Molecular Basis of Müllerian Agenesis Causing Congenital Uterine Factor Infertility—A Systematic Review
Abstract
1. Introduction
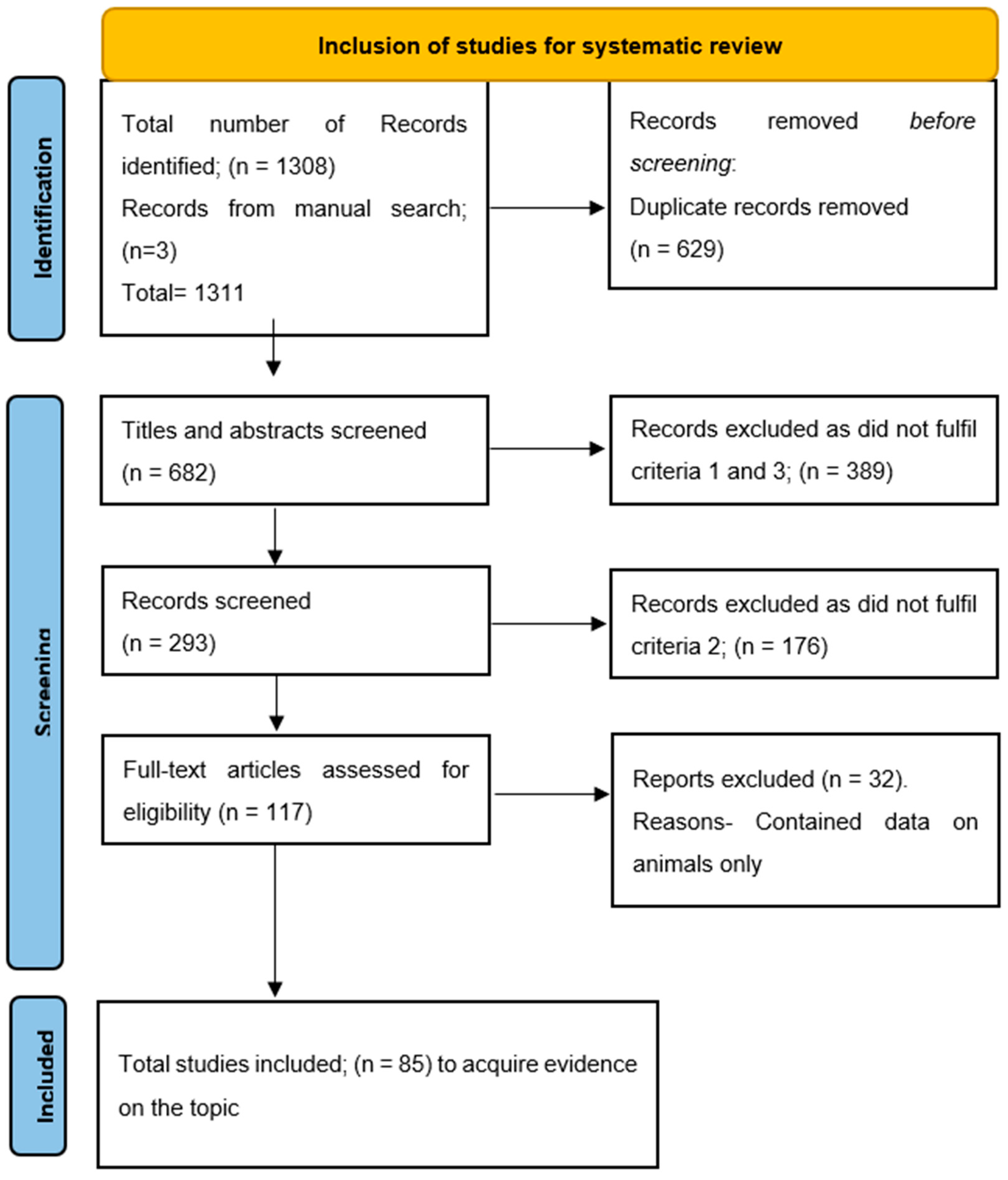
2. Methods
2.1. Search Strategy
2.2. Eligibility Criteria
2.2.1. Inclusion Criteria
- English language articles or where English translation is available;
- Full-text articles reporting on human genes, genome, genetics, or molecular bases;
- Containing information on Müllerian duct abnormality or Müllerian agenesis or Müllerian aplasia or uterine aplasia or Mayer–Rokitansky–Kuster–Hauser syndrome.
2.2.2. Exclusion Criteria
2.3. Study Selection
2.4. Data Collection
3. Results and Discussion
3.1. Genetic Basis of MA
3.2. Mechanisms of Genetic Changes
3.3. Fertility Options in Women with Müllerian Agenesis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization (WHO). International Classification of Diseases; 11th Revision (ICD-11); WHO: Geneva, Switzerland, 2018. [Google Scholar]
- Larsen, U. Research on infertility: Which definition should we use? Fertil. Steril. 2005, 83, 846–852. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO fact sheet on infertility. Glob. Reprod. Health 2021, 6, e52. [Google Scholar] [CrossRef]
- Assistance Médicale à la Procréation (AMP). Inserm-La Science Pour la Santé n.d. Available online: https://www.inserm.fr/information-en-sante/dossiers-information/assistance-medicale-procreation-amp (accessed on 7 August 2022).
- Dube, R.; Kar, S.S. Genital Microbiota and Outcome of Assisted Reproductive Treatment—A Systematic Review. Life 2022, 12, 1867. [Google Scholar] [CrossRef] [PubMed]
- Sallée, C.; Margueritte, F.; Marquet, P.; Piver, P.; Aubard, Y.; Lavoué, V.; Dion, L.; Gauthier, T. Uterine Factor Infertility, a Sys-tematic Review. J. Clin. Med. 2022, 11, 4907. [Google Scholar] [CrossRef] [PubMed]
- Brännström, M.; Johannesson, L.; Dahm-Kähler, P.; Enskog, A.; Mölne, J.; Kvarnström, N.; Diaz-Garcia, C.; Hanafy, A.; Lundmark, C.; Marcickiewicz, J.; et al. First clinical uterus transplantation trial: A six-month report. Fertil. Steril. 2014, 101, 1228–1236. [Google Scholar] [CrossRef] [PubMed]
- Hur, C.; Rehmer, J.; Flyckt, R.; Falcone, T. Uterine Factor Infertility: A Clinical Review. Clin. Obstet. Gynecol. 2019, 62, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Moncada-Madrazo, M.; Rodríguez Valero, C. Embryology, Uterus; Updated 25 July 2023; StatPearls: Treasure Island, FL, USA, 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK547748/ (accessed on 30 August 2023).
- Robbins, J.B.; Broadwell, C.; Chow, L.C.; Parry, J.P.; Sadowski, E.A. Müllerian duct anomalies: Embryological development, classification, and MRI assessment. J. Magn. Reson. Imaging 2015, 41, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ghofrani, M. Embryology. PathologyOutlines.com Website. Available online: https://www.pathologyoutlines.com/topic/ovarynontumorembryology.html (accessed on 28 September 2023).
