Abstract
A number of muscular disorders are hallmarked by the aggregation of misfolded proteins within muscle fibers. A specialized form of macroautophagy, termed aggrephagy, is designated to remove and degrade protein aggregates. This review aims to summarize what has been studied so far about the direct involvement of aggrephagy and the activation of the key players, among others, p62, NBR1, Alfy, Tollip, Optineurin, TAX1BP1 and CCT2 in muscular diseases. In the first part of the review, we describe the aggrephagy pathway with the involved proteins; then, we illustrate the muscular disorder histologically characterized by protein aggregates, highlighting the role of aggrephagy pathway abnormalities in these muscular disorders.
1. Introduction
The accumulation of misfolded proteins is a common pathological characteristic of degenerative diseases of the central nervous system, but it is also present in muscular disorders where the inhibition of autophagy leads to protein aggregates and/or vacuoles formation, causing muscle fiber degeneration and myopathy [1,2,3].
Autophagic vacuolar myopathies (AVMs) and protein aggregate myopathies (PAMs) represent a series of clinically heterogeneous myopathies characterized by lysosomal or extralysosomal accumulation of proteins or other substances, with varied ages of onset, progressive disease course and variable degree of severity [4,5,6].
AVMs, characterized by the presence of autophagic vacuoles, can be classified into three groups based on specific pathomechanical and morphological findings.
Specifically, the first group is characterized by the deficiency of the lysosomal α-1,4-glucosidase enzyme, causing Pompe disease, and the second by abnormal and functionally impaired lysosomes as in X-linked myopathy with excessive autophagy (XMEA) and Danon disease. The third group is defined by secondary lysosomal dysfunction leading to rimmed vacuoles, popcorn-like clear membrane-bound organelles with a densely blue rim by hematoxylin and eosin staining. This group comprises inclusion body myopathy with Paget disease and frontotemporal dementia (IBMPFD), hereditary and sporadic inclusion body myopathy (hIBM and sIBM) and oculopharyngeal muscular dystrophy (OPMD), among others [4,7]. Recently, a new AVM caused by mutations in the PLIN4 gene has been described [8].
PAMs are mainly identified by the presence of extralysosomal protein aggregates, and it has been hypothesized that abnormal protein synthesis, impaired extralysosomal degradation system or the integration of proteins in the intracellular structured constituents could contribute to the development of protein aggregation [5,6,9,10,11]. One type of PAMs could be classified as “catabolic”, where the aggregation is likely due to extralysosomal protein degradation failure, as observed in but not limited to myofibrillar myopathies (MFMs), cores diseases, sIBM and hIBM, reducing body myopathy and oculopharyngeal muscular dystrophy. In another subgroup, defined as “anabolic PAM” marked by actin filament and granular myosin aggregates, the accumulation is ascribable to synthetic/developmental defects of actin and myosin filaments, notably associated with onset often in early childhood. Nemaline myopathies and myosinopathies belong to this group [5,6,9,12,13].
Both AVMs and PAMs show some overlap due to similarities among the pathological mechanisms.
To date, no treatment is available for any of these disorders, except for the enzyme replacement therapy in Pompe disease [14,15].
This review focuses on aggrephagy, a specialized form of autophagy responsible for the removal of protein aggregates, a mechanism that may be relevant in specific muscle diseases and potentially representing a therapeutic target. Despite this, so far, only a few studies investigating aggrephagy in muscle diseases have been reported in the literature.
2. Protein Quality Control (PQC) System: Protein Misfolding and Aggrephagy
2.1. Protein Folding and Misfolding and Molecular Helpers
The folding of newly synthesized proteins is a highly demanding process, especially in the crowded cellular environment where protein content might reach concentrations of 300–400 g/L [16]. Folding is ruled by kinetic (the vectorial nature of translation) and thermodynamic (the need for energy minimization) factors. It can occur co-translationally, during ribosomal-assisted synthesis either on free ribosomes or on ribosomes that are bound to the sarcoplasmic/endoplasmic reticulum (SR/ER) [17], as well as post-translationally in the cytoplasm or in confined organelles (mitochondria and endoplasmic reticulum) [18].
The newly synthesized polypeptides, while reaching their native structures, fluctuate between unfolded and folded states, depicting a funnel-shaped energy landscape [19]. Moreover, while the energy landscape is smoother for small proteins (<100 amino acids in length) that can fold very rapidly within microseconds, larger proteins (>100 amino acids), which account for ~90% of the proteome in a cell, have a rougher path because of a higher tendency to collapse in the aqueous solvent [16,19]. Likewise, proteins might be intrinsically disordered within the cytoplasm, reaching a tri-dimensional state only upon binding with membrane surfaces or macromolecules [16,20,21]. In addition, this complex situation is further complicated by cellular unpredictable events such as genetic mutations, errors in transcription or translation and chemical or temperature stresses in the cellular environment.
