Genome-Wide Association Analysis of Heat Tolerance in F2 Progeny from the Hybridization between Two Congeneric Oyster Species
Abstract
:1. Introduction
2. Results
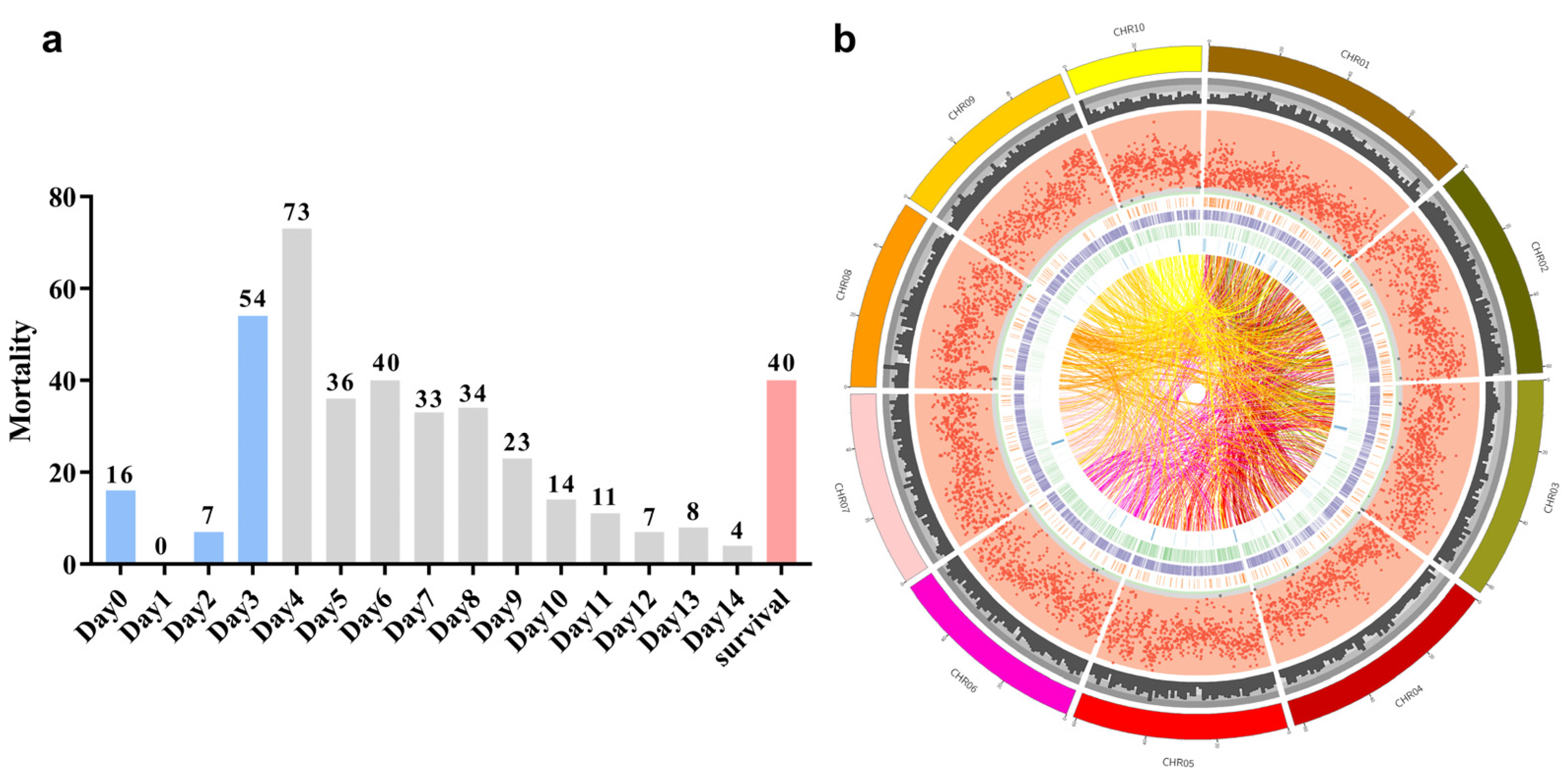
2.1. Heat Tolerance Differentiation
2.2. Sequencing and Genotyping
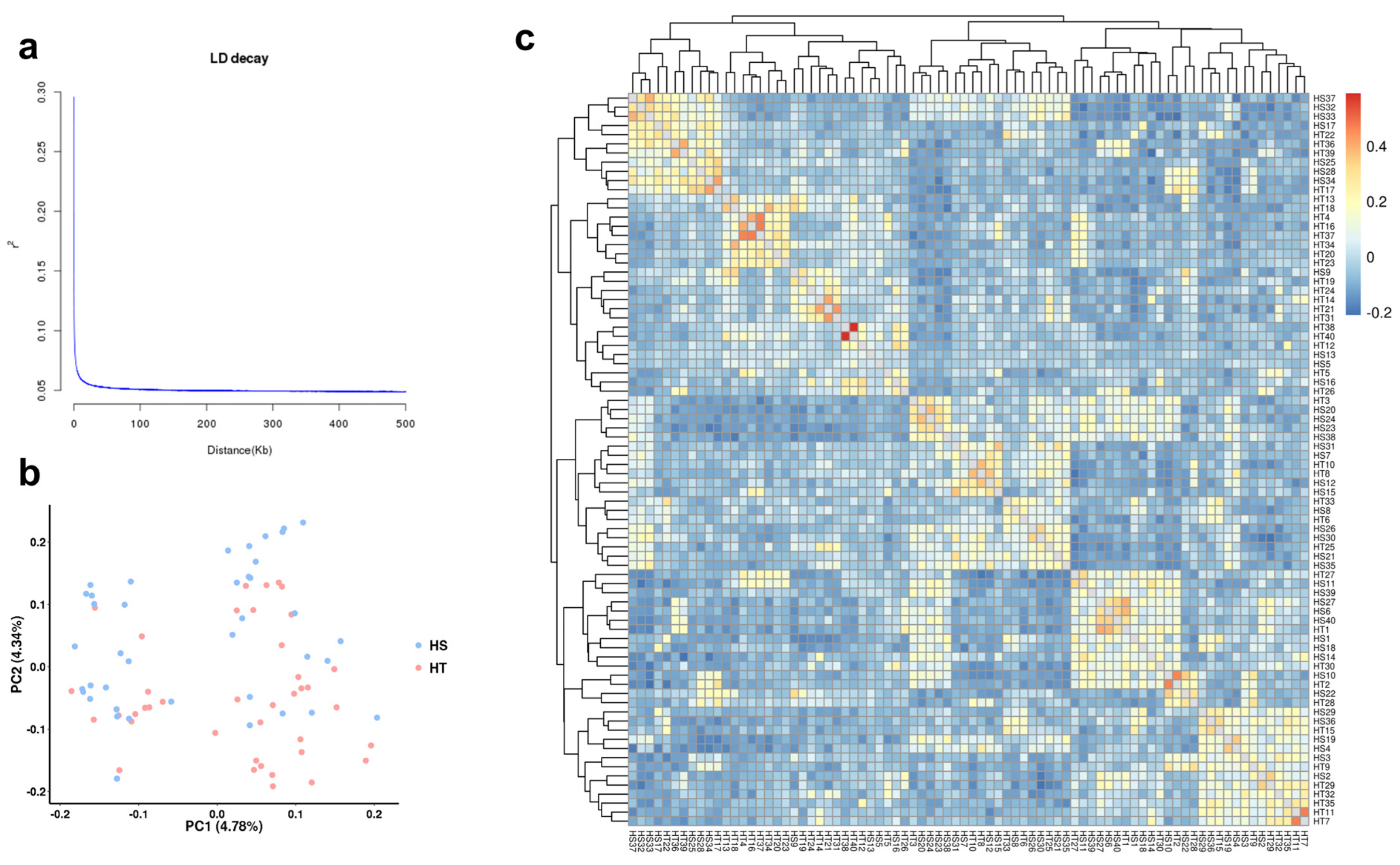
2.3. Linkage Disequilibrium (LD) and Population Structure
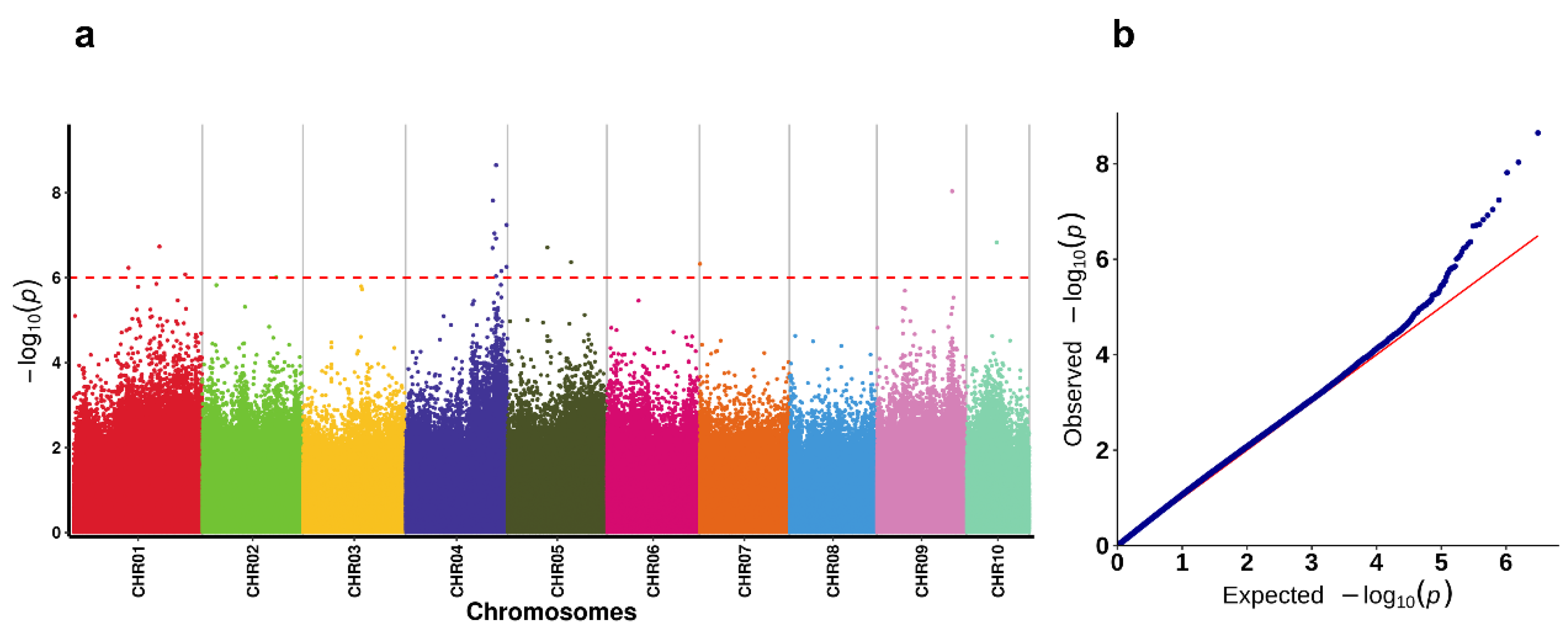
2.4. Genome-Wide Association Study (GWAS)
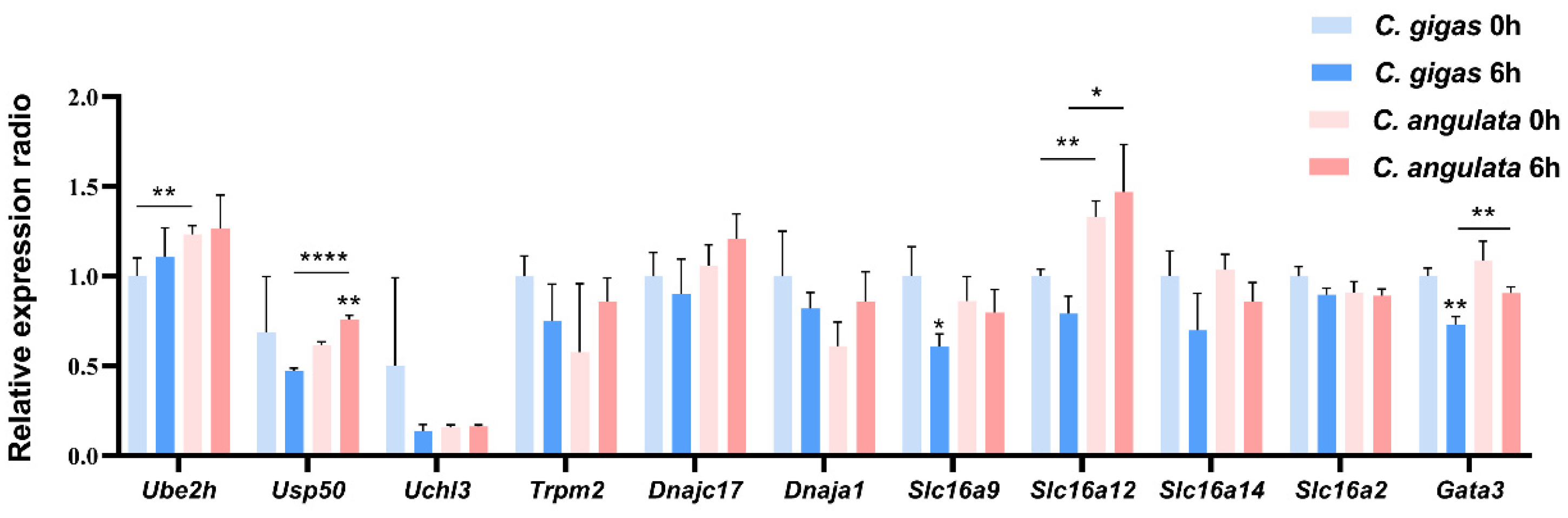
2.5. qRT-PCR Validation Results
3. Discussion
4. Materials and Methods
4.1. AG × AG F2 Populations Construction
4.2. Heat Stress Experiment and Sample Collection
4.3. Sequencing, Genotyping, and Quality Control
4.4. Linkage Disequilibrium and Population Structure Analysis
4.5. Genome-Wide Association Analysis and Candidate Genes Annotation
4.6. qRT-PCR Experiment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xiao, S.; Wong, N.K.; Li, J.; Lin, Y.; Zhang, Y.H.; Ma, H.T.; Mo, R.G.; Zhang, Y.; Yu, Z.N. Analysis of in situ Transcriptomes Reveals Divergent Adaptive Response to Hyper- and Hypo-Salinity in the Hong Kong Oyster, Crassostrea hongkongensis. Front. Physiol. 2018, 9, 1491. [Google Scholar] [CrossRef] [PubMed]
- Yuasa, M.; Kawabeta, K.; Eguchi, A.; Abe, H.; Yamashita, E.; Koba, K.; Tominaga, M. Characterization of taste and micronutrient content of rock oysters (Crassostrea nippona) and Pacific oysters (Crassostrea gigas) in Japan. Int. J. Gastron. Food Sci. 2018, 13, 52–57. [Google Scholar] [CrossRef]
- Guo, X.M. Use and exchange of genetic resources in molluscan aquaculture. Rev. Aquacult. 2009, 1, 251–259. [Google Scholar] [CrossRef]
- Guo, Z.X.; Zhao, F.J.Z.; Chen, H.; Tu, M.L.; Tao, S.F.; Wang, Z.Y.; Wu, C.; He, S.D.; Du, M. Heat treatments of peptides from oyster (Crassostrea gigas) and the impact on their digestibility and angiotensin I converting enzyme inhibitory activity. Food Sci. Biotechnol. 2020, 29, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.F.; Li, L.; Meng, J.; Qi, H.G.; Qu, T.; Xu, F.; Zhang, L.L. Molecular Basis for Adaptation of Oysters to Stressful Marine Intertidal Environments. Annu. Rev. Anim. Biosci. 2016, 4, 357–381. [Google Scholar] [CrossRef] [PubMed]
- McLeod, I.M.; Bostrom-Einarsson, L.; Creighton, C.; D’Anastasi, B.; Diggles, B.; Dwyer, P.G.; Firby, L.; Le Port, A.; Luongo, A.; Martinez-Baena, F.; et al. Habitat value of Sydney rock oyster (Saccostrea glomerata) reefs on soft sediments. Mar. Freshwater Res. 2020, 71, 771–781. [Google Scholar] [CrossRef]
- Zhang, G.F.; Fang, X.D.; Guo, X.M.; Li, L.; Luo, R.B.; Xu, F.; Yang, P.C.; Zhang, L.L.; Wang, X.T.; Qi, H.G.; et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 2012, 490, 49–54. [Google Scholar] [CrossRef]
- Walles, B.; Troost, K.; van den Ende, D.; Nieuwhof, S.; Smaal, A.C.; Ysebaert, T. From artificial structures to self-sustaining oyster reefs. J. Sea Res. 2016, 108, 1–9. [Google Scholar] [CrossRef]
- Richardson, M.A.; Zhang, Y.; Connolly, R.M.; Gillies, C.L.; McDougall, C. Some Like it Hot: The Ecology, Ecosystem Benefits and Restoration Potential of Oyster Reefs in Tropical Waters. Front. Mar. Sci. 2022, 9, 873768. [Google Scholar] [CrossRef]
- Hurst, N.R.; Locher, B.; Steinmuller, H.E.; Walters, L.J.; Chambers, L.G. Organic carbon dynamics and microbial community response to oyster reef restoration. Limnol. Oceanogr. 2022, 67, 1157–1168. [Google Scholar] [CrossRef]
- Yang, B.; Zhai, S.Y.; Li, X.; Tian, J.; Li, Q.; Shan, H.W.; Liu, S.K. Identification of Vibrio alginolyticus as a causative pathogen associated with mass summer mortality of the Pacific Oyster (Crassostrea gigas) in China. Aquaculture 2021, 535, 736363. [Google Scholar] [CrossRef]
