identification of AP2/ERF superfamily genes in Juglans mandshurica and expression analysis under cold stress. Int. J. Mol. Sci. 2022, 23, 15225. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, B.; Xin, G.; Zhang, X.; Li, J.; Wang, Y. Evaluation of cold tolerance of seven walnut varieties. Cryo Lett. 2022, 43, 74–82. [Google Scholar] [CrossRef]
- Han, L.; Ma, K.; Zhao, Y.; Mei, C.; Mamat, A.; Wang, J.; Qin, L.; He, T. The cold-stress responsive gene DREB1A involved in low-temperature tolerance in Xinjiang wild walnut. Peer J. 2022, 10, e14021. [Google Scholar] [CrossRef]
- Ebrahimi, A.; Lawson, S.S.; McKenna, J.R.; Jacobs, D.F. Morpho-physiological and genomic evaluation of Juglans species reveals regional maladaptation to cold stress. Front. Plant Sci. 2020, 11, 229. [Google Scholar] [CrossRef] [PubMed]
- Soveili, S.; Khadivi, A. Selecting the superior late-leafing genotypes of Persian walnut (Juglans regia L.) using morphological and pomological evaluations. BMC Plant Biol. 2023, 23, 379. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Tang, H.; Wang, B.; Yue, C.; Wang, L.; Zeng, J.; Yang, Y.; Wang, X. Integrative transcriptional and metabolic analyses provide insights into cold spell response mechanisms in young shoots of the tea plant. Tree Physiol. 2018, 38, 1655–1671. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Xu, Z.; Peng, S.; Sun, Y.; Jia, C.; Zhai, M. In planta characterization of a tau class glutathione S-transferase gene from Juglans regia (JrGSTTau1) involved in chilling tolerance. Plant Cell Rep. 2016, 35, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Ma, J.; Liu, H.; Ou, M.; Ye, H.; Zhao, P. Identification and characterization of wall-associated kinase (WAK) and WAK-like (WAKL) gene family in Juglans regia and its wild related species Juglans mandshurica. Genes 2022, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Hao, F.; Yang, G.; Zhou, H.; Yao, J.; Liu, D.; Zhao, P.; Zhang, S. Genome-wide identification and transcriptional expression profiles of transcription factor WRKY in common walnut (Juglans regia L.). Genes 2021, 12, 1444. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.S.; Deng, J.R.; Cheng, P.D.; Zhang, Z.L.; Wang, Y.H.; Chen, S.W.; Tang, Y.; Wang, T.Y.; Yang, G.Y. Transcriptome-wide identification of walnut PP2C family genes in response to external stimulus. BMC Genom. 2022, 23, 640. [Google Scholar]
- Liu, X.; Meng, P.; Yang, G.; Zhang, M.; Peng, S.; Zhai, M.Z. Genome-wide identification and transcript profiles of walnut heat stress transcription factor involved in abiotic stress. BMC Genom. 2020, 21, 474. [Google Scholar] [CrossRef]
- Hwarari, D.; Guan, Y.; Ahmad, B.; Movahedi, A.; Min, T.; Hao, Z.; Lu, Y.; Chen, J.; Yang, L. ICE-CBF-COR signaling cascade and its regulation in plants responding to cold stress. Int. J. Mol. Sci. 2022, 23, 1549. [Google Scholar] [CrossRef]
- Ye, K.; Li, H.; Ding, Y.; Shi, Y.; Song, C.; Gong, Z.; Yang, S. BRASSINOSTEROID-INSENSITIVE2 negatively regulates the stability of transcription factor ICE1 in response to cold stress in Arabidopsis. Plant Cell 2019, 31, 2682–2696. [Google Scholar] [CrossRef]
- Wu, C.L.; Lin, L.F.; Hsu, H.C.; Huang, L.F.; Hsiao, C.D.; Chou, M.L. Saussurea involucrata (Snow Lotus) ICE1 and ICE2 orthologues involved in regulating cold stress tolerance in transgenic Arabidopsis. Int. J. Mol. Sci. 2021, 22, 10850. [Google Scholar] [CrossRef] [PubMed]