It is no surprise then that proteostasis (the maintenance of protein homeostasis) involves numerous pathways composed of hundreds of proteins such as specialized chaperones and folding enzymes, facilitating folding and refolding, and degradation components like the ubiquitin-proteasome system (UPS) and autophagy for the removal of the permanently misfolded and aggregated proteins [16,22,23,24].
In human cells, over 300 chaperones and co-chaperones [25] are known to assist in de-novo folding, refolding of denatured proteins, assembly of protein complexes and protein degradation [26].
Heat shock proteins are a special family of chaperones that are upregulated upon thermal stress or other proteotoxic stresses that elevate the concentrations of aggregation-prone intermediates. They are classified based on their molecular weight (HSP40, HSP60, HSP70, HSP90, HSP100 and the small HSPs) [27].
The HSP70 family includes 47 proteins encoded by 17 genes in the human genome that can be stress-inducible, such as HSP70s, or constitutively expressed as the heat shock cognate HSC70 [28].
All HSP70 members share common structural features, having a 45 kDa N-terminal ATPase nucleotide binding domain (NTD) and a 25 kDa C-terminal substrate binding domain (SBD) that is able to recognize co-chaperones through its conserved motif EEVD [29,30,31]. The mechanism of action of HSP70 chaperones is based on their ability to cycle through conformational changes in an ATP-dependent manner under the regulation of the chaperones of the HSP40 (DnaJ) family and nucleotide exchange factors (NEF). Briefly, HSP40 chaperones recognize and deliver non-native proteins that expose hydrophobic regions to HSP70s, temporarily blocking their aggregation propensity. When ATP is hydrolyzed to ADP, the peptide is stably bound. The return of the ATP-bound state will allow the release of the substrate [30,31,32,33,34].
A member of the HSP70 family, well-conserved among eukaryotes, is BiP, which, along with lectin chaperones, is the master of ER quality control (ERQC). BiP recognizes nascent polypeptide chains of both glycosylated and non-glycosylated proteins and can bind to unfolded substrates in an ATP-dependent manner. This process of binding and release is assisted by co-factors such as the ER-localized DnaJ-like proteins (ERdjs) and two NEF named GRP170 and Sil1 [35,36].
HSP40 proteins, also known as JDPs (J domain proteins), are encoded by 50 genes in the human genome [27]. They are all characterized by the presence of a ~70 amino acid J domain highly conserved and have been divided into three classes, A, B and C (previously named class I, II and III). JDPs from class A and B also share a glycine-phenilalanine-rich region (G/F) and two sandwich domains at the C-terminal, while only class A present a Zn-binding domain. The class C proteins have little homology with one another, only sharing the J domain, which could be located at any place in the protein sequence [32,33,34].
HSP90/HSC family proteins are encoded by six genes in the human genomes and function as a homodimer. Each HSP90 monomer is composed of three highly conserved domains, an N-terminal domain (NTD) implicated in the binding of ATP, a middle domain (MD) necessary for the ATP hydrolysis and a C-terminal domain (CTD) with a MEEVD motif for co-chaperone coupling. Upon binding and hydrolysis of ATP, HSP90 cycle through different conformational states with different affinity for HSP70, the adaptor protein Hop/Sti1 and the protein client to fold [37,38]. About 10% of human proteins are HSP90 clients, depending on them for their maturation [30].
Chaperones of the HSP110 family are encoded in the human genome by four genes and are highly homologous to the HSP70 family. Three of these proteins (HSPH1-3 according to a revised nomenclature [27]) are both nuclear and cytosolic, while the fourth HSPH4, also known as GRP170 or HYOU1, is located in the SR/ER where it covers a fundamental role in the ERQC, also acting as NEF for BiP [30,39].
To date, 11 small HSPs chaperones have been found located in the cytosol and nucleus of mammalian cells and are highly upregulated under cellular stress. These small HSPs are characterized by a conserved crystallin domain, with N- and C-terminus that can vary and might oligomerize with different members of the family, thus increasing their chaperone specificity [27,40].
Additionally, two types of HSP60 proteins, also known as chaperonins, have been found in different subcellular locations. Group I members (HSP60 in eukaryotes, GroEL in prokaryotes) have a double-layered, 7-subunit ring configuration and are located in the eukaryotic mitochondria or chloroplasts. They can bind cofactor HSP10 (GroES in prokaryotes) in an ATP-dependent manner. Group II members are composed of double-layered, 8,9-subunit rings and are located in the cytosol (also known as TRiC or CCT). These large double-ring complexes can encapsulate their client molecule one at a time in the central cavity for folding, lowering the risk of aggregation [30,41,42].