- Ashton, E.C.; Guist, S.; Roberts, D.; Sigwart, J.D. Effects of Environmental Factors and Husbandry Practices on Summer Mortality Events in the Cultivated Pacific Oyster Crassostrea gigas in the North of Ireland. J. Shellfish. Res. 2020, 39, 13–20. [Google Scholar] [CrossRef]
- Ventilla, R.F. Recent Developments in the Japanese Oyster Culture Industry. Adv. Mar. Biol. 1984, 21, 1–57. [Google Scholar]
- Chávez-Villalba, J.; Arreola-Lizárraga, A.; Burrola-Sánchez, S.; Hoyos-Chairez, F. Growth, condition, and survival of the Pacific oyster Crassostrea gigas cultivated within and outside a subtropical lagoon. Aquaculture 2010, 300, 128–136. [Google Scholar] [CrossRef]
- Pernet, F.; Barret, J.; Le Gall, P.; Corporeau, C.; Degremont, L.; Lagarde, F.; Pepin, J.F.; Keck, N. Mass mortalities of Pacific oysters Crassostrea gigas reflect infectious diseases and vary with farming practices in the Mediterranean Thau lagoon, France. Aquacult Environ. Interact. 2012, 2, 215–237. [Google Scholar] [CrossRef]
- Samain, J.F.; Degremont, L.; Soletchnik, P.; Haure, J.; Bedier, E.; Ropert, M.; Moal, J.; Huvet, A.; Bacca, H.; Van Wormhoudt, A.; et al. Genetically based resistance to summer mortality in the Pacific oyster (Crassostrea gigas) and its relationship with physiological, immunological characteristics and infection processes. Aquaculture 2007, 268, 227–243. [Google Scholar] [CrossRef]
- Li, Y.; Qin, J.G.; Abbott, C.A.; Li, X.X.; Benkendorff, K. Synergistic impacts of heat shock and spawning on the physiology and immune health of Crassostrea gigas: An explanation for summer mortality in Pacific oysters. Am. J. Physiol. Integr. Comp. Physiol. 2007, 293, R2353–R2362. [Google Scholar] [CrossRef]
- Petton, B.; Pernet, F.; Robert, R.; Boudry, P. Temperature influence on pathogen transmission and subsequent mortalities in juvenile Pacific oysters Crassostrea gigas. Aquacult. Environ. Interac. 2013, 3, 257–273. [Google Scholar] [CrossRef]
- Crandall, G.; Thompson, R.E.; Eudeline, B.; Vadopalas, B.; Timmins-Schiffman, E.; Roberts, S. Proteomic response of early juvenile Pacific oysters to temperature. Peerj 2022, 10, e14158. [Google Scholar] [CrossRef]
- Pathirana, E.; Whittington, R.J.; Hick, P.M. Impact of seawater temperature on the Pacific oyster microbiome and susceptibility to disease associated with (OsHV-1). Anim. Prod. Sci. 2022, 62, 1040–1054. [Google Scholar] [CrossRef]
- Nell, J.A.; Hand, R.E. Evaluation of the progeny of second-generation Sydney rock oyster Saccostrea glomerata (Gould, 1850) breeding lines for resistance to QX disease Marteilia sydneyi. Aquaculture 2003, 228, 27–35. [Google Scholar] [CrossRef]
- Meng, L.X.; Li, Q.; Xu, C.X.; Liu, S.K.; Kong, L.F.; Yu, H. Hybridization improved stress resistance in the Pacific oyster: Evidence from physiological and immune responses. Aquaculture 2021, 545, 737227. [Google Scholar] [CrossRef]
- Li, Y.G.; Xu, C.X.; Li, Q. Physiological and gene expression responses of diploid and triploid Pacific oyster to heat acclimation. Aquac. Res. 2022, 53, 6641–6650. [Google Scholar] [CrossRef]
- Zhou, Q.; Chen, Y.D.; Lu, S.; Liu, Y.; Xu, W.T.; Li, Y.Z.; Wang, L.; Wang, N.; Yang, Y.M.; Chen, S.L. Development of a 50K SNP Array for Japanese Flounder and Its Application in Genomic Selection for Disease Resistance. Engineering 2021, 7, 406–411. [Google Scholar] [CrossRef]
- Qiu, L.L.; Zhou, P.; Wang, H.; Zhang, C.; Du, C.X.; Tian, S.J.; Wu, Q.Q.; Wei, L.T.; Wang, X.Y.; Zhou, Y.M.; et al. Photoperiod Genes Contribute to Daylength-Sensing and Breeding in Rice. Plants 2023, 12, 899. [Google Scholar] [CrossRef] [PubMed]