- Leblanc-Fournier, N.; Coutand, C.; Crouzet, J.; Brunel, N.; Lenne, C.; Moulia, B.; Julien, J.L. Jr-ZFP2, encoding a Cys2/His2-type transcription factor, is involved in the early stages of the mechano-perception pathway and specifically expressed in mechanically stimulated tissues in woody plants. Plant Cell Environ. 2008, 31, 715–726. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, W.; Ji, F.; Qiu, J.; Song, X.; Bu, D.; Pan, G.; Ma, Q.; Chen, J.; Huang, R.; et al. A high-quality walnut genome assembly reveals extensive gene expression divergences after whole-genome duplication. Plant Biotechnol. J. 2020, 18, 1848–1850. [Google Scholar] [CrossRef] [PubMed]
- Cuperus, J.T.; Fahlgren, N.; Carrington, J.C. Evolution and functional diversification of MIRNA genes. Plant Cell 2011, 23, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.; Sugita, K.; Qin, Y.; Rahman, A. Aux/IAA14 regulates microRNA-mediated cold stress response in Arabidopsis roots. Int. J. Mol. Sci. 2020, 21, 8441. [Google Scholar] [CrossRef] [PubMed]
- Moturu, T.R.; Sinha, S.; Salava, H.; Thula, S.; Nodzyński, T.; Vařeková, R.S.; Friml, J.; Simon, S. Molecular evolution and diversification of proteins involved in miRNA maturation pathway. Plants 2020, 9, 299. [Google Scholar] [CrossRef]
- Park, S.; Shi, A.; Mou, B. Genome-wide identification and expression analysis of the CBF/DREB1 gene family in lettuce. Sci. Rep. 2020, 10, 5733. [Google Scholar] [CrossRef]
- Wang, D.; Cui, B.; Guo, H.; Liu, Y.; Nie, S. Genome-wide identification and expression analysis of the CBF transcription factor family in Lolium perenne under abiotic stress. Plant Signal Behav. 2022, 18, 2086733. [Google Scholar] [CrossRef]
- Wang, X.; Li, Z.; Shi, Y.; Liu, Z.; Zhang, X.; Gong, Z.; Yang, S. Strigolactones promote plant freezing tolerance by releasing the WRKY41-mediated inhibition of CBF/DREB1 expression. EMBO J. 2023, 42, e112999. [Google Scholar] [CrossRef]
- Dong, X.; Yan, Y.; Jiang, B.; Shi, Y.; Jia, Y.; Cheng, J.; Shi, Y.; Kang, J.; Li, H.; Zhang, D.; et al. The cold response regulator CBF1 promotes Arabidopsis hypocotyl growth at ambient temperatures. EMBO J. 2020, 39, e103630. [Google Scholar] [CrossRef]
- Li, J.; Qin, R.Y.; Li, H.; Xu, R.F.; Yang, Y.C.; Ni, D.H.; Ma, H.; Li, L.; Wei, P.C.; Yang, J.B. Low-temperature-induced expression of rice ureidoglycolate amidohydrolase is mediated by a C-Repeat/Dehydration-Responsive element that specifically interacts with rice C-Repeat-Binding factor 3. Front. Plant Sci. 2015, 6, 1011. [Google Scholar] [CrossRef] [PubMed]
- Jaglo, K.R.; Kleff, S.; Amundsen, K.L.; Zhang, X.; Haake, V.; Zhang, J.Z.; Deits, T.; Thomashow, M.F. Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol. 2001, 127, 910–917. [Google Scholar] [CrossRef] [PubMed]
- Martínez-García, P.J.; Crepeau, M.W.; Puiu, D.; Gonzalez-Ibeas, D.; Whalen, J.; Stevens, K.A.; Paul, R.; Butterfield, T.S.; Britton, M.T.; Reagan, R.L.; et al. The walnut (Juglans regia) genome sequence reveals diversity in genes coding for the biosynthesis of non-structural polyphenols. Plant J. 2016, 87, 507–532. [Google Scholar] [CrossRef] [PubMed]
- Thamilarasan, S.K.; Park, J.I.; Jung, H.J.; Nou, I.S. Genome-wide analysis of the distribution of AP2/ERF transcription factors reveals duplication and CBFs genes elucidate their potential function in Brassica Oleracea. BMC Genom. 2014, 15, 422. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Gong, Y.; Sun, P.; Chen, S.; Ma, C. Genome-wide identification of CBF genes and their responses to cold acclimation in Taraxacum kok-saghyz. Peer J. 2022, 10, e13429. [Google Scholar] [CrossRef] [PubMed]