2.2. Alternative PQC: Autophagy and Aggrephagy
If the misfolded state of proteins becomes irreversible, then the ubiquitin-proteasome and the autophagy systems intervene for their degradation.
The initial step for both mechanisms is the ubiquitination of the cargoes that have to be recognized and targeted for degradation. Ubiquitin is a small regulatory protein (76 amino acids) [43] that was first described in 1975 by Goldstein and colleagues [44]. It is part of a family of proteins with diverse amino acid sequences but sharing similar folding structures [45,46]. Ubiquitin is found in all eukaryotes but not in prokaryotes, and its sequence is remarkably conserved between humans and yeast, with a difference of only three amino acids.
The cargoes’ ubiquitination occurs at lysine residues via an isopeptide bound with the C-terminal Gly of ubiquitin. Furthermore, ubiquitin itself has seven lysine residues (K6, K11, K27, K29, K33, K48 and K63) and an N-terminal methionine (M1) which in turn can be ubiquitinated. Moreover, ubiquitin can undergo ubiquitin-like modifications such as SUMOylation and NEDDylation, thus generating a wide range of signaling codes. Conjugation of ubiquitin occurs in a three-step cascade sequentially involving three classes of enzymes: the ubiquitin-activating enzyme E1, the ubiquitin-conjugating enzyme E2 and a substrate-specific ubiquitin-protein ligase E3. Another group of enzymes called deubiquitinating enzymes (DUBs) is responsible for the removal of ubiquitins from their substrates.
For a more complete overview of ubiquitin, its structure and modifications, we suggest some comprehensive reviews to look at in citations [46,47,48,49,50,51,52,53,54,55,56].
Notably, ubiquitination at Lys11 and Lys48 residues is usually a signal for the elimination of the targeted cargoes through the proteasome, a large multicomplex of 2.5 MDa composed of the 20S catalytic core particle (CP), and one or two 19S regulatory particles (RPs). For an in-depth description and characterization of the 26S proteasome, we refer to some key papers [47,51,57,58,59,60,61,62].
Before proteasome degradation could proceed, misfolded proteins should be unfolded and fit through the narrow pore of the 20S subunit. If this cannot occur, ubiquitinated proteins are targeted for autophagy degradation. In particular, the ubiquitination of Lys68 seems to be preferentially a signal for degradation through this pathway [63].
It is nowadays clear that UPS and autophagy are interconnected, sharing some of their components, and the perturbation of one system affects the other. For instance, proteasome inhibition induces activation of autophagy as a compensatory system in cell lines [64,65] as well as in in vivo experiments in genetically modified Drosophila melanogaster [66].
The term autophagy was first conceived by biochemist Christian de Duve, a Nobel Prize laureate in Physiology or Medicine of 1974, for his discovery of lysosome. In 2016, another Nobel Prize in Physiology or Medicine was awarded to the cell biologist Yoshinori Ohsumi for his in-depth studies on the autophagy machinery and the discovery of Autophagy-related genes (Atg) [67]. In mammalian cells, three types of autophagy are characterized, chaperone-mediated autophagy (CMA), microautophagy and macroautophagy. We will particularly focus on macroautophagy (henceforth autophagy) and its specialized form of selective removal and degradation of protein aggregates termed aggrephagy by Seglen and coworkers in 2007 [68].
Autophagy is a tightly regulated process necessary for the maintenance of proper cellular homeostasis by bulk degradation of cytoplasmic content during nutrient starvation or the removal of specific elements such as damaged organelles, lipids, pathogens and, more importantly, protein aggregates.
Activation of autophagy involves the formation of an isolation membrane, the phagophore, which expands into a double-membrane vesicle, the autophagosome, to engulf what needs to be recycled. The fusion with a lysosome generates an autolysosome in which the lytic enzymes can carry out degradation [69]. In brief, initiation of autophagy is achieved by the ULK complex comprised of, in mammals, ULK1or ULK2 as well as the mammalian homologs ATG13, ATG101 and RB1 inducible coiled-coil 1 (RB1CC1, also known as FIP200). The following nucleation step is controlled by the ATG14-containing class III phosphatidylinositol 3-kinase (PtdIns3K) complex, consisting of PIK3C3/VPS34, PIK3R4/VPS15, BECN1, the nuclear receptor binding factor 2 (NRBF2) and the membrane curvature sensor ATG14. The phagophore expansion step then can proceed with regulation from the ATG12-ATG5-ATG16L1 complex and the Atg8/LC3 complex composed of different subfamilies and their isoforms (LC3 family including LC3A, LC3B, LC3B2 and LC3C and GABARAP family including GABARAP, GABARAPL1 and GABARAPL2). Closure of the membrane and fusion with a lysosome will form a mature autolysosome [67,70,71,72,73].