- Zhan, A.B.; Bao, Z.M.; Hu, X.L.; Wang, S.; Peng, W.; Wang, M.L.; Hu, J.J.; Liang, C.Z.; Yue, Z.Q. Characterization of 95 novel microsatellite markers for Zhikong scallop Chlamys farreri using FIASCO-colony hybridization and EST database mining. Fish. Sci. 2008, 74, 516–526. [Google Scholar] [CrossRef]
- Liu, Y.; Lu, S.; Liu, F.; Shao, C.W.; Zhou, Q.; Wang, N.; Li, Y.Z.; Yang, Y.M.; Zhang, Y.P.; Sun, H.J.; et al. Genomic Selection Using BayesC and GBLUP for Resistance Against Edwardsiella tarda in Japanese Flounder (Paralichthys olivaceus). Mar. Biotechnol. 2018, 20, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.F.; Liu, X.A.; Jiang, F.; Guo, X.M.; Liu, B. Unusual conservation of mitochondrial gene order in Crassostrea oysters: Evidence for recent speciation in Asia. BMC Evol. Biol. 2010, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Y.; Qian, L.M.; Liu, X.A.; Zhang, G.F.; Guo, X.M. Classification of a Common Cupped Oyster from Southern China. J. Shellfish. Res. 2010, 29, 857–866. [Google Scholar] [CrossRef]
- Wang, H.Y.; Zhang, G.F.; Lio, X.; Guo, X.M. Classification of common oysters from North China. J. Shellfish. Res. 2008, 27, 495–503. [Google Scholar] [CrossRef]
- Li, A.; Li, L.; Zhang, Z.Y.; Li, S.M.; Wang, W.; Guo, X.M.; Zhang, G.F. Noncoding Variation and Transcriptional Plasticity Promote Thermal Adaptation in Oysters by Altering Energy Metabolism. Mol. Biol. Evol. 2021, 38, 5144–5155. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.G.; Li, A.; Cong, R.H.; Qi, H.G.; Wang, W.; Zhang, G.F.; Li, L. Cis- and Trans-variations of Stearoyl-CoA Desaturase Provide New Insights into the Mechanisms of Diverged Pattern of Phenotypic Plasticity for Temperature Adaptation in Two Congeneric Oyster Species. Mol. Biol. Evol. 2023, 40, msad015. [Google Scholar] [CrossRef]
- Wang, C.; Li, A.; Wang, W.; Cong, R.H.; Wang, L.P.; Zhang, G.F.; Li, L. Integrated Application of Transcriptomics and Metabolomics Reveals the Energy Allocation-Mediated Mechanisms of Growth-Defense Trade-Offs in Crassostrea gigas and Crassostrea angulata. Front. Mar. Sci. 2021, 8, 744626. [Google Scholar] [CrossRef]
- Ghaffari, H.; Wang, W.; Li, A.; Zhang, G.F.; Li, L. Thermotolerance Divergence Revealed by the Physiological and Molecular Responses in Two Oyster Subspecies of Crassostrea gigas in China. Front. Physiol. 2019, 10, 93–105. [Google Scholar] [CrossRef]
- Jiang, G.W.; Li, Y.; Cheng, G.; Jiang, K.Y.; Zhou, J.M.; Xu, C.X.; Kong, L.F.; Yu, H.; Liu, S.K.; Li, Q. Transcriptome Analysis of Reciprocal Hybrids Between Crassostrea gigas and C. angulata Reveals the Potential Mechanisms Underlying Thermo-Resistant Heterosis. Mar. Biotechnol. 2023, 25, 235–246. [Google Scholar] [CrossRef]
- Jiang, G.; Zhou, J.M.; Cheng, G.; Meng, L.X.; Chi, Y.; Xu, C.X.; Li, Q. Examination of survival, physiological parameters and immune response in relation to the thermo-resistant heterosis of hybrid oysters derived from Crassostrea gigas and C. angulata. Aquaculture 2022, 559, 738454. [Google Scholar] [CrossRef]
- Korte, A.; Farlow, A. The advantages and limitations of trait analysis with GWAS: A review. Plant Methods 2013, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.K.; Tang, W.Q.; Bu, S.H.; Wu, W.R. BRM: A statistical method for QTL mapping based on bulked segregant analysis by deep sequencing. Bioinformatics 2020, 36, 2150–2156. [Google Scholar] [CrossRef] [PubMed]