- Dube, R.; Al-Zuheiri, S.T.S.; Syed, M.; Harilal, L.; Zuhaira, D.A.L.; Kar, S.S. Prevalence, Clinico-Bacteriological Profile, and Antibiotic Resistance of Symptomatic Urinary Tract Infections in Pregnant Women. Antibiotics 2023, 12, 33. [Google Scholar] [CrossRef]
- Dube, R.; Kar, S.S. COVID-19 in pregnancy: The foetal perspective—A systematic review. BMJ Paediatr. Open 2020, 4, e000859. [Google Scholar] [CrossRef]
- AlZuheiri, S.T.; Dube, R.; Menezes, G.; Qasem, S. Clinical profile and outcome of Group B streptococcal colonization in moth-ers and neonates in Ras Al Khaimah, United Arab Emirates: A prospective observational study. Saudi J. Med. Med. Sci. 2021, 9, 235–240. [Google Scholar]
- Fontana, L.; Gentilin, B.; Fedele, L.; Gervasini, C.; Miozzo, M. Genetics of Mayer-Rokitansky-Küster-Hauser (MRKH) syn-drome. Clin. Genet. 2017, 91, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Morcel, K.; Camborieux, L.; Programme de Recherches sur les Aplasies Müllériennes (PRAM); Guerrier, D. May-er-Rokitansky-Küster-Hauser (MRKH) syndrome. Orphanet J. Rare Dis. 2007, 14, 13. [Google Scholar] [CrossRef] [PubMed]
- Oppelt, P.; Strissel, P.; Kellermann, A.; Seeber, S.; Humeny, A.; Beckmann, M.; Strick, R. DNA sequence variations of the entire anti-Müllerian hormone (AMH) gene promoter and AMH protein expression in patients with the Mayer–Rokitanski–Küster–Hauser syndrome. Hum. Reprod. 2005, 20, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Guerrier, D.; Mouchel, T.; Pasquier, L.; Pellerin, I. The Mayer-Rokitansky-Küster-Hauser syndrome (congenital absence of uterus and vagina)—Phenotypic manifestations and genetic approaches. J. Negat. Results Biomed. 2006, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Herlin, M.; Højland, A.T.; Petersen, M.B. Familial occurrence of Mayer-Rokitansky-Küster-Hauser syndrome: A case report and review of the literature. Am. J. Med. Genet. A 2014, 164A, 2276–2286. [Google Scholar] [CrossRef]
- Jones, B.P.; Ranaei-Zamani, N.; Vali, S.; Williams, N.; Saso, S.; Thum, M.Y.; Al-Memar, M.; Dixon, N.; Rose, G.; Testa, G.; et al. Options for acquiring motherhood in abso-lute uterine factor infertility; adoption, surrogacy and uterine transplantation. Obstet. Gynaecol. 2021, 23, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Ledig, S.; Schippert, C.; Strick, R.; Beckmann, M.W.; Oppelt, P.G.; Wieacker, P. Recurrent aberrations identified by array-CGH in patients with Mayer-Rokitansky-Küster-Hauser syndrome. Fertil. Steril. 2011, 95, 1589–1594. [Google Scholar] [CrossRef]
- Ma, D.; Marion, R.; Punjabi, N.P.; Pereira, E.; Samanich, J.; Agarwal, C.; Li, J.; Huang, C.-K.; Ramesh, K.H.; Cannizzaro, A.L.; et al. A de novo 10.79 Mb interstitial deletion at 2q13q14.2 involving PAX8 causing hypothyroidism and mullerian agenesis: A novel case report and literature review. Mol. Cytogenet. 2014, 7, 85. [Google Scholar] [CrossRef]
- Chen, M.-J.; Wei, S.-Y.; Yang, W.-S.; Wu, T.-T.; Li, H.-Y.; Ho, H.-N.; Yang, Y.-S.; Chen, P.-L. Concurrent exome-targeted next generation sequencing and single nucleotide polymorphism array to identify the causative genetic aberrations of isolated Mayer-Rokitansky-Kuster-Hauser syndrome. Hum. Reprod. 2015, 30, 1732–1742. [Google Scholar] [CrossRef]
- Lischke, J.H.; Curtis, C.H.; Lamb, E.J. Discordance of vaginal agenesis in monozygotic twins. Obstet. Gynecol. 1973, 41, 920–924. [Google Scholar]
- Rall, K.; Eisenbeis, S.; Barresi, G.; Rückner, D.; Walter, M.; Poths, S.; Wallwiener, D.; Riess, O.; Bonin, M.; Brucker, S. May-er-Rokitansky-Küster-Hauser syndrome discordance in monozygotic twins: Matrix metalloproteinase 14, low-density lip-oprotein receptor–related protein 10, extracellular matrix, and neoangiogenesis genes identified as candidate genes in a tissue-specific mosaicism. Fertil. Steril. 2015, 103, 494–502.e3. [Google Scholar] [PubMed]
- Heidenreich, W.; Pfeiffer, A.; Kumbnani, H.K.; Scholz, W.; Zeuner, W. Disordant monozygotic twins with Mayer Rokitansky Kütser syndrome (author’s transl). Geburtshilfe Und Frauenheilkd. 1977, 37, 221–223. [Google Scholar]
- Steinkampf, M.P.; Dharia, S.P.; Dickerson, R.D. Monozygotic twins discordant for vaginal agenesis and bilateral tibial longi-tudinal deficiency. Fertil. Steril. 2003, 80, 643–645. [Google Scholar] [CrossRef] [PubMed]
- Regenstein, A.C.; Berkeley, A.S. Discordance of müllerian agenesis in monozygotic twins. A case report. J. Reprod. Med. 1991, 36, 396–397. [Google Scholar] [PubMed]
- Duru, U.A.; Laufer, M.R. Discordance in Mayer-von Rokitansky-Küster-Hauser Syndrome Noted in Monozygotic Twins. J. Pediatr. Adolesc. Gynecol. 2009, 22, e73–e75. [Google Scholar] [CrossRef] [PubMed]
- Milsom, S.R.; Ogilvie, C.M.; Jefferies, C.; Cree, L. Discordant Mayer-Rokitansky-Kuster-Hauser (MRKH) syndrome in identi-cal twins—A case report and implications for reproduction in MRKH women. Gynecol. Endocrinol. 2015, 31, 684–687. [Google Scholar] [CrossRef] [PubMed]
- Maniglio, P.; Ricciardi, E.; Laganà, A.S.; Triolo, O.; Caserta, D. Epigenetic modifications of primordial reproductive tract: A common etiologic pathway for Mayer-Rokitansky-Kuster-Hauser Syndrome and endometriosis? Med. Hypotheses 2016, 90, 4–5. [Google Scholar] [CrossRef]
- Rall, K.; Barresi, G.; Walter, M.; Poths, S.; Haebig, K.; Schaeferhoff, K.; Schoenfisch, B.; Riess, O.; Wallwiener, D.; Bonin, M.; et al. A combination of transcriptome and methylation analyses reveals embryologically-relevant candidate genes in MRKH patients. Orphanet J. Rare Dis. 2011, 6, 32. [Google Scholar] [CrossRef]
- Ray, R.; Kar, S.S.; Mahapatro, S.; Tripathy, R.; Mohanty, R. A Rare cause of seizure-dysgenesis of corpus callosum. Indian J. Pract. Pediatr. 2009, 11, 101–104. [Google Scholar]
- Theisen, A. Microarray-based Comparative Genomic Hybridization (aCGH). Nat. Educ. 2008, 1, 45. [Google Scholar]
- Eleanor, G.; Seaby, R.J.; Pengelly, S.E. Exome sequencing explained: A practical guide to its clinical application. Brief. Funct. Genom. 2016, 15, 374–384. [Google Scholar] [CrossRef]
- Van Dijk, E.L.; Jaszczyszyn, Y.; Naquin, D.; Thermes, C. The Third Revolution in Sequencing Technology. Trends Genet. 2018, 34, 666–681. [Google Scholar] [CrossRef] [PubMed]
- Sahyouni, J.K.; Odeh, L.B.M.; Mulla, F.; Junaid, S.; Kar, S.S.; Almarri, N.M.J.A.B. Infantile Sandhoff disease with ventricular septal defect: A case report. J. Med. Case Rep. 2022, 16, 317. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Zhao, S.; Jolly, A.; Wang, L.; Pan, H.; Yuan, J.; Chen, S.; Koch, A.; Ma, C.; Tian, W.; et al. Perturbations of genes essential for Mullerian duct and Wolffian duct development in Mayer-Rokitansky-Kuster-Hauser syndrome. Am. J. Hum. Genet. 2021, 108, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Mikhael, S.; Dugar, S.; Morton, M.; Chorich, L.P.; Tam, K.B.; Lossie, A.C.; Kim, H.-G.; Knight, J.; Taylor, H.S.; Mukherjee, S.; et al. Genetics of agenesis/hypoplasia of the uterus and vagina: Narrowing down the number of candidate genes for Mayer–Rokitansky–Küster–Hauser Syndrome. Qual. Life Res. 2021, 140, 667–680. [Google Scholar] [CrossRef]
- Williams, L.S.; Kim, H.G.; Kalscheuer, V.M.; Tuck, J.M.; Chorich, L.P.; Sullivan, M.E.; Falkenstrom, A.; Reindollar, R.H.; Layman, L.C. A balanced chromosomal translocation involving chromosomes 3 and 16 in a patient with Mayer-Rokitansky-Kuster-Hauser syndrome reveals new candidate genes at 3p22.3 and 16p13.3. Mol. Cytogenet. 2016, 9, 57. [Google Scholar] [CrossRef]
- Pan, H.-X.; Luo, G.-N.; Wan, S.-Q.; Qin, C.-L.; Tang, J.; Zhang, M.; Du, M.; Xu, K.-K.; Shi, J.-Q. Detection of de novo genetic var-iants in Mayer–Rokitansky–Küster–Hauser syndrome by whole genome sequencing. Eur. J. Obstet. Gynecol. Reprod. Biol. X 2019, 4, 100089. [Google Scholar]
- Thomson, E.; Tran, M.; Robevska, G.; Ayers, K.; van der Bergen, J.; Bhaskaran, P.G.; Haan, E.; Cereghini, S.; Vash-Margita, A.; Margetts, M.; et al. Functional genomics analysis identifies loss of HNF1B function as a cause of Mayer-Rokitansky-Küster-Hauser syndrome. Hum. Mol. Genet. 2023, 32, 1032–1047. [Google Scholar] [CrossRef]
- Backhouse, B.; Hanna, C.; Robevska, G.; Bergen, J.V.D.; Pelosi, E.; Simons, C.; Koopman, P.; Juniarto, A.Z.; Grover, S.; Faradz, S.; et al. Identification of Candidate Genes for Mayer-Rokitansky-Küster-Hauser Syndrome Using Genomic Approaches. Sex. Dev. 2019, 13, 26–34. [Google Scholar] [CrossRef]