Molecular regulation of autophagy involves a complex yet beautifully harmonized cascade of events that is initiated by inhibition of the mechanistic target or rapamycin mTOR, which is composed of two complexes, mTOR complex1 and mTOR complex2 (mTORC1 and mTORC2). Likewise, the energy-sensing AMP-activated protein kinase (AMPK) is also involved in the initiation of autophagy by the inhibition of mTOR and activation of ULK1 through its phosphorylation. Many detailed studies illustrate in depth these mechanisms [70,74,75,76,77,78,79,80,81].
Notably, autophagy in skeletal muscle is regulated by transcription factors such as Runx1 and the members of the Forkhead Box O family (FoxO1, 3 and 4), which control the expression of autophagy genes like LC3 and VPS34, among others. FoxO transcription factors are, in turn, downstream targets of the kinase Akt [11,82,83]. Very recently, a new FoxO-dependent regulator of autophagy in skeletal muscle has been identified by Leduc-Gaudet and colleagues. This regulator, named MYTHO (Macroautophagy and YouTH Optimizer) by the authors, was found to co-localize with LC3 and, in part, with LAMP2. Moreover, with knockdown experiments, MYTHO proved to be essential for autophagy, leading, upon long-term silencing, to severe myopathic features with inflammation, degeneration/regeneration, mitochondria swelling and tubular aggregates. Furthermore, the authors showed that MYTHO’s expression is also downregulated in skeletal muscle biopsies from dystrophy type 1 (DM1) patients, which are characterized by similar myopathic histopathological features [84].
It is well understood that autophagy is not solely a non-selective invagination of cytoplasmic portions induced upon starvation but also could be a highly specialized mechanism able to remove many different selected elements within the cell, relying on cargo receptors. These processes have been named based on the element that is targeted for degradation, and they include, among others, the elimination of damaged or old mitochondria (mitophagy), nuclei (nucleophagy), ribosomes (ribophagy), lipids (lipophagy), lysosomes (lysophagy) and also protein aggregates (aggrephagy) [85,86,87,88,89,90].
When misfolding becomes irreversible, proteins might form oligomers and interact with other unfolded proteins, thus producing larger insoluble protein aggregates within the cytoplasm. These aggregates can organize into histologically diverse structures such as spheres, tangles or threads, either amorphous or structured, similar to amyloids [91,92]. These aggregates, named aggresomes by Kopito and colleagues [93], have been found to localize in a region near the nuclei, the microtubule-organizing center (MTOC), surrounded by a structure resembling a cage constituted by the intermediate filament vimentin [92,93,94]. Protein aggregates are delivered to the aggresome by the motor protein dynein that, facilitated by the mediation of the histone deacetylase 6 (HDAC6), moves the polyubiquitinated proteins along the microtubules [95,96].
2.3. Key Proteins in Aggrephagy
Since the first descriptions of the aggresomes and aggrephagy pathway as a specialized form of autophagy, the interest of many researchers has focused on the identification of the specific players that could possibly orchestrate this fine mechanism (Figure 1). Structures, functions and interactors described in this paragraph are summarized in Table 1.
2.3.1. HDAC6
One of the first studied proteins involved in the removal of aggregates is the cytoplasmic histone deacetylase 6 (HDAC6), a microtubule-associate deacetylase [97] containing a ubiquitin-binding domain (BUZ) and two catalytic domains (DD1 and DD2). Kawaguchi and colleagues [95] showed that, in an in vitro system, HDAC6 interacts with polyubiquitin-positive aggresomes via its BUZ domain. It can also associate with the motor protein dynein through its dynein motor binding domain (DMB). Moreover, it was demonstrated that HDAC6 mediates the recruitment of lysosomes to the MTOC, thus allowing the removal of aggregated Huntingtin in vitro [98]. It was also discovered that, in Drosophila melanogaster, HDAC6 is crucial for the compensatory role of autophagy when the UPS system is impaired [66]. HDAC6 also mediates the aggresomes removal, facilitating autophagosomes and lysosomes fusion by activating cortactin (through its deacetylation), an actin-remodeling factor that stimulates the local assembly of an F-actin network [99]. HDAC6 has been found to accumulate in Lewy bodies of brain sections of Parkinson’s disease (PD) patients, along with α-synuclein and ubiquitin [95].