- Uleberg, E.; Meuwissen, T.H.E. Fine mapping of multiple QTL using combined linkage and linkage disequilibrium mapping—A comparison of single QTL and multi QTL methods. Genet. Sel. Evol. 2007, 39, 285–299. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wang, Q.C.; Zhang, Q.; Luo, Z.; Wang, Y.; Zhang, X.J.; Huang, H.; Xiang, J.H.; Li, F.H. Genome Scan for Genomic Regions and Genes Associated with Growth Trait in Pacific White Shrimp Litopeneaus vannamei. Mar. Biotechnol. 2019, 21, 374–383. [Google Scholar] [CrossRef]
- Oikonomou, S.; Kazlari, Z.; Papapetrou, M.; Papanna, K.; Papaharisis, L.; Manousaki, T.; Loukovitis, D.; Dimitroglou, A.; Kottaras, L.; Gourzioti, E.; et al. Genome Wide Association (GWAS) Analysis and genomic heritability for parasite resistance and growth in European seabass. Aquacult. Rep. 2022, 24, 101178. [Google Scholar] [CrossRef]
- Huang, P.P.; Guo, W.J.; Wang, Y.H.; Xiong, Y.; Ge, S.; Gong, G.R.; Lin, Q.H.; Xu, Z.; Gui, J.F.; Mei, J. Genome-wide association study reveals the genetic basis of growth trait in catfish with sexual size. Genomics 2022, 114, 110380. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, L.; Meng, J.; Song, K.; Huang, B.Y.; Wang, W.; Zhang, G.F. Association and Functional Analyses Revealed That PPP1R3B Plays an Important Role in the Regulation of Glycogen Content in the Pacific Oyster Crassostrea gigas. Front. Genet. 2019, 10, 106. [Google Scholar] [CrossRef] [PubMed]
- Sodeland, M.; Gaarder, M.; Moen, T.; Thomassen, M.; Kjoglum, S.; Kent, M.; Lien, S. Genome-wide association testing reveals quantitative trait loci for fillet texture and fat content in Atlantic salmon. Aquaculture 2013, 408, 169–174. [Google Scholar] [CrossRef]
- Zhang, K.X.; Wang, J.P.; Ding, F.F.; Shi, R.H.; Wang, W.; Zhang, G.F.; Li, L. Identification of Distant Regulatory Elements Using Expression Quantitative Trait Loci Mapping for Heat-Responsive Genes in Oysters. Genes 2021, 12, 1040. [Google Scholar] [CrossRef]
- Ko, C.B.; Woo, Y.M.; Lee, D.J.; Lee, M.C.; Kim, C.S. Enhanced tolerance to heat stress in transgenic plants expressing the GASA4 gene. Plant Physiol. Biochem. 2007, 45, 722–728. [Google Scholar] [CrossRef]
- Grover, A.; Mittal, D.; Negi, M.; Lavania, D. Generating high temperature tolerant transgenic plants: Achievements and challenges. Plant Sci. 2013, 205, 38–47. [Google Scholar] [CrossRef]
- Ahmad, M.; Waraich, E.A.; Skalicky, M.; Hussain, S.; Zulfiqar, U.; Anjum, M.Z.; Rahman, M.H.U.; Brestic, M.; Ratnasekera, D.; Lamilla-Tamayo, L.; et al. Adaptation Strategies to Improve the Resistance of Oilseed Crops to Heat Stress Under a Changing Climate: An Overview. Front. Plant Sci. 2021, 12, 767150. [Google Scholar] [CrossRef]
- Jiang, G.W.; Li, Q.; Xu, C.X.; Liu, S.K.; Kong, L.F.; Yu, H. Reciprocal hybrids derived from Crassostrea gigas and C. angulata exhibit high heterosis in growth, survival and thermotolerance in northern China. Aquaculture 2021, 545, 737173. [Google Scholar] [CrossRef]
- Kema, G.H.J.; Verstappen, E.C.P.; Waalwijk, C. Avirulence in the wheat septoria tritici leaf blotch fungus Mycosphaerella graminicola is controlled by a single locus. Mol. Plant-Microbe Interact. 2000, 13, 1375–1379. [Google Scholar] [CrossRef]
- O’Quin, C.T.; Drilea, A.C.; Conte, M.A.; Kocher, T.D. Mapping of pigmentation QTL on an anchored genome assembly of the cichlid fish, Metriaclima zebra. BMC Genom. 2013, 14, 287. [Google Scholar] [CrossRef] [PubMed]
- Cuevas, H.E.; Song, H.; Staub, J.E.; Simon, P.W. Inheritance of beta-carotene-associated flesh color in cucumber (Cucumis sativus L.) fruit. Euphytica 2010, 171, 301–311. [Google Scholar] [CrossRef]
- Qiu, B.Y.; Zhang, T.F.; Zhang, S.Q.; Qu, Q.R.; Zhu, Z.C.; Liu, S.S.; Song, Z.F.; Xia, L.Q.; Yang, Z.Z.; Zhang, Q.; et al. BSA-seq and quantitative trait locus mapping reveals a major effective QTL for carpel number in watermelon (Citrullus lanatus). Plant Breed. 2022, 141, 460–470. [Google Scholar] [CrossRef]