- Chu, C.; Li, L.; Li, S.; Zhou, Q.; Zheng, P.; Zhang, Y.-D.; Duan, A.-H.; Lu, D.; Wu, Y.-M. Variants in genes related to develop-ment of the urinary system are associated with Mayer–Rokitansky–Küster–Hauser syndrome. Hum. Genom. 2022, 16, 10. [Google Scholar] [CrossRef]
- Pontecorvi, P.; Bernardini, L.; Capalbo, A.; Ceccarelli, S.; Megiorni, F.; Vescarelli, E.; Bottillo, I.; Preziosi, N.; Fabbretti, M.; Perniola, G.; et al. Protein–protein interaction network analysis applied to DNA copy number profiling suggests new per-spectives on the aetiology of Mayer–Rokitansky–Küster–Hauser syndrome. Sci. Rep. 2021, 11, 448. [Google Scholar] [CrossRef] [PubMed]
- Nodale, C.; Ceccarelli, S.; Giuliano, M.; Cammarota, M.; D’Amici, S.; Vescarelli, E.; Maffucci, D.; Bellati, F.; Panici, P.B.; Ro-mano, F.; et al. Gene Expression Profile of Patients with Mayer-Rokitansky-Küster-Hauser Syndrome: New Insights into the Potential Role of Developmental Pathways. PLoS ONE 2014, 9, e91010. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cheroki, C.; Krepischi, A.; Szuhai, K.; Brenner, V.; Kim, C.; Otto, P.A.; Rosenberg, C. Genomic imbalances associated with mullerian aplasia. J. Med. Genet. 2007, 45, 228–232. [Google Scholar] [CrossRef] [PubMed]
- McGowan, R.; Tydeman, G.; Shapiro, D.; Craig, T.; Morrison, N.; Logan, S.; Balen, A.H.; Ahmed, S.F.; Deeny, M.; Tolmie, J.; et al. DNA copy number variations are important in the complex genetic architecture of müllerian disorders. Fertil. Steril. 2015, 103, 1021–1030.e1. [Google Scholar] [CrossRef] [PubMed]
- Ledig, S.; Wieacker, P. Clinical and genetic aspects of Mayer–Rokitansky–Küster–Hauser syndrome. Med. Genet. 2018, 30, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Tewes, A.-C.; Rall, K.K.; Römer, T.; Hucke, J.; Kapczuk, K.; Brucker, S.; Wieacker, P.; Ledig, S. Variations in RBM8A and TBX6 are associated with disorders of the müllerian ducts. Fertil. Steril. 2015, 103, 1313–1318. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Li, Y.; Wang, M.; Li, H.; Su, T.; Wang, S. Associations of Polymorphisms in WNT9B and PBX1 with May-er-Rokitansky-Küster-Hauser Syndrome in Chinese Han. PLoS ONE 2015, 10, e0130202. [Google Scholar]
- Wang, M.; Li, Y.; Ma, W.; Li, H.; He, F.; Pu, D.; Su, T.; Wang, S. Analysis of WNT9B mutations in Chinese women with Mayer–Rokitansky–Küster–Hauser syndrome. Reprod. Biomed. Online 2014, 28, 80–85. [Google Scholar] [CrossRef]
- Buchert, R.; Schenk, E.; Hentrich, T.; Weber, N.; Rall, K.; Sturm, M.; Kohlbacher, O.; Koch, A.; Riess, O.; Brucker, S.Y.; et al. Ge-nome Sequencing and Transcriptome Profiling in Twins Discordant for Mayer-Rokitansky-Küster-Hauser Syndrome. J. Clin. Med. 2022, 11, 5598. [Google Scholar] [CrossRef]
- Waschk, D.; Tewes, A.-C.; Römer, T.; Hucke, J.; Kapczuk, K.; Schippert, C.; Hillemanns, P.; Wieacker, P.; Ledig, S. Mutations in WNT9B are associated with Mayer-Rokitansky-Küster-Hauser syndrome. Clin. Genet. 2016, 89, 590–596. [Google Scholar] [CrossRef]
- Nik-Zainal, S.; Strick, R.; Storer, M.; Huang, N.; Rad, R.; Willatt, L.; Fitzgerald, T.; Martin, V.; Sandford, R.; Carter, N.P.; et al. High incidence of recurrent copy number variants in patients with isolated and syndromic Mullerian aplasia. J. Med. Genet. 2011, 48, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Sandbacka, M.; Laivuori, H.; Freitas, É.; Halttunen, M.; Jokimaa, V.; Morin-Papunen, L.; Rosenberg, C.; Aittomäki, K. TBX6, LHX1 and copy number variations in the complex genetics of Müllerian aplasia. Orphanet J. Rare Dis. 2013, 8, 125. [Google Scholar] [CrossRef] [PubMed]
- Eksi, D.D.; Shen, Y.; Erman, M.; Chorich, L.P.; Sullivan, M.E.; Bilekdemir, M.; Yılmaz, E.; Luleci, G.; Kim, H.-G.; Alper, O.M.; et al. Copy number variation and regions of homozygosity analysis in patients with MÜLLERIAN aplasia. Mol. Cytogenet. 2018, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Tewes, A.-C.; Hucke, J.; Römer, T.; Kapczuk, K.; Schippert, C.; Hillemanns, P.; Wieacker, P.; Ledig, S. Sequence Variants in TBX6 Are Associated with Disorders of the Müllerian Ducts: An Update. Sex. Dev. 2019, 13, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Chen, N.; Jolly, A.; Zhao, S.; Coban-Akdemir, Z.; Tian, W.; Kang, J.; Ye, Y.; Wang, Y.; Koch, A.; et al. Functional characteristics of a broad spectrum of TBX6 variants in Mayer-Rokitansky-Küster-Hauser syndrome. Genet. Med. Off. J. Am. Coll. Med. Genet. 2022, 24, 2262–2273. [Google Scholar] [CrossRef] [PubMed]
- Bernardini, L.; Gimelli, S.; Gervasini, C.; Carella, M.; Baban, A.; Frontino, G.; Barbano, G.; Divizia, M.T.; Fedele, L.; Novelli, A.; et al. Recurrent microdeletion at 17q12 as a cause of Mayer-Rokitansky-Kuster-Hauser (MRKH) syndrome: Two case reports. Orphanet J. Rare Dis. 2009, 4, 25. [Google Scholar] [CrossRef] [PubMed]
- Ledig, S.; Brucker, S.; Barresi, G.; Schomburg, J.; Rall, K.; Wieacker, P. Frame shift mutation of LHX1 is associated with May-er-Rokitansky-Kuster-Hauser (MRKH) syndrome. Hum. Reprod. 2012, 27, 2872–2875. [Google Scholar] [CrossRef]
- Hinkes, B.; Hilgers, K.F.; Bolz, H.J.; Goppelt-Struebe, M.; Amann, K.; Nagl, S.; Bergmann, C.; Rascher, W.; Eckardt, K.-U.; Jaco-bi, J. A complex microdeletion 17q12 phenotype in a patient with recurrent de novo membranous nephropathy. BMC Nephrol. 2012, 13, 27. [Google Scholar] [CrossRef]
- Ledig, S.; Tewes, A.; Hucke, J.; Römer, T.; Kapczuk, K.; Schippert, C.; Hillemanns, P.; Wieacker, P. Array-comparative genomic hybridization analysis in patients with Müllerian fusion anomalies. Clin. Genet. 2018, 93, 640–646. [Google Scholar] [CrossRef]
- Mencarelli, M.A.; Katzaki, E.; Papa, F.T.; Sampieri, K.; Caselli, R.; Uliana, V.; Pollazzon, M.; Canitano, R.; Mostardini, R.; Grosso, S.; et al. Private inherited microdeletion/microduplications: Implications in clinical practice. Eur. J. Med. Genet. 2008, 51, 409–416. [Google Scholar] [CrossRef]
- Lindner, T.H.; Njølstad, P.R.; Horikawa, Y.; Bostad, L.; Bell, G.I.; Søvik, O. A novel syndrome of diabetes mellitus, renal dys-function and genital malformation associated with a partial deletion of the pseudo-POU domain of hepatocyte nuclear fac-tor-1beta. Hum. Mol. Genet. 1999, 8, 2001–2008. [Google Scholar] [CrossRef] [PubMed]
- Williams, L.S.; Eksi, D.D.; Shen, Y.; Lossie, A.C.; Chorich, L.P.; Sullivan, M.E.; Phillips, J.A.; Erman, M.; Kim, H.-G.; Alper, O.M.; et al. Genetic analysis of Mayer-Rokitansky-Kuster-Hauser syndrome in a large cohort of families. Fertil. Steril. 2017, 108, 145–151.e2. [Google Scholar] [CrossRef] [PubMed]
- Xia, M.; Zhao, H.; Qin, Y.; Mu, Y.; Wang, J.; Bian, Y.; Ma, J.; Chen, Z.-J. LHX1 mutation screening in 96 patients with müllerian duct abnormalities. Fertil. Steril. 2012, 97, 682–685. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhou, X.; Liu, L.; Zhu, Y.; Liu, C.; Pan, H. Identification and functional analysis of a novel LHX1 mutation associ-ated with congenital absence of the uterus and vagina. Oncotarget 2017, 8, 8785–8790. [Google Scholar] [CrossRef] [PubMed]
- Oram, R.A.; Edghill, E.L.; Blackman, J.; Taylor, M.J.; Kay, T.; Flanagan, S.E.; Ismail-Pratt, I.; Creighton, S.M.; Ellard, S.; Hatters-ley, A.T.; et al. Mutations in the hepatocyte nuclear factor-1_ (HNF1B) gene are common with combined uterine and renal malformations but are not found with isolated uterine malformations. Am. J. Obstet. Gynecol. 2010, 203, 364.e1–364.e5. [Google Scholar] [CrossRef] [PubMed]
- Bingham, C.; Ellard, S.; Cole, T.R.; Jones, K.E.; Allen, L.I.; Goodship, J.A.; Goodship, T.H.; Bakalinova-Pugh, D.; Russell, G.I.; Woolf, A.; et al. Solitary functioning kidney and diverse genital tract malformations associated with hepatocyte nuclear factor-1 mutations. Kidney Int. 2002, 61, 1243–1251. [Google Scholar] [CrossRef] [PubMed]
- Sundaram, U.T.; McDonald-McGinn, D.M.; Huff, D.; Emanuel, B.S.; Zackai, E.H.; Driscoll, D.A.; Bodurtha, J. Primary amenor-rhea and absent uterus in the 22q11.2 deletion syndrome. Am. J. Med. Genet. Part A 2007, 143, 2016–2018. [Google Scholar] [CrossRef]
- Morcel, K.; Watrin, T.; Pasquier, L.; Rochard, L.; Le Caignec, C.; Dubourg, C.; Loget, P.; Paniel, B.-J.; Odent, S.; David, V.; et al. Utero-vaginal aplasia (Mayer-Rokitansky-Küster-Hauser syndrome) associated with deletions in known DiGeorge or Di-George-like loci. Orphanet J. Rare Dis. 2011, 6, 9. [Google Scholar] [CrossRef]