2.3.2. P62/SQSTM1
Sequestosome 1, also known as p62, is the first selective autophagy receptor that was described to be capable of binding polyubiquitinated proteins [100] as well as binding directly to LC3 and other human homologues of Atg8 [101]. Mutations in the p62 gene have been linked to diverse human diseases such as amyotrophic lateral sclerosis (ALS), frontotemporal dementia (FD), neurodegeneration with ataxia and also distal myopathy with rimmed vacuoles and Paget disease of the bone [102,103,104,105,106]. P62 is ubiquitously expressed and is able to shuttle between the nucleus and cytoplasm [107] and is characterized by specific domains that shape its functions. It contains a Phox1 and Bem1p (PB1) domain necessary for homo or hetero-oligomerization, thus mediating the formation of protein aggregates, a ZZ-type zinc finger (ZZ) domain, two nuclear localization signal (NLS) sequences and a nuclear export signal (NES) sequence, an LC3-interacting region (LIR) and a C-terminal ubiquitin-associated (UBA) domain. Homodimerization via the UBA domain keeps p62 inactive [108], while phosphorylation of the Ser407 is able to reverse it to an active monomeric form [109]. Thus, through its PB1 domain, p62 is able to bind ubiquitinated proteins, forming long helical filaments [110] that are next targeted for autophagy degradation via the binding to LC3 with its LIR domain [101,111]. Two independent studies [112,113] have demonstrated that in in vitro experiments, p62 is able to phase separate ubiquitinated proteins and that the p62-decorated aggregates have liquid-like features and are able to exchange their components.
2.3.3. NBR1 (Neighbor of BRCA1 Gene 1)
NBR1 is a soluble selective autophagy receptor that was discovered soon after p62 for its ability to bind to LC3 and the presence of a similar structural organization [114,115]. It is evolutionarily conserved and precedes the existence of p62 since most non-metazoans only contain NBR1 but not p62 [116]. Its structure contains a PB1 domain which is able to bind to p62, a ZZ domain, an LIR motif for the interaction with LC3 and a C-terminal UBA domain [114,117]. Moreover, unlike p62, it contains four tryptophan (FW) domains allowing its binding to microtubule-associated protein MAP1B and TAX1BP1 [118], as well as an amphipathic helix (HA) domain that, together with the UBA domain, is necessary for co-localization with LAMP2 [119]. During the removal of aggregates, NBR1 uses its PB1 domain to bind p62 and its UBA domain to enhance the binding to ubiquitinated cargoes. Meanwhile, TAX1BP1 is recruited through its FW domain [113,120]. In the muscle, NBR1 directly interacts with the serine/threonine kinase domain (TK) of the protein titin, a domain that regulates muscle gene expression. Two mutations have been identified in this domain, causing disruption of the interaction between NBR1 and titin and leading to core myopathy, probably the most common form of congenital myopathies in childhood [121,122,123,124].
2.3.4. WDFY3 (Alfy)
The link between Alfy (autophagy-linked FYVE protein) and protein aggregates’ removal was first demonstrated in 2004 [125]. Alfy is a 400 kDa protein, evolutionarily conserved and expressed in all tissues. It contains five WD40 repeats and a PH-BEACH domain assemblage, and a C terminal phosphatidylinositol-3-phosphate (PI3P)-binding FYVE domain; therefore, it is also known as WDFY3. Alfy was found to co-localize with the p62-positive aggregates, and, in vitro, this interaction was found to be mediated by the region comprising amino acids 170–206. It is required for aggrephagy but not for starvation-induced bulk autophagy [90,126,127,128]. Due to its large size, it is considered a scaffold protein facilitating the assembly of p62 bodies and their incorporation in autophagosomes. It was demonstrated that overexpression of Alfy decreases the level of polyglutamine aggregates in a primary neuronal HD model and in an in vivo model of eye polyglutamine disease in Drosophila [126], thus highlighting its potential therapeutic value.
2.3.5. TOLLIP (TOLL-Interacting Protein)
Tollip is the mammalian homolog of the yeast Cue5. It is a member of the CUET family of proteins and therefore is characterized by a ubiquitin-binding CUE domain and an LC3-interacting region (LIR). It was demonstrated that Cue5 and Tollip are able to promote the clearance of the aggregate of human huntingtin mutant and that Tollip is more efficient than p62 in the clearance of aggregates in HeLa cells [129].
2.3.6. Optineurin
Optineurin is a cytosolic protein ubiquitously expressed and with numerous cellular functions such as vesicle trafficking, cell division control, and autophagy. It is linked to numerous diseases, like amyotrophic lateral sclerosis and primary open angle glaucoma (POAG) [130,131,132,133,134]. Optineurin is constituted by a NEMO-like domain, an LC3-interacting (LIR) domain, an LZ domain, a ubiquitin-binding domain (UBD), coil-coiled (CC) domains, and a zinc-finger-like (ZnF) domain. It was first demonstrated [135] that lack of optineurin in HeLa cells increases aggregation of SOD1 G93C and huntingtin mutant, revealing its involvement in the aggregates’ clearance. Moreover, Shen and colleagues demonstrated that [136], in transgenic mice, the co-localization of optineurin with huntingtin mutant inclusion bodies requires its UBD domain. The overexpression of optineurin caused a reduction in inclusion bodies through polyubiquitin-mediated autophagy, while mutation of the UBD domain caused inclusion body accumulation.