- Wessler, S.R. Homing into the origin of the AP2 DNA binding domain. Trends Plant Sci. 2005, 10, 54–56. [Google Scholar] [CrossRef] [PubMed]
- Magnani, E.; Sjölander, K.; Hake, S. From endonucleases to transcription factors: Evolution of the AP2 DNA binding domain in plants. Plant Cell 2004, 16, 2265–2277. [Google Scholar] [CrossRef]
- Jofuku, K.D.; Boer, B.G.W.D.; Okamuro, M.J.K. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 1994, 6, 1211–1225. [Google Scholar]
- Feng, K.; Hou, X.L.; Xing, G.M.; Liu, J.X.; Duan, A.Q.; Xu, Z.S.; Li, M.Y.; Zhuang, J.; Xiong, A.S. Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef]
- Xie, W.; Ding, C.; Hu, H.; Dong, G.; Zhang, G.; Qian, Q.; Ren, D. Molecular events of rice AP2/ERF transcription factors. Int. J. Mol. Sci. 2022, 23, 12013. [Google Scholar] [CrossRef]
- Chen, C.H.; Lin, H.Y.; Pan, C.L.; Chen, F.C. The plausible reason why the length of 5’ untranslated region is unrelated to organismal complexity. BMC Res. Notes. 2011, 4, 312. [Google Scholar] [CrossRef] [PubMed]
- Leppek, K.; Das, R.; Barna, M. Functional 5’ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 2018, 19, 158–174. [Google Scholar] [CrossRef] [PubMed]
- Mukheriee, D.; Saha, D.; Acharya, D.; Mukheriee, A.; Chakraborty, S.; Ghosh, T.C. The role of introns in the conservation of the metabolic genes of Arabidopsis thaliana. Genomics. 2018, 110, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Ku, Y.S.; Sintaha, M.; Cheung, M.Y.; Lam, H.M. Plant Hormone Signaling Crosstalks between Biotic and Abiotic Stress Responses. Int. J. Mol. Sci. 2018, 19, 3206. [Google Scholar] [CrossRef] [PubMed]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, M.; Hao, Y.; Kapoor, A.; Dong, C.H.; Fujii, H.; Zheng, X.; Zhu, J.K. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2006, 281, 37636–37645. [Google Scholar] [CrossRef] [PubMed]
- Ishitani, M.; Xiong, L.; Lee, H.; Stevenson, B.; Zhu, J.K. HOS1, a genetic locus involved in cold-responsive gene expression in Arabidopsis. Plant Cell 1998, 10, 1151–1161. [Google Scholar] [CrossRef]
- Dong, C.H.; Agarwal, M.; Zhang, Y.; Xie, Q.; Zhu, J.K. The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA 2006, 103, 8281–8286. [Google Scholar] [CrossRef]
- Jung, J.H.; Park, J.H.; Lee, S.; To, T.K.; Kim, J.M.; Seki, M.; Park, C.M. The cold signaling attenuator HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 activates FLOWERING LOCUS C transcription via chromatin remodeling under short-term cold stress in Arabidopsis. Plant Cell 2013, 25, 4378–4390. [Google Scholar] [CrossRef]
- Zhang, L.; Jiang, X.; Liu, Q.; Ahammed, G.J.; Lin, R.; Wang, L.; Shao, S.; Yu, J.; Zhou, Y. The HY5 and MYB15 transcription factors positively regulate cold tolerance in tomato via the CBF pathway. Plant Cell Environ. 2020, 43, 2712–2726. [Google Scholar] [CrossRef]