- Cheroki, C.; Krepischi-Santos, A.C.; Rosenberg, C.; Jehee, F.S.; Mingroni-Netto, R.C.; Filho, I.P.; Filho, S.Z.; Kim, C.A.; Bagnoli, V.R.; Mendonca, B.B.; et al. Report of a del22q11 in a patient with Mayer-Rokitansky-Küster-Hauser (MRKH) anomaly and exclusion ofWNT-4,RAR-gamma, andRXR-alpha as major genes determining MRKH anomaly in a study of 25 affected women. Am. J. Med. Genet. Part A 2006, 140, 1339–1342. [Google Scholar] [CrossRef]
- AlSubaihin, A.; Vandermeulen, J.; Harris, K.; Duck, J.; McCready, E. Müllerian Agenesis in Cat Eye Syndrome and 22q11 Chromosome Abnormalities: A Case Report and Literature Review. J. Pediatr. Adolesc. Gynecol. 2018, 31, 158–161. [Google Scholar] [CrossRef]
- Philibert, P.; Biason-Lauber, A.; Gueorguieva, I.; Stuckens, C.; Pienkowski, C.; Lebon-Labich, B.; Paris, F.; Sultan, C. Molecular analysis of WNT4 gene in four adolescent girls with mullerian duct abnormality and hyperandrogenism (atypical Mayer-Rokitansky-Küster-Hauser syndrome). Fertil. Steril. 2011, 95, 2683–2686. [Google Scholar] [CrossRef] [PubMed]
- Ragitha, T.S.; Sunish, K.S.; Gilvaz, S.; Daniel, S.; Varghese, P.R.; Raj, S.; Francis, J.; Kumar, R.S. Mutation analysis of WNT4 gene in SRY negative 46,XX DSD patients with Mullerian agenesis and/or gonadal dysgenesis-An Indian study. Gene 2023, 861, 147236. [Google Scholar] [CrossRef] [PubMed]
- Biason-Lauber, A.; Konrad, D.; Navratil, F.; Schoenle, E.J. A WNT4 Mutation Associated with Müllerian-Duct Regression and Virilization in a 46,XXWoman. N. Engl. J. Med. 2004, 351, 792–798. [Google Scholar] [CrossRef] [PubMed]
- Kyei Barffour, I.; Kyei Baah Kwarkoh, R. GREB1L as a candidate gene of Mayer-Rokitansky-Kuster-Hauser Syndrome. Eur. J. Med. Genet. 2021, 64, 104158. [Google Scholar] [CrossRef] [PubMed]
- Herlin, M.K.; Le-Quy, V.; Højland, A.T.; Ernst, A.; Okkels, H.; Petersen, A.C.; Petersen, M.B.; Pedersen, I.S. Whole-exome se-quencing identifies a GREB1L variant in a three-generation family with Müllerian and renal agenesis: A novel candidate gene in Mayer–Rokitansky–Küster–Hauser (MRKH) syndrome. A case report. Hum. Reprod. 2019, 34, 1838–1846. [Google Scholar] [CrossRef] [PubMed]
- Jacquinet, A.; Boujemla, B.; Fasquelle, C.; Thiry, J.; Josse, C.; Lumaka, A.; Brischoux-Boucher, E.; Dubourg, C.; David, V.; Pas-quier, L.; et al. GREB1L variants in familial and sporadic hereditary urogenital adysplasia and May-er-Rokitansky-Kuster-Hauser syndrome. Clin. Genet. 2020, 98, 126–137. [Google Scholar] [CrossRef]
- Xing, Q.; Xu, Z.; Zhu, Y.; Wang, X.; Wang, J.; Chen, D.; Xu, Y.; He, X.; Xiang, H.; Wang, B.; et al. Genetic analysis of DACT1 in 100 Chinese Han women with Müllerian duct anomalies. Reprod. Biomed. Online 2016, 32, 420–426. [Google Scholar] [CrossRef][Green Version]
- Ravel, C.; Bashamboo, A.; Bignon-Topalovic, J.; Siffroi, J.-P.; McElreavey, K.; Darai, E. Polymorphisms in DLGH1 and LAMC1 in Mayer–Rokitansky–Kuster–Hauser syndrome. Reprod. Biomed. Online 2012, 24, 462–465. [Google Scholar] [CrossRef]
- Brucker, S.Y.; Frank, L.; Eisenbeis, S.; Henes, M.; Wallwiener, D.; Riess, O.; van Eijck, B.; Schöller, D.; Bonin, M.; Rall, K.K. Se-quence variants in ESR1 and OXTR are associated with Mayer-Rokitansky-Küster-Hauser syndrome. Acta Obstet. Gynecol. Scand. 2017, 96, 1338–1346. [Google Scholar] [CrossRef]
- Brucker, S.Y.; Eisenbeis, S.; Konig, J.; Lamy, M.; Salker, M.S.; Zeng, N.; Seeger, H.; Henes, M.; Scholler, D.; Schonfisch, B.; et al. Decidualization is Impaired in Endometrial Stromal Cells from Uterine Rudiments in Mayer-Rokitansky-Kuster-Hauser Syndrome. Cell Physiol. Biochem. 2017, 41, 1083–1097. [Google Scholar] [CrossRef]
- Miyamoto, Y.; Taniguchi, H.; Hamel, F.; Silversides, D.W.; Viger, R.S. A GATA4/WT1 cooperation regulates transcription of genes required for mammalian sex determination and differentiation. BMC Mol Biol. 2008, 9, 44. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liao, S.; Luo, G.; Li, H.; Wang, S.; Li, Z.; Luo, X. An Association between EMX2 Variations and May-er-Rokitansky-Küster-Hauser Syndrome: A Case-Control Study of Chinese Women. J. Healthc. Eng. 2022, 2022, 9975369. [Google Scholar] [PubMed]
- Liu, S.; Gao, X.; Qin, Y.; Liu, W.; Huang, T.; Ma, J.; Simpson, J.L.; Chen, Z.-J. Nonsense mutation of EMX2 is potential causative for uterus didelphysis: First molecular explanation for isolated incomplete müllerian fusion. Fertil. Steril. 2015, 103, 769–774.e2. [Google Scholar] [CrossRef] [PubMed]
- Smol, T.; Ribero-Karrouz, W.; Edery, P.; Gorduza, D.B.; Catteau-Jonard, S.; Manouvrier-Hanu, S.; Ghoumid, J. May-er-Rokitansky-Kunster-Hauser syndrome due to 2q12.1q14.1 deletion: PAX8 the causing gene? Eur. J. Med. Genet. 2020, 63, 103812. [Google Scholar] [CrossRef]
- Gervasini, C.; Grati, F.R.; Lalatta, F.; Tabano, S.; Gentilin, B.; Colapietro, P.; De Toffol, S.; Frontino, G.; Motta, F.; Maitz, S.; et al. SHOX duplications found in some cases with type I Mayer-Rokitansky-Kuster-Hauser syndrome. Genet. Med. 2010, 12, 634–640. [Google Scholar] [CrossRef]
- Ma, J.; Qin, Y.; Liu, W.; Duan, H.; Xia, M.; Chen, Z.-J. Analysis of PBX1 mutations in 192 Chinese women with Müllerian duct abnormalities. Fertil. Steril. 2011, 95, 2615–2617. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/gene/7849 (accessed on 10 September 2023).
- Timmreck, L.S.; Gray, M.R.; Handelin, B.; Allito, B.; Rohlfs, E.; Davis, A.J.; Gidwani, G.; Reindollar, R.H. Analysis of cystic fi-brosis transmembrane conductance regulator gene mutations in patients with congenital absence of the uterus and vagina. Am. J. Med. Genet. 2003, 120, 72–76. [Google Scholar] [CrossRef]
- Drummond, J.B.; Rezende, C.F.; Peixoto, F.C.; Carvalho, J.S.; Reis, F.M.; De Marco, L. Molecular analysis of the beta-catenin gene in patients with the Mayer-Rokitansky-Küster-Hauser syndrome. J. Assist. Reprod. Genet. 2008, 25, 511–514. [Google Scholar] [CrossRef][Green Version]
- Lalwani, S.; Wu, H.-H.; Reindollar, R.H.; Gray, M.R. HOXA10 mutations in congenital absence of uterus and vagina. Fertil. Steril. 2008, 89, 325–330. [Google Scholar] [CrossRef]
- Takahashi, K.; Hayano, T.; Sugimoto, R.; Kashiwagi, H.; Shinoda, M.; Nishijima, Y.; Suzuki, T.; Suzuki, S.; Ohnuki, Y.; Kondo, A.; et al. Exome and copy number variation analyses of Mayer–Rokitansky–Küster–Hauser syndrome. Hum. Genome Var. 2018, 5, 27. [Google Scholar] [CrossRef]
- Petrozza, J.C.; Gray, M.R.; Davis, A.J.; Reindollar, R.H. Congenital absence of the uterus and vagina is not commonly transmitted as a dominant genetic trait: Outcomes of surrogate pregnancies. Fertil. Steril. 1997, 67, 387–389. [Google Scholar] [CrossRef] [PubMed]
- Hentrich, T.; Koch, A.; Weber, N.; Kilzheimer, A.; Maia, A.; Burkhardt, S.; Rall, K.; Casadei, N.; Kohlbacher, O.; Riess, O.; et al. The Endometrial Transcription Landscape of MRKH Syndrome. Front. Cell Dev. Biol. 2020, 8, 572281. [Google Scholar] [CrossRef] [PubMed]
- Brakta, S.; Hawkins, Z.A.; Sahajpal, N.; Seman, N.; Kira, D.; Chorich, L.P.; Kim, H.G.; Xu, H.; Phillips, J.A., 3rd; Kolhe, R.; et al. Rare structural variants, aneuploidies, and mosaicism in in-dividuals with Mullerian aplasia detected by optical genome mapping. Hum Genet. 2023, 142, 483–494. [Google Scholar] [CrossRef] [PubMed]
- Murry, J.B.; Santos, X.M.; Wang, X.; Wan, Y.-W.; Veyver, I.B.V.D.; Dietrich, J.E. A genome-wide screen for copy number altera-tions in an adolescent pilot cohort with müllerian anomalies. Fertil. Steril. 2015, 103, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhao, H.; Sun, M.; Li, Y.; Chen, Z.-J. PAX2 in 192 Chinese women with Müllerian duct abnormalities: Mutation analysis. Reprod. Biomed. Online 2012, 25, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.; Qin, Y.; Xu, C.; Li, G.; Zhao, X.; Chen, Z.-J. Mutations in WNT4 are not responsible for Müllerian duct abnormali-ties in Chinese women. Reprod. Biomed. Online 2012, 24, 630–633. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liatsikos, S.A.; Grimbizis, G.F.; Georgiou, I.; Papadopoulos, N.; Lazaros, L.; Bontis, J.N.; Tarlatzis, B.C. HOX A10 and HOX A11 mutation scan in congenital malformations of the female genital tract. Reprod. Biomed. Online 2010, 21, 126–132. [Google Scholar] [CrossRef]
- Hofstetter, G.; Concin, N.; Marth, C.; Rinne, T.; Erdel, M.; Janecke, A. Genetic analyses in a variant of May-er-Rokitansky-Kuster-Hauser syndrome (MURCS association). Wien. Klin. Wochenschr. 2008, 120, 435–439. [Google Scholar] [CrossRef]