2.3.7. TAX1BP1 (Tax1-Binding Protein 1)
TAX1BP1 involvement in the clearance of aggregates was first discovered in 2012 by Newman and colleagues [137], and it was more recently demonstrated that loss of TAX1BP1 induces the formation of proteotoxic aggregates in the brain [138]. TAX1BP1 has an N-terminal SKIP carboxyl homology (SKICH) domain, a central oligomerization domain containing three coiled-coil (CC) regions, and a C-terminal ubiquitin binding domain (UBD) containing two zinc fingers [139]. Its direct interaction with NBR1 allows its recruitment to the aggregates and, in turn, TAX1BP1 is able to recruit the autophagy factor FIP200 (an ULK-interacting protein also known as RB1-inducible coiled-coil protein 1, RBCC1) via the Claw domain, allowing autophagosome formation [120].
2.3.8. CCT2 (Chaperonin Containing TCP1 Subunit 2)
Very recently, Ma and colleagues [140] identified a new function of the chaperonin subunit CCT2 in the elimination of aggregates. The chaperonin TCP-1 ring complex (TRiC) is constituted by eight subunits (CCT1-8) and each one contains an equatorial domain responsible for the binding of ATP as well as an apical domain responsible for substrate binding. In this work, the authors highlighted a newly discovered function of the CCT2 subunit as an aggrephagy receptor, facilitating protein aggregates’ engulfment into the autophagosome and their subsequent clearance. This function is independent from other aggrephagy proteins such as p62, NBR1 and TAX1BP1 and is mediated by the interaction with the ATG8 proteins. The mechanism of action and the ability to switch between functions, from protein-folding to autophagy, is conveyed by its monomeric form, which exposes the ATG8-interaction motif. The authors speculate that under physiological conditions, CCT2 work as a subunit of TRiC to carry out protein folding function. However, in the presence of excessive aggregation-prone proteins, CCT2 becomes monomeric to promote aggregate removal through autophagy. CCT2 does not bind poly-ubiquitinated proteins.
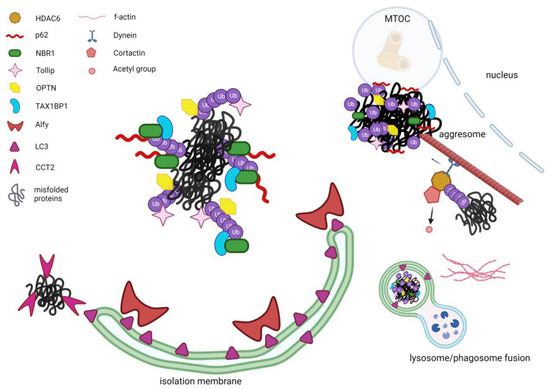
Figure 1.
Schematic representation of the known aggrephagy modulators. Misfolded ubiquitinated proteins are recognized by the selective aggrephagy receptors such as p62 and NBR1 which jointly associate through their UBA domains to the ubiquitins that tag misfolded proteins. Moreover, via their LIR domains they interact with LC3. Tollip, optineurin and TAX1BP1 can also recognize ubiquitinated aggregates, with TAX1BP1 being mobilized to the aggregates by NBR1. The large protein Alfy can act as a scaffold, stabilizing the forming phagophore and interacting with LC3 and p62. HDAC6 is also facilitating the selective aggregates’ removal by bridging the ubiquitinated proteins to the microtubule motor dynein, which can guide the aggregates toward the microtubule organization centre (MTOC) where aggresomes are localized. Furthermore, HDAC6 recruits and de-acetylates cortactin which, once activated, can assemble an F-actin network surrounding aggregates and lysosome, thus facilitating the fusion between lysosome and utophagosome. Likewise, CCT2 in its monomeric form can recognize aggregated proteins independent of ubiquitin-binding receptors and direct them towards the nascent autophagosome through interaction with LC3 via its VLIR motif. (Figure created with Biorender.com, Toronto, ON, Canada).

Table 1.
Summary of the aggrephagy key players and their different cellular functions.
Table 1.