- Wang, X.; Ding, Y.; Li, Z.; Shi, Y.; Wang, J.; Hua, J.; Gong, Z.; Zhou, J.M.; Yang, S. PUB25 and PUB26 promote plant freezing tolerance by degrading the cold signaling negative regulator MYB15. Dev. Cell. 2019, 51, 222–235. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Kim, H.S.; Bahk, S.; An, J.; Yoo, Y.; Kim, J.Y.; Chung, W.S. Phosphorylation of the transcriptional repressor MYB15 by mitogen-activated protein kinase 6 is required for freezing tolerance in Arabidopsis. Nucleic Acids Res. 2017, 45, 6613–6627. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Song, J.; Lin, R.; Tang, M.; Shao, S.; Yu, J.; Zhou, Y. Tomato SlMYB15 transcription factor targeted by sly-miR156e-3p positively regulates ABA-mediated cold tolerance. J. Exp. Bot. 2022, 73, 7538–7551. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Jin, J.B.; Lee, J.; Yoo, C.Y.; Stirm, V.; Miura, T.; Ashworth, E.N.; Bressan, R.A.; Yun, D.J.; Hasegawa, P.M. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 2007, 19, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhou, J.; Li, Y.; Wu, J.; Ma, C.; Chen, Y.; Sun, X.; Wu, L.; Liang, X.; Fu, Q.; et al. AP2/EREBP pathway plays an important role in Chaling wild rice tolerance to cold stress. Int. J. Mol. Sci. 2023, 24, 14441. [Google Scholar] [CrossRef] [PubMed]
- Siddiqua, M.; Nassuth, A. Vitis CBF1 and Vitis CBF4 differ in their effect on Arabidopsis abiotic stress tolerance, development and gene expression. Plant Cell Environ. 2011, 34, 1345–1359. [Google Scholar] [CrossRef] [PubMed]
- Mehdi, S.M.M.; Krishnamoorthy, S.; Szczesniak, M.W.; Ludwików, A. Identification of novel miRNAs and their target genes in the response to abscisic acid in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 7153. [Google Scholar] [CrossRef]
- Lu, Q.; Guo, F.; Xu, Q.; Cang, J. LncRNA improves cold resistance of winter wheat by interacting with miR398. Funct. Plant Biol. 2020, 47, 544–557. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression of RNA-seq data at the gene level–the DESeq package. Mol. Biol. 2012, 11, 106. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer-Verlag: New York, NY, USA, 2016. [Google Scholar]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Penelope, C.; Eberhardt, R.Y.; Eddy, S.R.; Jaina, M.; Mitchell, A.L.; Potter, S.C.; Marco, P.; Matloob, Q.; Amaia, S.V. The Pfam protein family’s database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Doerks, T.; Bork, P. Smart 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinf. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Yang, Z.; Nielsen, R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000, 17, 32–43. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, C.P.; Hernandez-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. EggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Zuo, D.; Ye, H.; Yan, Y.; Li, M.; Zhao, P. Genome-wide identification, characterization, and expression pattern of the late embryogenesis abundant (LEA) gene family in Juglans regia and its wild relatives. J. Mandshurica BMC Plant Biol. 2023, 23, 80. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Yan, F.; Hao, F.; Ye, H.; Yue, M.; Woeste, K.; Zhao, P.; Zhang, S. Pan-genome and transcriptome analyses provide insights into genomic variation and differential gene expression profiles related to disease resistance and fatty acid biosynthesis in eastern black walnut (Juglans nigra). Hortic. Res. 2023, 10, uhad015. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Wang, L.; You, F.M.; Rodriguez, J.C.; Deal, K.R.; Chen, L.; Li, J.; Chakraborty, S.; Balan, B.; Jiang, C.Z.; et al. Sequencing a Juglans regia × J. microcarpa hybrid yields high-quality genome assemblies of parental species. Hortic. Res. 2019, 6, 55. [Google Scholar] [CrossRef]