- Tian, C.; Gregersen, P.K.; Seldin, M.F. Accounting for ancestry: Population substructure and genome-wide association studies. Hum. Mol. Genet. 2008, 17, R143–R150. [Google Scholar] [CrossRef] [PubMed]
- Campbell, C.D.; Ogburn, E.L.; Lunetta, K.L.; Lyon, H.N.; Freedman, M.L.; Groop, L.C.; Altshuler, D.; Ardlie, K.G.; Hirschhorn, J.N. Demonstrating stratification in a European American population. Nat. Genet. 2005, 37, 868–872. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Zhou, T.; Geng, X.; Liu, S.; Chen, A.; Yao, J.; Jiang, C.; Tan, S.; Su, B.; Liu, Z. A genome-wide association study of heat stress-associated SNPs in catfish. Anim. Genet. 2017, 48, 233–236. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Cong, R.; Wang, Y.; Li, L.; Zhang, G. Construction and analysis of the chromosome-level haplotype-resolved genomes of two crassostrea oyster congeners: Crassostrea angulata and Crassostrea gigas. Gigascience 2022, 12, giad077. [Google Scholar] [CrossRef]
- Liu, W.W.; Mazor, O.; Wilson, R.I. Thermosensory processing in the Drosophila brain. Nature 2015, 519, 353–357. [Google Scholar] [CrossRef]
- Ciaglia, T.; Vestuto, V.; Bertamino, A.; Gonzalez-Muniz, R.; Gomez-Monterrey, I. On the modulation of TRPM channels: Current perspectives and anticancer therapeutic implications. Front. Oncol. 2023, 12, 1065935. [Google Scholar] [CrossRef]
- Togashi, K.; Hara, Y.; Tominaga, T.; Higashi, T.; Konishi, Y.; Mori, Y.; Tominaga, M. TRPM2 activation by cyclic ADP-ribose at body temperature is involved in insulin secretion. EMBO J. 2006, 25, 1804–1815. [Google Scholar] [CrossRef]
- Hertweck, A.; de Mucha, M.V.; Barber, P.R.; Dagil, R.; Porter, H.; Ramos, A.; Lord, G.M.; Jenner, R.G. The T(H)1 cell lineage-determining transcription factor T-bet suppresses T(H)2 gene expression by redistributing GATA3 away from T(H)2 genes. Nucleic Acids Res. 2022, 50, 4557–4573. [Google Scholar] [CrossRef] [PubMed]
- Lacoste, A.; Jalabert, F.; Malham, S.; Cueff, A.; Gelebart, F.; Cordevant, C.; Lange, M.; Poulet, S.A. A Vibrio splendidus strain is associated with summer mortality of juvenile oysters Crassostrea gigas in the Bay of Morlaix (North Brittany, France). Dis. Aquat. Organ. 2001, 46, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Q.; Song, X.R.; Wang, W.L.; Wang, L.L.; Yi, Q.L.; Jiang, S.; Jia, Z.H.; Du, X.Y.; Qiu, L.M.; Song, L.S. The hematopoiesis in gill and its role in the immune response of Pacific oyster Crassostrea gigas against secondary challenge with Vibrio splendidus. Dev. Comp. Immunol. 2017, 71, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Yue, F.; Zhou, Z.; Wang, L.L.; Wang, M.Q.; Song, L.S. A conserved zinc finger transcription factor GATA involving in the hemocyte production of scallop Chlamys farreri. Fish Shellfish. Immun. 2014, 39, 125–135. [Google Scholar] [CrossRef]
- Song, S.L.; Tan, J.; Miao, Y.Y.; Zhang, Q. Crosstalk of ER stress-mediated autophagy and ER-phagy: Involvement of UPR and the core autophagy machinery. J. Cell. Physiol. 2018, 233, 3867–3874. [Google Scholar] [CrossRef]
- Peron, M.; Bonvini, P.; Rosolen, A. Effect of inhibition of the Ubiquitin-Proteasome System and Hsp90 on growth and survival of Rhabdomyosarcoma cells in vitro. BMC Cancer 2012, 12, 233. [Google Scholar] [CrossRef]
- Bozaykut, P.; Ozer, N.K.; Karademir, B. Regulation of protein turnover by heat shock proteins. Free Radic. Biol. Med. 2014, 77, 195–209. [Google Scholar] [CrossRef]
- Vourc’h, P.; Martin, I.; Bonnet-Brilhault, F.; Marouillat, S.; Barthelemy, C.; Muh, J.P.; Andres, C. Mutation screening and association study of the UBE2H gene on chromosome 7q32 in autistic disorder. Psychiatr. Genet. 2003, 13, 221–225. [Google Scholar] [CrossRef]