Summary of the aggrephagy key players and their different cellular functions.
| Protein | Function/Structure | Main Functions in Aggrephagy | Interactors |
|---|---|---|---|
| HDAC6 | Microtubule-associate deacetylase. BUZ domain; DD1 and DD2 catalytic domains. |
|
|
| P62/SQSTM1 | Receptor for polyubiquitinated proteins. PB1 and ZZ domains; Two NLS and one NES sequences; LIR motif; C-terminal UBA domain. |
|
|
| NBR1 | Soluble selective autophagy receptor. PB1, ZZ and C-terminal UBA domains; LIR motif; FW and HA domains. |
|
|
| WDFY3 (ALFY) | Scaffold protein, evolutionarily conserved and expressed in all tissues. Five WD40 repeats PH-BEACH domain assemblage; C terminal PI3P-binding FYVE domain. |
|
|
| TOLLIP | Member of the CUET family of protein. Ubiquitin-binding CUE domain; LIR motif. |
|
|
| OPTINEURIN | Cytosolic protein ubiquitously expressed implies in cellular functions such as vesicle trafficking, cell division control, and autophagy. NEMO-like, LZ, UBD, CC and ZnF domains; LIR motif. |
|
|
| TAX1BP1 | Protein implies in regulation of cell death and inflammatory signalling pathways. N-terminal SKICH domain; Central oligomerization domain containing three CC regions; C-terminal UBD containing two zinc fingers. |
|
|
| CCT2 | Chaperonin subunit Implies in protein-folding and autophagy ATG8-interaction motif |
|
|
BUZ: Ubiquitin-Binding BUZ, PB1: Phox1 And Bem1p, ZZ: Zz-Type Zinc Finger, NLS: Nuclear Localization Signal, NES: Nuclear Export Signal, LIR: Lc3-Interacting Region, UBA: Ubiquitin-Associated, FW: Four Tryptophan, HA: Amphipatic Helix, PI3P: Phosphati-Dylinositol-3-Phosphate, UBD: Ubiquitin-Binding, Cc: Coil-Coiled, ZnF: Zinc-Finger-Like, SKICH: Skip Carboxyl Homology.
2.4. Aggrephagy Players as Therapeutic Targets
Defective autophagy is known to be linked to various human diseases, such as neurodegenerative, cardiovascular, metabolic, autoimmune diseases and cancer, as well as aging [141,142,143,144,145,146]. Various drugs targeting macroautophagy exist, and although their pharmacological effects are non-specific, they have been approved in the clinical practice for diseases other than myopathies. Their mechanism of action and use in clinical settings or in preclinical studies have been recently reviewed [147,148]. Among the activators of autophagy, rapamycin (known as sirolimus) and its analogs (rapalogs), such as everolimus, tacrolimus and temsirolimus, to name some, act by allosterically inhibiting the kinase activity of mTORC1. These compounds have already been approved to prevent transplant rejection [149], for the treatment of some types of cancers (like pancreatic neuroendocrine tumors and renal cell carcinoma) [150] and moreover, a phase II randomized multicenter clinical trial is undergoing to test the efficacy of the use of rapamycin in combination with riluzole for the treatment of amyotrophic lateral sclerosis (NCT03359538). Other compounds, Torin1 and 2, found to activate autophagy by targeting the ATP binding site of both mTORC1 and 2, are promising drugs to be used in the clinic [151,152]. Similarly, inhibitory drugs have also been developed. Among these are chloroquine and hydroxychloroquine, two lysosomotropic agents that, in combination with anticancer drugs, radiotherapy or surgery, are used for the treatment of solid and hematological tumors [153].
Despite many compounds being known to modulate autophagy and, therefore, could be potential drugs for those diseases in which autophagy is impaired, more studies are needed to fully clarify their exact mechanism of action and, moreover, to reduce the tissue’s toxicity, especially in chronic administration.
To date, no treatments specifically targeting the aggrephagy exist. Therefore, the development of such selective drugs that might increase efficacy and/or reduce side effects represents a fundamental and undeniable urgency.
However, pharmacological discovery in this field shows some challenges, including the lack of aggrephagy-specific biomarkers and the need for appropriate in vivo models.
Thus far, only a few molecules have been designed and tested in vitro to inhibit autophagy for the treatment of various types of cancers. Specifically, p62 has been identified as a possible target, and two different molecules have been developed. Kalid and colleagues [154] showed that PTX80, a compound that was retrospectively identified as a ligand to p62, causes proteotoxic stress-inducing cell apoptosis by reducing the solubility of p62 and, as a consequence, its ability to co-localize with ubiquitinated proteins which will therefore accumulate in the cytoplasm. In another work by Lee and colleagues [155], a small molecule named YOK-1104, able to bind to the ZZ domain of p62, was shown to be a promising drug to be used in combination with irradiation for the induction of apoptosis in those cancer cells that are resistant to cell death.