- Burel, A.; Mouchel, T.; Odent, S.; Tiker, F.; Knebelmann, B.; Pellerin, I.; Guerrier, D. Role of HOXA7 to HOXA13 and PBX1 genes in various forms of MRKH syndrome (congenital absence of uterus and vagina). J. Negat. Results Biomed. 2006, 5, 4. [Google Scholar] [CrossRef]
- Acién, P.; Galán, F.; Manchón, I.; Ruiz, E.; Acién, M.; Alcaraz, A.L. Hereditary renal adysplasia, pulmonary hypoplasia and Mayer-Rokitansky-Küster-Hauser (MRKH) syndrome: A case report. Orphanet J. Rare Dis. 2010, 5, 6. [Google Scholar] [CrossRef]
- Resendes, B.L.; Sohn, S.H.; Stelling, J.R.; Tineo, R.; Davis, A.J.; Gray, M.R.; Reindollar, R.H. Role for anti-Müllerian hormone in congenital absence of the uterus and vagina. Am. J. Med. Genet. 2001, 98, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Clément-Ziza, M.; Khen-Dunlop, N.; Gonzales, J.; Crétolle-Vastel, C.; Picard, J.-Y.; Tullio-Pelet, A.; Besmond, C.; Munnich, A.; Lyonnet, S.; Nihoul-Fékété, C. Exclusion ofWNT4 as a major gene in Rokitansky-Küster-Hauser anomaly. Am. J. Med. Genet. Part A 2005, 137, 98–99. [Google Scholar] [CrossRef] [PubMed]
- Zenteno, J.C.; Carranza-Lira, S.; Kofman-Alfaro, S. Molecular analysis of the anti-Müllerian hormone, the anti-Müllerian hormone receptor, and galactose-1-phosphate uridyl transferase genes in patients with the May-er-Rokitansky-Küster-Hauser syndrome. Arch. Gynecol. Obstet. 2004, 269, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Klipstein, S.; Bhagavath, B.; Topipat, C.; Sasur, L.; Reindollar, R.; Gray, M. The N314D polymorphism of the GALT gene is not associated with congenital absence of the uterus and vagina. Mol. Hum. Reprod. 2003, 9, 171–174. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Plevraki, E.; Kita, M.; Goulis, D.G.; Hatzisevastou-Loukidou, H.; Lambropoulos, A.F.; Avramides, A. Bilateral ovarian agen-esis and the presence of the testis-specific protein 1-Y-linked gene: Two new features of Mayer-Rokitansky-Küster-hauser syndrome. Fertil. Steril. 2004, 81, 689–692. [Google Scholar] [CrossRef] [PubMed]
- Cramer, D.W.; Goldstein, D.P.; Fraer, C.; Reichardt, J. Vaginal agenesis (Mayer-Rokitansky-Kuster-Hauser Syndrome) asso-ciated with the N314D mutation of galactose-1-phosphate uridyl transferase (GALT). Mol. Hum. Reprod. 1996, 2, 145–148. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stévant, I.; Nef, S. Genetic Control of Gonadal Sex Determination and Development. Trends Genet. 2019, 35, 346–358. [Google Scholar] [CrossRef]
- Rotgers, E.; Jørgensen, A.; Yao, H.H. At the Crossroads of Fate-Somatic Cell Lineage Specification in the Fetal Gon-ad. Endocr. Rev. 2018, 39, 739–759. [Google Scholar] [CrossRef]
- Orvis, G.D.; Behringer, R.R. Cellular mechanisms of Müllerian duct formation in the mouse. Dev. Biol. 2007, 306, 493–504. [Google Scholar] [CrossRef]
- Mullen, R.D.; Behringer, R.R. Molecular genetics of Müllerian duct formation, regression and differentiation. Sex. Dev. 2014, 8, 281–296. [Google Scholar] [CrossRef]
- Kobayashi, A.; Shawlot, W.; Kania, A.; Behringer, R.R. Requirement of Lim1 for female reproductive tract development. Development 2004, 131, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Fujino, A.; MacLaughlin, D.T.; Manganaro, T.F.; Szotek, P.P.; Arango, N.A.; Teixeira, J.; Donahoe, P.K. Müllerian inhibiting substance regulates its receptor/SMAD signaling and causes mesenchymal transition of the coe-lomic epithelial cells early in Müllerian duct regression. Development 2006, 133, 2359–2369. [Google Scholar] [CrossRef] [PubMed]
- Arango, N.A.; Kobayashi, A.; Wang, Y.; Jamin, S.P.; Lee, H.H.; Orvis, G.D.; Behringer, R.R. A mesenchymal per-spective of Müllerian duct differentiation and regression in Amhr2-lacZ mice. Mol. Reprod. Dev. 2008, 75, 1154–1162. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.J.; Harding, M.; Moayedi, Y.; Mardon, G. Mouse Dach1 and Dach2 are redundantly required for Müllerian duct development. Genesis 2008, 46, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.C.; Orvis, G.D.; Kwan, K.M.; Behringer, R.R. Lhx1 is required in Müllerian duct epithelium for uterine development. Dev. Biol. 2014, 389, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Vainio, S.; Heikkila, M.; Kispert, A.; Chin, N.; McMahon, A.P. Female development in mammals is regulated by Wnt-4 signaling. Nature 1999, 397, 405–409. [Google Scholar] [CrossRef]
- Carroll, T.J.; Park, J.S.; Hayashi, S.; Majumdar, A.; McMahon, A.P. Wnt9b plays a central role in the regulation of mesen-chymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev.Cell 2005, 9, 283–292. [Google Scholar] [CrossRef]
- Arango, N.A.; Lovell-Badge, R.; Behringer, R.R. Targeted mutagenesis of the endogenous mouse Mis gene promot-er: In vivo definition of genetic pathways of vertebrate sexual development. Cell 1999, 99, 409–419. [Google Scholar] [CrossRef]
- Lasala, C.; Schteingart, H.F.; Arouche, N.; Bedecarrás, P.; Grinspon, R.P.; Picard, J.Y.; Josso, N.; di Clemente, N.; Rey, R.A. SOX9 and SF1 are involved in cyclic AMP-mediated upregulation of anti-Mullerian gene expression in the testicular prepubertal Sertoli cell line SMAT1. American journal of physiology. Endocrinol. Metab. 2011, 301, E539–E547. [Google Scholar] [CrossRef]
- Barrionuevo, F.; Bagheri-Fam, S.; Klattig, J.; Kist, R.; Taketo, M.M.; Englert, C.; Scherer, G. Homozygous Inactivation of Sox9 Causes Complete XY Sex Reversal in Mice. Biol. Reprod. 2006, 74, 195–201. [Google Scholar] [CrossRef]
- Kim, Y.; Bingham, N.; Sekido, R.; Parker, K.L.; Lovell-Badge, R.; Capel, B. Fibroblast growth factor receptor 2 regulates proliferation and Sertoli differentiation during male sex determination. Proc. Natl. Acad. Sci. USA 2007, 104, 16558–16563. [Google Scholar] [CrossRef] [PubMed]
- Bagheri-Fam, S.; Sim, H.; Bernard, P.; Jayakody, I.; Taketo, M.M.; Scherer, G.; Harley, V.R. Loss of Fgfr2 leads to par-tial XY sex reversal. Dev. Biol. 2008, 314, 71–83. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Carré, G.A.; Greenfield, A. Characterising novel pathways in testis determination using mouse genetics. Sex. Dev. 2014, 8, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Sutton, E.; Hughes, J.; White, S.; Sekido, R.; Tan, J.; Arboleda, V.; Rogers, N.; Knower, K.; Rowley, L.; Eyre, H.; et al. Identification of SOX3 as an XX male sex reversal gene in mice and humans. J. Clin. Investig. 2011, 121, 328–341. [Google Scholar] [CrossRef] [PubMed]
- Ottolenghi, C.; Omari, S.; García-Ortiz, J.E.; Uda, M.; Crisponi, L.; Forabosco, A.; Pilia, G.; Schlessinger, D. Foxl2 is required for commitment to ovary differentiation. Hum. Mol. Genet. 2005, 14, 2053–2062. [Google Scholar] [CrossRef]
- Wilhelm, D.; Martinson, F.; Bradford, S.; Wilson, M.J.; Combes, A.N.; Beverdam, A.; Bowles, J.; Mizusaki, H.; Koopman, P. Sertoli cell differentiation is induced both cell-autonomously and through prostaglandin signaling during mammalian sex determination. Dev. Biol. 2005, 287, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kobayashi, A.; Sekido, R.; DiNapoli, L.; Brennan, J.; Chaboissier, M.-C.; Poulat, F.; Behringer, R.R.; Lov-ell-Badge, R.; Capel, B. Fgf9 and Wnt4 Act as Antagonistic Signals to Regulate Mammalian Sex Determination. PLoS Biol. 2006, 4, e187. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, A.; Stewart, C.A.; Wang, Y.; Fujioka, K.; Thomas, N.C.; Jamin, S.P.; Behringer, R.R. β-Catenin is essential for Müllerian duct regression during male sexual differentiation. Development 2011, 138, 1967–1975. [Google Scholar] [CrossRef]
- Deutscher, E.; Hung-Chang Yao, H. Essential roles of mesenchyme-derived beta-catenin in mouse Müllerian duct morphogenesis. Dev. Biol. 2007, 307, 227–236. [Google Scholar] [CrossRef][Green Version]
- Ungewitter, E.K.; Yao, H.H. How to make a gonad: Cellular mechanisms governing formation of the testes and ova-ries. Sex Dev. 2013, 7, 7–20. [Google Scholar] [CrossRef]
- Schrader, T.J. Encyclopedia of Food Sciences and Nutrition, 2nd ed.; Academic Press: New York, NY, USA, 2003; pp. 4059–4067. [Google Scholar] [CrossRef]
- Liu, J.L.C. Coding or Noncoding, the Converging Concepts of RNAs. Front. Genet. 2019, 10, 496. [Google Scholar] [CrossRef]
- Candidate Gene. Available online: https://www.genome.gov/genetics-glossary/Candidate-Gene (accessed on 20 August 2023).