- Guo, X.L.; Ma, A.J.; Huang, Z.H.; Wang, X.A.; Yang, K.; Liu, Z.F.; Zhang, J.S.; Cui, W.X. Molecular characterization of ubiquitin-conjugating enzyme gene ube2h and siRNA-mediated regulation on targeting p53 in turbot, Scophthalmus maximus. J. Therm. Biol. 2021, 99, 102938. [Google Scholar] [CrossRef]
- Nishi, R.; Wijnhoven, P.W.G.; Kimura, Y.; Matsui, M.; Konietzny, R.; Wu, Q.; Nakamura, K.; Blundell, T.L.; Kessler, B.M. The deubiquitylating enzyme UCHL3 regulates Ku80 retention at sites of DNA damage. Sci. Rep. 2018, 8, 17891. [Google Scholar] [CrossRef]
- Li, R.Q.; Li, X.Q.; Zhao, J.; Meng, F.D.; Yao, C.; Bao, E.S.; Sun, N.; Chen, X.; Cheng, W.P.; Hua, H.; et al. Mitochondrial STAT3 exacerbates LPS-induced sepsis by driving CPT1a-mediated fatty acid oxidation. Theranostics 2022, 12, 976–998. [Google Scholar] [CrossRef]
- Dumas, A.; Liao, K.L.; Jeffries, K.M. Mathematical modeling and analysis of the heat shock protein response during thermal stress in fish and HeLa cells. Math. Biosci. 2022, 346, 108692. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Yuan, Z.H.; Li, Y.F.; Ma, Z. Genome-wide comparative analysis of DNAJ genes and their co-expression patterns with HSP70s in aestivation of the sea cucumber Apostichopus japonicus. Funct. Integr. Genom. 2022, 22, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Abisambra, J.F.; Jinwal, U.K.; Suntharalingam, A.; Arulselvam, K.; Brady, S.; Cockman, M.; Jin, Y.; Zhang, B.; Dickey, C.A. DnaJA1 Antagonizes Constitutive Hsp70-Mediated Stabilization of Tau. J. Mol. Biol. 2012, 421, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.D.; Zhou, Z.X.; Pan, Y.; Zhao, J.; Bai, H.Q.; Chen, B.H.; Zhang, X.Y.; Pu, F.; Chen, J.; Xu, P. GWAS identified candidate variants and genes associated with acute heat tolerance of large yellow croaker. Aquaculture 2021, 540, 736696. [Google Scholar] [CrossRef]
- Ishii, Y.; Sonezaki, S.; Iwasaki, Y.; Tauchi, E.; Shingu, Y.; Okita, K.; Ogawa, H.I.; Kato, Y.; Kondo, A. Single-step purification and characterization of MBP (maltose binding protein)-DnaJ fusion protein and its utilization for structure-function analysis. J. Biochem. 1998, 124, 842–847. [Google Scholar] [CrossRef] [PubMed]
- Li, G.L.; Zhao, H.; Guo, H.B.; Wang, Y.; Cui, X.P.; Li, H.; Xu, B.H.; Guo, X.Q. Analyses of the function of DnaJ family proteins reveal an underlying regulatory mechanism of heat tolerance in honeybee. Sci. Total Environ. 2020, 716, 137036. [Google Scholar] [CrossRef]
- Zhou, Z.Y.; Chen, G.; Shen, M.J.; Li, J.X.; Liu, K.; Liu, M.; Shi, S.; Chen, W.; Chen, S.X.; Yin, Y.X.; et al. Genome-scale CRISPR-Cas9 knockout screening in nasopharyngeal carcinoma for radiosensitive and radioresistant genes. Transl. Oncol. 2023, 30, 101625. [Google Scholar] [CrossRef]
- Li, A.; Li, L.; Wang, W.; Zhang, G.F. Evolutionary trade-offs between baseline and plastic gene expression in two congeneric oyster species. Biol. Lett. 2019, 15, 20190202. [Google Scholar] [CrossRef]
- Sokolova, I.M.; Frederich, M.; Bagwe, R.; Lannig, G.; Sukhotin, A.A. Energy homeostasis as an integrative tool for assessing limits of environmental stress tolerance in aquatic invertebrates. Mar. Environ. Res. 2012, 79, 1–15. [Google Scholar] [CrossRef]
- Duan, Q.X.; Zhang, S.; Wang, Y.; Lu, D.M.; Sun, Y.M.; Wu, Y.Y. Proton-coupled monocarboxylate transporters in cancer: From metabolic crosstalk, immunosuppression and anti-apoptosis to clinical applications. Front. Cell Dev. Biol. 2022, 10, 1069555. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.; Pedersen, B.; Guma, M. Solute carrier nutrient transporters in rheumatoid arthritis fibroblast-like synoviocytes. Front. Immunol. 2022, 13, 984408. [Google Scholar] [CrossRef] [PubMed]