- Yan, F.; Xi, R.M.; She, R.X.; Chen, P.P.; Yan, Y.J.; Yang, G.; Dang, M.; Yue, M.; Pei, D.; Woeste, K.; et al. Improved de novo chromosome-level genome assembly of the vulnerable walnut tree Juglans mandshurica reveals gene family evolution and possible genome basis of resistance to lesion nematode. Mol. Ecol. Resour. 2021, 21, 2063–2076. [Google Scholar] [CrossRef]
- Stevens, K.A.; Woeste, K.; Chakraborty, S.; Crepeau, M.W.; Leslie, C.A.; Martínez-García, P.J.; Puiu, D.; Romero-Severson, J.; Coggeshall, M.; Dandekar, A.M.; et al. Genomic variation among and within six Juglans species. G3-Genes Genom. Genet. 2018, 8, 2153–2165. [Google Scholar]
- Huang, Y.; Xiao, L.; Zhang, Z.; Zhang, R.; Wang, Z.; Huang, C.; Huang, R.; Luan, Y.; Fan, T.; Wang, J.; et al. The genomes of pecan and Chinese hickory provide insights into Carya evolution and nut nutrition. Gigascience 2019, 8, giz036. [Google Scholar] [CrossRef]
- Xing, Y.; Liu, Y.; Zhang, Q.; Nie, X.; Sun, Y.; Zhang, Z.; Li, H.; Fang, K.; Wang, G.; Huang, H.; et al. Hybrid de novo genome assembly of Chinese chestnut (Castanea mollissima). Gigascience 2019, 8, giz112. [Google Scholar] [CrossRef] [PubMed]
- Plomion, C.; Aury, J.M.; Amselem, J.; Leroy, T.; Murat, F.; Duplessis, S.; Faye, S.; Francillonne, N.; Labadie, K.; Le Provost, G.; et al. Oak genome reveals facets of long lifespan. Nat. Plants. 2018, 4, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yu, S.; Tong, C.; Zhao, Y.; Liu, Y.; Song, C.; Zhang, Y.; Zhang, X.; Wang, Y.; Hua, W.; et al. Genome sequencing of the high oil crop sesame provides insight into oil biosynthesis. Genome Biol. 2014, 15, R39. [Google Scholar] [CrossRef] [PubMed]
- Velasco, R.; Zharkikh, A.; Affourtit, J.; Dhingra, A.; Cestaro, A.; Kalyanaraman, A.; Fontana, P.; Bhatnagar, S.K.; Troggio, M.; Pruss, D.; et al. The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 2010, 42, 833–839. [Google Scholar] [CrossRef] [PubMed]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [PubMed]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. French-Italian Public Consortium for Grapevine Genome Characterization. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar] [PubMed]
- Cruz, F.; Julca, I.; Gómez-Garrido, J.; Loska, D.; Marcet-Houben, M.; Cano, E.; Galán, B.; Frias, L.; Ribeca, P.; Derdak, S.; et al. Genome sequence of the olive tree, Olea europaea. Gigascience 2016, 5, 29. [Google Scholar] [CrossRef]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef]
- Argout, X.; Salse, J.; Aury, J.M.; Guiltinan, M.J.; Droc, G.; Gouzy, J.; Allegre, M.; Chaparro, C.; Legavre, T.; Maximova, S.N.; et al. The genome of Theobroma cacao. Nat. Genet. 2011, 43, 101–108. [Google Scholar] [CrossRef]
- Jain, R.; Jenkins, J.; Shu, S.; Chern, M.; Martin, J.A.; Copetti, D.; Duong, P.Q.; Pham, N.T.; Kudrna, D.A.; Talag, J.; et al. Genome sequence of the model rice variety KitaakeX. BMC Genom. 2019, 20, 905. [Google Scholar] [CrossRef]
- Tian, T.; Wang, S.; Yang, S.; Yang, Z.; Liu, S.; Wang, Y.; Gao, H.; Zhang, S.; Yang, X.; Jiang, C.; et al. Genome assembly and genetic dissection of a prominent drought-resistant maize germplasm. Nat Genet. 2023, 55, 496–506. [Google Scholar] [CrossRef]
- Zhu, T.; Wang, L.; Rimbert, H.; Rodriguez, J.C.; Deal, K.R.; De Oliveira, R.; Choulet, F.; Keeble-Gagnère, G.; Tibbits, J.; Rogers, J.; et al. Optical maps refine the bread wheat Triticum aestivum cv. Chinese Spring genome assembly. Plant J. 2021, 107, 303–314. [Google Scholar] [CrossRef]