- Brewer, C.; Holloway, S.; Zawalnyski, P.; Schinzel, A.; FitzPatrick, D. A chromosomal deletion map of human malfor-mations. Am. J. Hum. Genet. 1998, 63, 1153–1159. [Google Scholar] [CrossRef] [PubMed]
- Clancy, S.; Shaw, K. DNA deletion and duplication and the associated genetic disorders. Nat. Educ. 2008, 1, 23. [Google Scholar]
- Cheng, Z.; Zhu, Y.; Su, D.; Wang, J.; Cheng, L.; Chen, B.; Wei, Z.; Zhou, P.; Wang, B.; Ma, X.; et al. A novel mutation of HOXA10 in a Chinese woman with a Mullerian duct anomaly. Hum. Reprod. 2011, 26, 3197–3201. [Google Scholar] [CrossRef] [PubMed]
- Ekici, A.B.; Strissel, P.L.; Oppelt, P.G.; Renner, S.P.; Brucker, S.; Beckmann, M.W.; Strick, R. HOXA10 and HOXA13 sequence variations in human female genital malformations including congenital absence of the uterus and vagina. Gene 2013, 518, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Biason-Lauber, A.; De Filippo, G.; Konrad, D.; Scarano, G.; Nazzaro, A.; Schoenle, E. WNT4 deficiency—A clinical phenotype distinct from the classic Mayer–Rokitansky–Kuster–Hauser syndrome: A Case Report. Hum. Reprod. 2007, 22, 224–229. [Google Scholar] [CrossRef]
- Philibert, P.; Biason-Lauber, A.; Rouzier, R.; Pienkowski, C.; Paris, F.; Konrad, D.; Schoenle, E.; Sultan, C. Identification and Functional Analysis of a New WNT4 Gene Mutation among 28 Adolescent Girls with Primary Amenorrhea and Muüllerian Duct Abnormalities: A French Collaborative Study. J. Clin. Endocrinol. Metab. 2008, 93, 895–900. [Google Scholar] [CrossRef]
- Ravel, C.; Lorenço, D.; Dessolle, L.; Mandelbaum, J.; McElreavey, K.; Darai, E.; Siffroi, J.P. Mutational analysis of the WNT gene family in women with May-er-Rokitansky-Kuster-Hauser syndrome. Fertil Steril 2009, 91 (Suppl. S4), 1604–1607. [Google Scholar] [CrossRef]
- Mansouri, A.; Chowdhury, K.; Gruss, P. Follicular cells of the thyroid gland require Pax8 gene function. Nat. Genet. 1998, 19, 87–90. [Google Scholar] [CrossRef]
- Sauter, C.N.; McDermid, R.L.; Weinberg, A.L.; Greco, T.L.; Xu, X.; Murdoch, F.E.; Fritsch, M.K. Differentiation of murine embryonic stem cells in-duces progesterone receptor gene expression. Exp. Cell Res. 2005, 311, 251–264. [Google Scholar] [CrossRef]
- Lappin, T.R.; Grier, D.G.; Thompson, A.; Halliday, H.L. HOX genes: Seductive science, mysterious mechanisms. Ulster Med. J. 2006, 75, 23–31, Erratum in Ulster Med. J. 2006, 75, 135. [Google Scholar] [PubMed]
- Taylor, H.S. Endocrine disruptors affect developmental programming of HOX gene expression. Fertil Steril 2008, 89, e57–e58. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Aubin, J.; Lemieux, M.; Tremblay, M.; Behringer, R.R.; Jeannotte, L. Transcriptional interferences at the Hoxa4/Hoxa5 locus: Importance of correct Hoxa5 expression for the proper specification of the axial skeleton. Dev Dyn. 1998, 212, 141–156. [Google Scholar] [CrossRef]
- Yi, C.H.; Terrett, J.A.; Li, Q.Y.; Ellington, K.; Packham, E.A.; Armstrong-Buisseret, L.; McClure, P.; Slingsby, T.; Brook, J.D. Identification, mapping, and phylogenomic analysis of four new human members of the T box gene family: EOMES, TBX6, TBX18, and TBX19. Genomics 1999, 55, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Coffinier, C.; Barra, J.; Babinet, C.; Yaniv, M. Expression of the vHNF1/HNF1beta homeoprotein gene during mouse organ-ogenesis. Mech. Dev. 1999, 89, 211–213. [Google Scholar] [CrossRef] [PubMed]
- Lokmane, L.; Heliot, C.; Garcia-Villalba, P.; Fabre, M.; Cereghini, S. vHNF1 functions in distinct regulatory circuits to con-trol ureteric bud branching and early nephrogenesis. Development 2010, 137, 347–357. [Google Scholar] [CrossRef]
- Anant, M.; Raj, N.; Yadav, N.; Prasad, A.; Kumar, S.; Saxena, A.K. Two Distinctively Rare Syndromes in a Case of Primary Amenorrhea: 18p Deletion and Mayer–Rokitansky–Kuster–Hauser Syndromes. J. Pediatr. Genet. 2020, 9, 193–197. [Google Scholar] [CrossRef] [PubMed]
- De Tomasi, L.; David, P.; Humbert, C.; Silbermann, F.; Arrondel, C.; Tores, F.; Fouquet, S.; Desgrange, A.; Niel, O.; Bole-Feysot, C.; et al. Mutations in GREB1L Cause Bilateral Kidney Agenesis in Humans and Mice. Am. J. Hum. Genet. 2017, 101, 803–814. [Google Scholar] [CrossRef]
- Mason, H.R.; Lake, A.C.; Wubben, J.E.; Nowak, R.A.; Castellot, J.J., Jr. The growth arrest-specific gene CCN5 is deficient in human leiomyomas and inhibits the proliferation and motility of cultured human uterine smooth muscle cells. Mol. Hum. Reprod. 2004, 10, 181–187. [Google Scholar] [CrossRef]
- Chen, G.; Shinka, T.; Kinoshita, K.; Yan, H.T.; Iwamoto, T.; Nakahori, Y. Roles of estrogen receptor alpha (ER alpha) in the regulation of the human Müllerian inhibitory substance (MIS) promoter. J. Med. Investig. 2003, 50, 192–198. [Google Scholar]
- Dell’Edera, D.; Allegretti, A.; Ventura, M.; Mercuri, L.; Mitidieri, A.; Cuscianna, G.; Epifania, A.A.; Morizio, E.; Alfonsi, M.; Guanciali-Franchi, P. Mayer-Rokitansky-Küster-Hauser syndrome with 22q11.21 microduplication: A case report. J. Med. Case Rep. 2021, 15, 208. [Google Scholar] [CrossRef] [PubMed]
- Wottgen, M.; Brucker, S.; Renner, S.P.; Strissel, P.L.; Strick, R.; Kellermann, A.; Wallwiener, D.; Beckmann, M.W.; Oppelt, P. Higher incidence of linked malformations in siblings of May-er-Rokitansky-Küster-Hauser-syndrome patients. Hum. Reprod. 2008, 23, 1226–1231. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shawlot, W.; Behringer, R.R. Requirement for LIml in head-organizer function. Nature 1995, 374, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Aydos, S.; Tükün, A.; Bökesoy, I. Gonadal dysgenesis and the Mayer-Rokitansky-Kuster-Hauser syndrome in a girl with 46,X,del(X)(pter-->q22:). Arch. Gynecol. Obstet. 2003, 267, 173–174. [Google Scholar] [CrossRef] [PubMed]
- Kucheria, K.; Taneja, N.; Kinra, G. Autosomal translocation of chromosomes 12q & 14q in mullerian duct failure. Indian J. Med. Res. 1988, 87, 290–292. [Google Scholar] [PubMed]
- Amesse, L.; Yen, F.F.; Weisskopf, B.; Hertweck, S.P. Vaginal uterine agenesis associated with amastia in a phenotypic female with a de novo 46,XX,t(8;13)(q22.1;q32.1) translocation. Clin. Genet. 1999, 55, 493–495. [Google Scholar]
- ACTR3B. Available online: https://www.proteinatlas.org/ENSG00000133627-ACTR3B/tissue (accessed on 15 February 2023).
- Leacock, S.W. Available online: https://bio.libretexts.org/@go/page/102481?pdf (accessed on 4 October 2023).