- Merezhinskaya, N.; Fishbein, W.N. Monocarboxylate transporters: Past, present, and future. Histol. Histopathol. 2009, 24, 243–264. [Google Scholar] [PubMed]
- Jomura, R.; Akanuma, S.; Tachikawa, M.; Hosoya, K. SLC6A and SLC16A family of transporters: Contribution to transport of creatine and creatine precursors in creatine biosynthesis and distribution. BBA-Biomembranes 2022, 1864, 183840. [Google Scholar] [CrossRef]
- Kenkel, C.D.; Meyer, E.; Matz, M.V. Gene expression under chronic heat stress in populations of the mustard hill coral (Porites astreoides) from different thermal environments. Mol. Ecol. 2013, 22, 4322–4334. [Google Scholar] [CrossRef]
- Wang, C.; Du, M.; Jiang, Z.; Cong, R.; Wang, W.; Zhang, G.; Li, L. Comparative proteomic and phosphoproteomic analysis reveals differential heat response mechanism in two congeneric oyster species. Ecotoxicol. Environ. Saf. 2023, 263, 115197. [Google Scholar] [CrossRef]
- Qi, H.G.; Li, L.; Zhang, G.F. Construction of a chromosome-level genome and variation map for the Pacific oyster Crassostrea gigas. Mol. Ecol. Resour. 2021, 21, 1670–1685. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.A.M.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, s13742-015. [Google Scholar] [CrossRef]
- Yang, J.A.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A Tool for Genome-wide Complex Trait Analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.S.; Xu, J.Y.; He, W.M.; Yang, T.L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef] [PubMed]
| Gene ID | Start | End | Gene Name | Gene Annotation | Classification | |
|---|---|---|---|---|---|---|
| CHR01 | g01534 | 33,529,362 | 33,551,395 | Megf10 | Multiple epidermal growth factor-like domains protein | - |
| g01537 | 33,606,084 | 33,608,607 | Cav2 | Caveolin-2 | - | |
| g03038 | 68,399,030 | 68,404,722 | Trpm2 | Transient receptor potential cation channel M member 2 | temperature sensors | |
| CHR02 | g06281 | 45,304,077 | 45,345,630 | Comp | Cartilage oligomeric matrix protein | - |
| g06282 | 45,365,916 | 45,367,651 | Dnajc17 | DnaJ homolog subfamily C member 17 | heat shock proteins | |
| g06283 | 45,373,304 | 45,375,343 | Dnaja1 | DnaJ homolog subfamily A member 1 | heat shock proteins | |
| g06284 | 45,376,540 | 45,393,197 | Mettl3 | N6-adenosine-methyltransferase catalytic subunit | - | |
| g06286 | 45,393,469 | 45,403,190 | Cpsf6 | Cleavage and polyadenylation specificity factor subunit 6 | - | |
| CHR04 | g12537 | 53,406,861 | 53,433,358 | C1ql4 | Complement C1q-like protein 4 | - |
| g12538 | 53,437,493 | 53,454,981 | Ube2h | Ubiquitin-conjugating enzyme E2 H | ubiquitination processes | |
| g12549 | 53,638,138 | 53,654,138 | Cttnbp2 | Cortactin-binding protein 2 | - | |
| g12592 | 54,489,394 | 54,502,007 | Usp50 | Ubiquitin carboxyl-terminal hydrolase50 | ubiquitination processes | |
| g12597 | 54,554,609 | 54,560,671 | Uchl3 | Ubiquitin carboxyl-terminal hydrolase isozyme L3 | ubiquitination processes | |
| g12599 | 54,566,743 | 54,573,426 | Prpf18 | Pre-mRNA-splicing factor 18 | - | |
| g12652 | 55,507,413 | 55,508,138 | C1qtnf4 | Complement C1q tumor necrosis factor-related protein 4 | - | |
| g12653 | 55,509,496 | 55,514,690 | Grhpr | Glyoxylate reductase/hydroxypyruvate reductase | - | |
| g12658 | 55,571,045 | 55,583,506 | Nek4 | Serine/threonine-protein kinase Nek4 | - | |
| g12825 | 58,764,950 | 58,784,477 | Gata3 | Transcription factor GATA-3 | transcription factors | |