| Gene Name 1 | No. of Amnio Acids | Mol. Wt (kDa) | Isoelectric Point (pI) | Instability Index (II) | Aliphatic Index | Grand Average of Hydropathicity (GRAVY) | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| JrCBF1 | 212 | 23,251.1 | 5.26 | 52.72 | 61.37 | −0.503 | Nucleus |
| JrCBF2 | 214 | 23,955.9 | 6.20 | 56.34 | 66.17 | −0.563 | Nucleus |
| JrCBF3 | 218 | 24,001.2 | 8.59 | 54.02 | 71.24 | −0.477 | Nucleus |
| JrCBF4 | 249 | 27,626.1 | 5.53 | 57.99 | 72.13 | −0.482 | Nucleus |
| JreCBF1 | 249 | 27,626.1 | 5.53 | 57.99 | 72.13 | −0.482 | Nucleus |
| JreCBF2 | 253 | 28,821.2 | 5.42 | 56.81 | 65.61 | −0.743 | Nucleus |
| JreCBF3 | 266 | 29,507.2 | 6.19 | 55.73 | 70.49 | −0.538 | Nucleus |
| JreCBF4 | 212 | 23,251.1 | 5.26 | 52.72 | 61.37 | −0.503 | Nucleus |
| JreCBF5 | 150 | 16,684.8 | 10.00 | 59.52 | 60.60 | −0.744 | Nucleus |
| JreCBF6 | 265 | 29,260.9 | 6.19 | 60.5 | 68.94 | −0.486 | Nucleus |
| JreCBF7 | 218 | 24,001.2 | 8.59 | 54.02 | 71.24 | −0.477 | Nucleus |
| JsCBF1 | 212 | 23,251.1 | 5.26 | 52.72 | 61.37 | −0.503 | Nucleus |
| JsCBF2 | 214 | 24,026.0 | 6.75 | 54.69 | 62.52 | −0.581 | Nucleus |
| JsCBF3 | 253 | 28,893.2 | 5.33 | 56.32 | 65.61 | −0.755 | Nucleus |
| JsCBF4 | 250 | 27,799.2 | 5.21 | 56.64 | 70.28 | −0.516 | Nucleus |
| JsCBF5 | 218 | 24,011.3 | 8.59 | 54.9 | 71.24 | −0.481 | Nucleus |
| JrICE1a | 534 | 57,836.5 | 5.36 | 46.26 | 74.01 | −0.516 | Nucleus |
| JrICE1b | 539 | 58,329.4 | 5.03 | 56.61 | 78.89 | −0.423 | Nucleus |
| JrICE1c | 537 | 58,102.2 | 4.98 | 56.64 | 78.99 | −0.420 | Nucleus |
| JrICE1d | 540 | 58,754.7 | 5.16 | 56.23 | 72.46 | −0. 542 | Nucleus |
| JrICE1e | 552 | 60,506.9 | 5.73 | 57.26 | 72.30 | −0. 530 | Nucleus |
| JrICE1f | 492 | 54,205.0 | 5.77 | 46.78 | 77.36 | −0. 515 | Nucleus |
| JreICE1 | 645 | 70,822.6 | 6.35 | 49.98 | 83.32 | −0. 305 | Nucleus |
| JreICE1a | 541 | 58,832.7 | 5.76 | 48.38 | 72.87 | −0.563 | Nucleus |
| JreICE1b | 560 | 61,275.6 | 5.22 | 58.34 | 75.09 | −0.521 | Nucleus |
| JreICE1c | 648 | 70,913.0 | 5.18 | 59.24 | 83.98 | −0.330 | Nucleus |
| JsICE1a | 539 | 58,395.5 | 5.14 | 55.74 | 78.89 | −0.441 | Nucleus |
| JsICE1b | 542 | 59,042.0 | 5.22 | 55.92 | 72.20 | −0.550 | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, H.; Ma, J.; Liu, H.; Zhao, P. Genome-Wide Identification of the CBF Gene Family and ICE Transcription Factors in Walnuts and Expression Profiles under Cold Conditions. Int. J. Mol. Sci. 2024, 25, 25. https://doi.org/10.3390/ijms25010025
Zhou H, Ma J, Liu H, Zhao P. Genome-Wide Identification of the CBF Gene Family and ICE Transcription Factors in Walnuts and Expression Profiles under Cold Conditions. International Journal of Molecular Sciences. 2024; 25(1):25. https://doi.org/10.3390/ijms25010025
Chicago/Turabian StyleZhou, Huijuan, Jiayu Ma, Hengzhao Liu, and Peng Zhao. 2024. "Genome-Wide Identification of the CBF Gene Family and ICE Transcription Factors in Walnuts and Expression Profiles under Cold Conditions" International Journal of Molecular Sciences 25, no. 1: 25. https://doi.org/10.3390/ijms25010025
APA StyleZhou, H., Ma, J., Liu, H., & Zhao, P. (2024). Genome-Wide Identification of the CBF Gene Family and ICE Transcription Factors in Walnuts and Expression Profiles under Cold Conditions. International Journal of Molecular Sciences, 25(1), 25. https://doi.org/10.3390/ijms25010025