- Dhar, G.A.; Saha, S.; Mitra, P.; Nag Chaudhuri, R. DNA methylation and regulation of gene expression: Guardian of our health. Nucleus 2021, 64, 259–270. [Google Scholar] [CrossRef]
- Sandbacka, M.; Bruce, S.; Halttunen, M.; Puhakka, M.; Lahermo, P.; Hannula-Jouppi, K.; Lipsanen-Nyman, M.; Kere, J.; Ait-tomäki, K.; Laivuori, H. Methylation of H19 and its imprinted control region (H19 ICR1) in Müllerian aplasia. Fertil. Steril. 2011, 95, 2703–2706. [Google Scholar] [CrossRef]
- Eggermann, T.; Ledig, S.; Begemann, M.; Elbracht, M.; Kurth, I.; Wieacker, P. Search for altered imprinting marks in Mayer–Rokitansky–Küster–Hauser patients. Mol. Genet. Genom. Med. 2018, 6, 1225–1228. [Google Scholar] [CrossRef]
- Saso, S.; Clarke, A.; Bracewell-Milnes, T.; Saso, A.; Al-Memar, M.; Thum, M.-Y.; Yazbek, J.; Del Priore, G.; Hardiman, P.; Ghaem-Maghami, S.; et al. Psychological issues associated with absolute uterine factor infertility and attitudes of patients toward uterine transplantation. Prog Transpl. 2016, 26, 28–39. [Google Scholar] [CrossRef]
- Herlin, M.K.; Petersen, M.B.; Brännström, M. Mayer-Rokitansky-Küster-Hauser (MRKH) syndrome: A comprehensive update. Orphanet J. Rare Dis. 2020, 15, 214. [Google Scholar] [CrossRef] [PubMed]
- Hodson, N.; Townley, L.; Earp, B.D. Removing harmful options: The law and ethics of international commercial surrogacy. Med. Law Rev. 2019, 27, 597–622. [Google Scholar] [CrossRef] [PubMed]
- International Federation of Fertility Societies (IFFS). IFFS surveillance 2016. Glob. Reprod. Health 2016, 1, 1–143. [Google Scholar] [CrossRef]
- White, P.M. Commercialization, altruism, clinical practice: Seeking explanation for similarities and differences in Califor-nian and Canadian gestational surrogacy outcomes. Womens Health Issues 2018, 28, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Saran, J.; Padubidri, J.R. New laws ban commercial surrogacy in India. Med. Leg. J. 2020, 88, 148–150. [Google Scholar] [CrossRef]
- Bröannström, M.; Johannesson, L.; Bokström, H.; Kvarnströom, N.; Mölne, J.; Dahm-Kähler, P.; Enskog, A.; Milenkovic, M.; Ekberg, J.; Diaz-Garcia, C.; et al. Livebirth after uterus transplantation. Lancet 2015, 385, 607–616. [Google Scholar] [CrossRef]
- Jones, B.P.; Saso, S.; Bracewell-Milnes, T.; Thum, M.Y.; Nicopoullos, J.; Diaz-Garcia, C.; Friend, P.; Ghaem-Maghami, S.; Testa, G.; Johannesson, L.; et al. Human uterine transplantation: A review of outcomes from the first 45 cases. BJOG 2019, 126, 1310–1319. [Google Scholar] [CrossRef]
- Johannesson, L.; Enskog, A.; Mölne, J.; Diaz-Garcia, C.; Hanafy, A.; Dahm-Kähler, P.; Tekin, A.; Tryphonopoulos, P.; Morales, P.; Rivas, K.; et al. Preclinical report on allogeneic uterus transplanta-tion in non-human primates. Hum. Reprod. 2013, 28, 189–198. [Google Scholar] [CrossRef]
- 178 Möolne, J.; Broecker, V.; Ekberg, J.; Nilsson, O.; Dahm-Köahler, P.; Brännström, M. Monitoring of human uterus transplantation with cervical biopsies: A provisional scoring system for rejection. Am. J. Transplant. 2017, 17, 1628–1636. [Google Scholar] [CrossRef]
- London, N.J.; Farmery, S.M.; Will, E.J.; Davison, A.M.; Lodge, J.P. Risk of neoplasia in renal transplant patients. Lancet 1995, 346, 403–406. [Google Scholar] [CrossRef]
- Catsanos, R.; Rogers, W.; Lotz, M. The ethics of uterus transplantation. Bioethics 2013, 27, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Arora, K.S.; Blake, V. Uterus transplantation: Ethical and regulatory challenges. J. Med. Ethics. 2014, 40, 396–400. [Google Scholar] [CrossRef] [PubMed]
- Daar, J.; Klipstein, S. Refocusing the ethical choices in womb transplantation. J. Law Biosci. 2016, 3, 383–388. [Google Scholar] [CrossRef]
| Genes and Location in Chromosome | Reference | Method Used | Associations |
|---|---|---|---|
| MEFV and IL-32-16p13.3 CMTM7-3p22.3 CCR4 3p22.3 | [40] | CGH and RT-qPCR | IL32 and MEFV gene mutations associated with Mediterranean fever. MRKHS |
| BAZ2B and SLC4A10-2q24.2 KLHL18-3p21.31 PIK3CD-1p36.22 TNK2-3q29 | [41] | WGS | MRKHS |
| LAMC1-1q25.3 RARA-17q21.2 HOXA10-7p15.2 PAX2-10q24–25 MMP14 and LRP10-14q11.2 | [39] | WES, confirmed by Sanger sequencing | MRKHS |
| IFTP57, HHLA2 and MYH15-3q13.13 PLA2R1-2q23-q24 ITGB6 and RBMS1-2q24.2 | [42] | SNP microarray analysis | MRKHS |
| LRP10-14q11.2 FRAS1-4q21.21 CC2D2A-4p15.32 KIF14-1q32.1 RSPO4-20p13 MKKS-20p12.2 NPHP3-3q22.1 DYNC2H1-11q22.3 SPECC1L-22q11 VWF-12p13.31 | [43] | WES | MRKHS |
| TBC1D1-4p14 KMT2D-12q13.12 HOXD3-2q31-37 DLG5-10q22.3 GLI3-7p14.1 HIRA-22q11.21 GATA3-10p14 LIFR-5p13.1 CLIP1-12q24.31 | [44] | Sanger sequencing | MRKHS |
| PRKX-Xp22.33 HOXC8-12q13.13 | [45] | RT-qPCR | MRKHS, Urinary malformations, skeletal malformations, and/or hearing defects. |
| MUC1-1q22 | [45,46] | Array analysis, RT-qPCR | MRKHS |
| RBM8A-1q21 | [21,47,48,49,50] | Array CGH, MLPA | TAR syndrome (thrombocytopenia, absence of radius) [21,47,48,49] |
| WNT9B-17q21 | [38,39,49,51,52,53,54] | Array CGH, WES | MA, renal abnormalities, and cervicothoracic somite dysplasia [38] |
| TBX6-16p11.2 | [38,39,43,48,49,50,55,56,57,58,59] | Array CGH. WES | Autism spectrum disorders, neurological disorders, unaffected persons [49] |
| ACTR3B-Pseudogenes chromosomes 2, 4, 10, 16, 22 and Y | [53] | WES | MA |
| LHX1-17q12 | [21,47,49,54,55,56,60,61,62,63,64,65,66,67,68] | Array CGH, gene sequencing | MA, Anomalies in the body axis formation [49], diabetes [61], learning disability [61] |
| TCF2/HNF1B-17q12 | [21,47,55,56,60,61,62,63,64,65,66,67,69,70] | Array CGH, DNA sequencing | MA, Renal cysts [49] diabetes [49,61], learning disability [61] |
| TBX1-22q11 | [21,47,48,55,71,72,73,74] | Array CGH, FISH, MLPA | DiGeorge syndrome, heart defect, hypocalcemia, immunodeficiency, typical facial malformations, cognitive and behavioral disorders |
| WNT4-1p36.12 | [39,66,75,76,77] | PCR sequencing, CGH | Hyperandrogenism (Atypical MRKHS), Gonadal dysgenesis |
| GREB1L-18q11.1-q11.2 | [45,53,78,79,80] | WES, CGH | MRKHS type 2 with kidney abnormalities, Twins discordant for MRKHS [45] |
| DOCK4-7q31.1 | [43,63] | MRKHS | |
| ZNF277-7q31.1 | [63] | MRKHS | |
| DACT1-14q23.1 | [81] | MRKHS | |
| DLGH1-3q29 | [82] | Direct sequencing | Unilateral agenesis, pelvic kidney |
| OXTR-3p25.3 | [83] | DNA sequence analysis | MRKHS |
| ESR1-6q25 | [84] | DNA sequence analysis | MRKHS |
| WT1-11p13 | [85] | PCR | |
| GATA4-8p23.1 | [85] | PCR | |
| EMX2-10q26 | [38,86,87] | Sequence analysis | MRKHS, other Müllerian fusion abnormalities |
| SHOX-Pseudoautosomal region Xp22. 3 | [39,88,89] | CGH | MRKHS |
| PBX1-1q23.3 | [90] | MRKHS | |
| PAX8-2q14.1 | [22,38,53,88] | Array CGH. WES | Mutations have been associated with thyroid dysgenesis, thyroid follicular carcinomas, and atypical follicular thyroid adenomas [91], MRKHS |
| Chromosome Number | Mechanism [Reference] | Possible Gene Involved |
|---|---|---|
| 1 | 1q31.1 Duplication (size 0.4 Mb) [55] 1q44 Deletion (size 0.32 Mb) [23,48] 1p36.12 mutations 1p36.21 deletion [23] Mutation p.(Glu226Gly) [77] Mutation c.1026C>T [73] Mutation p.(Arg83Cys) [143] Mutation c.35C>T p.(Leu12Pro) [144] Mutation c.483C>T [145] Mutation c.697G>A p.(Ala233Thr) [75] Mutation g.200583493A>T [43] 1q21 Deletion (size 0.4 Mb to 4.6 Mb) [21,23,48] 1q21 Duplication (size 0.26 Mb to 0.36 Mb) [47,48] | 1. KIF14 [43] 2. WNT4 is responsible for sex determination and affects the invagination of coelomic epithelial cells [121]. WNT4 mutation inhibits repression of ovarian steroid enzymes and causes abnormal expression of 17α hydroxylase and hyperandrogenism [75] 3. OR4M2, ZNF816 and PDE11A [23] |