| CHR05 | g15391 | 38,958,891 | 38,967,233 | Slc16a9 | Monocarboxylate transporter 9 | transmembrane transporters |
| g15392 | 38,973,035 | 38,985,570 | Slc16a14 | Monocarboxylate transporter 14 | transmembrane transporters | |
| g15393 | 38,988,858 | 38,999,293 | Slc16a12 | Monocarboxylate transporter 12 | transmembrane transporters | |
| g15394 | 39,005,142 | 39,014,052 | Slc16a2 | Monocarboxylate transporter 2 | transmembrane transporters | |
| CHR07 | g20019 | 305,324 | 309,093 | Yabd | Deoxyribonuclease YabD | - |
| g20021 | 330,650 | 333,412 | Tatdn2 | Deoxyribonuclease TATDN2 | - | |
| CHR09 | g28376 | 46,302,989 | 46,305,847 | Scop1 | Serine rich endogenous peptide 1 | - |
| g28379 | 46,334,670 | 46,354,389 | Itih4 | Inter alpha-trypsin inhibitor, heavy chain 4 | - |
| SNP | Chr | Pos | A1 | A0 | FDR | Beta | p Value |
|---|---|---|---|---|---|---|---|
| CHR01_33558322 | CHR01 | 33,558,322 | A | T | 0.130237 | −0.34032 | 5.87 × 10−7 |
| CHR01_52587047 | CHR01 | 52,587,047 | C | T | 0.062233 | 0.524834 | 1.85 × 10−7 |
| CHR01_68449911 | CHR01 | 68,449,911 | T | G | 0.163389 | 0.405082 | 8.42 × 10−7 |
| CHR02_45368198 | CHR01 | 45,368,198 | C | A | 0.169521 | −0.50435 | 9.82 × 10−7 |
| CHR04_53398632 | CHR04 | 53,398,632 | C | G | 0.062233 | −0.52111 | 2 × 10−7 |
| CHR04_53638776 | CHR04 | 53,638,776 | T | G | 0.015796 | −0.41998 | 1.53 × 10−8 |
| CHR04_54528444 | CHR04 | 54,528,444 | C | A | 0.056362 | −0.33407 | 9.08 × 10−8 |
| CHR04_55502224 | CHR04 | 55,502,224 | C | T | 0.168788 | −0.6035 | 9.24 × 10−7 |
| CHR04_55503359 | CHR04 | 55,503,359 | A | T | 0.062003 | −0.62427 | 1.2 × 10−7 |
| CHR04_55554566 | CHR04 | 55,554,566 | G | A | 0.006984 | 0.408852 | 2.25 × 10−9 |
| CHR04_58789597 | CHR04 | 58,789,597 | A | T | 0.145299 | −0.38664 | 7.02 × 10−7 |
| CHR04_62006586 | CHR04 | 62,006,586 | A | T | 0.044535 | 0.35255 | 5.74 × 10−8 |
| CHR04_62006599 | CHR04 | 62,006,599 | A | T | 0.130237 | 0.329153 | 5.56 × 10−7 |
| CHR05_24461145 | CHR05 | 24,461,145 | T | C | 0.062233 | 0.422095 | 1.95 × 10−7 |
| CHR05_38969828 | CHR05 | 38,969,828 | G | C | 0.122076 | −0.46043 | 4.33 × 10−7 |
| CHR07_343905 | CHR07 | 343,905 | C | A | 0.122917 | 0.59167 | 4.75 × 10−7 |
| CHR09_46322969 | CHR09 | 46,322,969 | T | C | 0.014379 | 0.443599 | 9.26 × 10−9 |
| CHR10_18611994 | CHR10 | 18,611,994 | T | C | 0.062233 | −0.61093 | 1.48 × 10−7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, M.; Jiang, Z.; Wang, C.; Wei, C.; Li, Q.; Cong, R.; Wang, W.; Zhang, G.; Li, L. Genome-Wide Association Analysis of Heat Tolerance in F2 Progeny from the Hybridization between Two Congeneric Oyster Species. Int. J. Mol. Sci. 2024, 25, 125. https://doi.org/10.3390/ijms25010125
Du M, Jiang Z, Wang C, Wei C, Li Q, Cong R, Wang W, Zhang G, Li L. Genome-Wide Association Analysis of Heat Tolerance in F2 Progeny from the Hybridization between Two Congeneric Oyster Species. International Journal of Molecular Sciences. 2024; 25(1):125. https://doi.org/10.3390/ijms25010125
Chicago/Turabian StyleDu, Mingyang, Zhuxiang Jiang, Chaogang Wang, Chenchen Wei, Qingyuan Li, Rihao Cong, Wei Wang, Guofan Zhang, and Li Li. 2024. "Genome-Wide Association Analysis of Heat Tolerance in F2 Progeny from the Hybridization between Two Congeneric Oyster Species" International Journal of Molecular Sciences 25, no. 1: 125. https://doi.org/10.3390/ijms25010125
APA StyleDu, M., Jiang, Z., Wang, C., Wei, C., Li, Q., Cong, R., Wang, W., Zhang, G., & Li, L. (2024). Genome-Wide Association Analysis of Heat Tolerance in F2 Progeny from the Hybridization between Two Congeneric Oyster Species. International Journal of Molecular Sciences, 25(1), 125. https://doi.org/10.3390/ijms25010125