| 2 | 2p14 Duplication (size-0.23 Mb) [21] 2p14 Mutation c.1315G>A, p.Ala439Thr [53] 2p23.1 Duplication (size-0.21 Mb) [55] 2p24 Deletion (size-4.6 Mb) [55] 2q11.2 Duplication (size-1.3 Mb) [55] 2q24.2 Duplication [42] 2q13 Deletion (size-0.12 Mb) [21] | 1. The PAX8 gene encodes a homeodomain signaling molecule, strongly expressed in the MD [146]. 2. Duplication at 2q24.2 of proband MRKHS involved PLA2R1, ITGB6 and RBMS1 |
| 3 | 3p21 Duplication 0.10 Mb [60] 3p21 Mutation c.861G>A [145] 3q13 Duplication at 3q13. [42] 3q29 Deletion 0.05 Mb [60] Mutation g.132403615G>A [43] | 1. WNT7A encodes secreted signaling proteins, and is involved in the development of the anterior–posterior axis in the female reproductive tract. Coded proteins are responsible for patterning during embryogenesis. Their role in MA is uncertain. 2. May involve DLGH1, OXTR, NPHP3 |
| 4 | 4q28 Deletion 0.11 Mb [60] 4q32 Deletion 0.34 Mb [60] 4q35.2 Deletion 1.1 Mb [54] Mutation g79204031G>A and g.15542618C>T [43] | 1. FRAS1 (g79204031G>A) [43] 2. CC2D2A (g.15542618C>T) [43] |
| 5 | 5p11 Deletion 0.40 Mb [62] 5q14.3 Deletion 0.40 Mb [62] | |
| 6 | 6p21 Duplication 0.17 Mb [60] 6q25.1 Duplication 0.42 Mb [60] 6q25.2 Duplication 0.44 Mb [60] 6q11.1 Duplication 0.41 Mb [62] | |
| 7 | 7p15.2 Hypomethylation [32] 7p14 Duplication 1.75 Mb [60] 7q31.2 Deletion 1.8 Mb [54] Mutation g111503593C>T [43] | 1. HOXA5 is a transcriptional regulator of p53 and progesterone receptor (PGR). Hypomethylation leads to overexpression, which causes overexpression of PGR [147]. Ectopic HOXA5 expression at the 5′end of the cluster might prevent normal differentiation of the MD or even regression. 2. It most likely involves Abdominal B (AbdB) homeobox genes (HOX-A9, A10, A11, and A13), required for differentiation and segmental patterning of MD [148] 3. HOXA9 is expressed in the region that becomes the oviduct [149]. Ectopic expression of HOXA5 or HOXA9 inhibits MD differentiation [150]. 4. DOCK4 |
| 8 | 8p23.1 Hypomethylation [85] 8p23.1 Activating mutations [32] | GATA binding protein 4 promotes AMH production and regulates sex determination and differentiation [85]. Overproduction of AMH leads to MA. |
| 10 | 10q24 Duplication 0.05 Mb [60] | |
| 11 | 11p11.12 Deletion 0.76 Mb (45) 11p 13 Hypomethylation [85] 11p 13 Activating mutation [32] Mutation g.102985987C>T [43] | 1. WT1 is a regulatory factor important for the transcription of anti-Müllerian hormone (AMH) genes. It promotes AMH expression and regulates sex determination and differentiation [85]. Activating the mutation of the gene for the AMH receptor, or the receptor, causes excessive production of AMH, leading to MRKHS [32] 2. DYNC2H1 |
| 12 | 12q13.13 Duplication [42] 12q23 Duplication 0.16 Mb [60] 12q24 Duplication 0.12 Mb [60] Mutation g.6085324G>A [43] | VWF |
| 13 | 13q21 Deletion 0.41 Mb [54] | |
| 14 | 14q32.33 Deletion 0.46 Mb [62] 14q32.33 mutation g.23345412G>A [43] | LRP10 |
| 15 | 15q21.1 Deletion 0.28 Mb [62] 15q26.3 Deletion 0.54 Mb [54] Deletions at 15q11.2 [23] | |
| 16 | 16p13.3 Increased expression [40] 16q11.2 Duplication 0.20 Mb [55] 16p11.2 Deletion (size 0.55 Mb to 0.6 Mb) [55] Splice site mutation c.622-2A>T (g.30100162 T>A) [56] Mutation c.484G>A(rs56098093) p.Gly162Ser [56] Mutation c.815G>A p.Arg272Gln [56] Mutation c.815G>A [54] | 1. Genes for IL32 and MEFV. 2. TBX6 is involved in paraxial mesoderm formation and somitogenesis in human embryos [151]. Deletion induces MRKHS due to the loss of the transcription factor. |
| 17 | 17q12 Deletion (size 1.2 to 1.9 Mb) [21,42,47,48,55,60,62] 17q12 Missense mutation of LHX1 [21,61] c.790C>G p.(Arg264Gly) c. c.25dup p.(Arg9Lysfs*25) 17q21-22 mutations c.28G>T p.Ala10Ser c.205C>T, p.Arg69Trp [53] c.*158C>T 17q21-22 Five Missense mutations [54] c.472C>G p.(Gln158Glu) c.665G>A p.(Arg222His) c.722G>A p.(Arg24His) c.974G>A p.(Arg325His) c.1029C>A p.(Cys343*) | 1. Involves the LHX1(LIM homeobox protein 1) gene, which is a transcription factor necessary for the formation of the Müllerian duct-derived uterine and vaginal epithelia [116]. 2. HNF1B is [Pit–Oct–Unc homeodomain-containing transcription factor that is frequently expressed in the Müllerian duct during development [152]. It positively regulates the expression of LHX1, PAX2, and WNT9B [153]. 3. Involves mutations in WNT9B, which acts upstream of another Wnt4. It is responsible for the caudal extension of the Müllerian duct and the organization of the urogenital system [122]. |
| 18 | 18q23 Duplication 0.20 Mb [62] 18p Deletion [154] 18q11.1-q11.2 mutation c.4665T>A, p.Tyr1555 | GREB1L is a target gene in the retinoic acid signaling pathway, which is highly expressed in the developing fetal human kidney and involved in the early metanephros and genital development [155]. |
| 19 | 19q13.31 deletion [23] | OR2T2 [23] |
| 20 | 20q13.12 Deletion Mutations g.941074G>A and g.10393439C>A [43] | 1. WISP2 is significant in smooth muscle cell proliferation and migration, and is induced by estrogen in the uterus [156]. Estrogen regulates AMH expression levels [157], and overexposure to estrogen during development activates AMH promotors [32]. 2. RSPO4 3. MKKS |
| 22 | 22q11 Deletion (size 0.39 Mb to 2.6 Mb) [55,61,71,73] Duplication (0.6 Mb–3.5 Mb) [47,61,158] Mutations g.24720297G>A and g.24718408G>A [43] | 1. TBX1 2. SPECC1L |
| X | Xp11.1 Deletion 0.12 Mb [60] Xp11.3 Duplication 0.24 Mb [60] Xp22 Duplication (0.07 to 0.36) [21,45,47,48,159] Xq21.31 Deletion 1 Mb [160] Xq deletion [161] Xq22.3 Duplication 0.09 Mb [60] Xq22.3 Microdeletion at Xp22.33 [42] | 1. May involve the PRKX gene, encoding for a serine/threonine kinase implicated in renal epithelium morphogenesis [45]. 2. May involve the SHOX gene, which encodes a transcription factor responsible for skeletal growth. The exact mechanism is unknown. |
| 8,13 | t(8;13) (q12;q14) translocation | Translocation causes MRKHSS with or without renal hypoplasia [162]. |
| t(8;13)(q22.1;q32.1) translocation | Translocation causes MRKHS with limb, breast, and urinary functional defects [163] | |
| 3,16 | t(3;16) (p22.3;p13.3) translocation | Translocation causes MRKHS [40,98] |
| 7,14 | t(7;14)(q32;q32) translocation | Translocation seen in MRKHS [98] |
| 2, 4, 10, 16, 22 and Y | c.1066G>A, p.Gly356Arg [53] | ACTR3B encodes a member of the actin-related proteins and plays a role in the organization of the actin cytoskeleton [164]. ACTR3B can have pseudogenes in more than one chromosome. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dube, R.; Kar, S.S.; Jhancy, M.; George, B.T. Molecular Basis of Müllerian Agenesis Causing Congenital Uterine Factor Infertility—A Systematic Review. Int. J. Mol. Sci. 2024, 25, 120. https://doi.org/10.3390/ijms25010120
Dube R, Kar SS, Jhancy M, George BT. Molecular Basis of Müllerian Agenesis Causing Congenital Uterine Factor Infertility—A Systematic Review. International Journal of Molecular Sciences. 2024; 25(1):120. https://doi.org/10.3390/ijms25010120
Chicago/Turabian StyleDube, Rajani, Subhranshu Sekhar Kar, Malay Jhancy, and Biji Thomas George. 2024. "Molecular Basis of Müllerian Agenesis Causing Congenital Uterine Factor Infertility—A Systematic Review" International Journal of Molecular Sciences 25, no. 1: 120. https://doi.org/10.3390/ijms25010120
APA StyleDube, R., Kar, S. S., Jhancy, M., & George, B. T. (2024). Molecular Basis of Müllerian Agenesis Causing Congenital Uterine Factor Infertility—A Systematic Review. International Journal of Molecular Sciences, 25(1), 120. https://doi.org/10.3390/ijms25010120