3. Aggrephagy Involvement in Muscle Diseases
Aggrephagy involvement has been studied and described in a few individual cases or a small case series of genetic and sporadic muscle diseases. Clinical phenotypes, age of onset, disease course and systemic involvement reported in these cases are heterogeneous and summarized in Table 2.
3.1. Myotilinopathies and Desminopathies
Myofibrillar myopathies (MFMs) represent a broad hereditary group of muscle disorders characterized by myofibrillar alterations, Z-disk disorganization, sarcoplasmic buildup of myofibrillar degradation products, often with the presence of rimmed vacuoles, and accumulation of multiple proteins in the muscle tissue (Figure 2a,b). Therefore, MFMs diagnosis is based on histological features since the clinical spectrum is relatively wide.
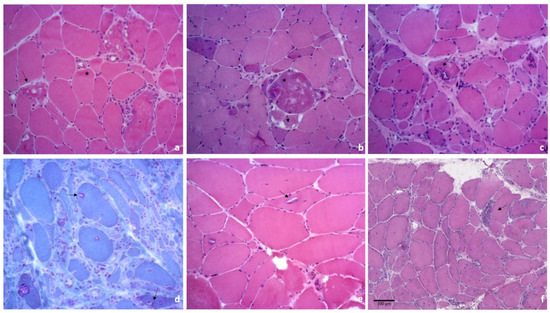
Figure 2.
Histology from muscle biopsies of patients affected by muscle diseases with aggrephagy involvement. (a) hematoxylin/eosin (H&E) staining of a muscle biopsy from a patient affected by a mutation in the myotilin gene highlighting fiber size variability, intracitoplasmatic and subsarcolemmal eosinophilic areas (*), vacuoles, either rimmed or not (arrows); (b) H&E of a muscle biopsy from a patient with a mutation in the desmin gene showing fiber size variability, with concomitant atrophy and hypertrophy, some fibers present basophilic and granular cytoplasmic material (*) and vacuoles (arrows); (c,d) histological findings in a muscle biopsy of an sIBM patient, showing atrophic and hypertrophic muscle fibers, with scattered and groups of small angulated fibers, fiber with rimmed vacuoles and vacuoles with a rim of granular basophilia in H&E (c), that is stained in red with Gomori trichrome (d); (e) muscle biopsy from a patient affected by OPMD, demonstrating variability in fibers size, rare internal nuclei, scattered atrophic and hypotrophic fibers and rimmed vacuoles (arrows); (f) muscle biopsy from a patient affected by PLIN4-related myopathy demonstrating numerous degenerated fibers, frequent central nuclei, fibers splitting and vacuoles (arrow).
Genetically, the cause of MFMs is highly heterogeneous and associated with genes encoding for Z-disk-related proteins, such as desmin, aB-crystallin, myotilin, ZASP, filamin c and BAG3 [156,157,158].
Recently, due to the use of next-generation sequencing, patients with MFM phenotypes have been found to carry mutations in new genes, such as SVIL [159] and UNC45B [160]. Additionally, known genes related to different disorders, such as FHL1, DNAJB6, VCP, HSBP8, TTN, ACTA1, LMNA, PLEC, KY, PYROXD1 and p62 + TIA1, have now been associated with MFM phenotypes [5,105,161,162,163,164].
Protein aggregates in MFMs have been found to contain a great number of different proteins, including phosphorylated tau (p-tau) and β-amyloid, and have also been associated with increased immunoreactivity for the proteasome 19S and 20S subunits [165,166]. Prompted by these observations, Olivé and colleagues [167] examined and first reported in 2008 the presence of p62 and UBB + 1, a mutant form of ubiquitin B known to accumulate in neurodegenerative disorders such as Alzheimer’s and Huntington’s disease, in a cohort of myotilinopathy and desminopathy patients. In their analyzed samples, twelve myotilin and five desmin-mutated patients, the muscle biopsies showed the presence of protein aggregates within the cytoplasm or in the subsarcolemmal region of the muscle fibers as revealed by the modified Gomori trichrome staining. All the myotilinopathies muscles also displayed a large number of vacuoles as well as spheroid bodies, strongly positive for Thioflavin T staining, while in the desminopathies muscles, only a few cytoplasmic bodies and vacuoles were visible, and no Thioflavin T positivity was present. In this study, all biopsy samples showed immunoreactivity for p62, with stronger signals present in myotilinopathy samples than in desminopathy samples. UBB + 1 was also found to be activated even though to a lesser extent and with a higher variability between patients. Similar to p62, fainter signals were found in desminopathy samples compared to myotilinopathy ones. These findings led the authors to conclude that the pathogenic mechanism in these MFM might indeed be due to an impairment of the UPS, while p62 upregulation could be interpreted as a compensatory response to the possibly toxic protein aggregates.
3.2. Sporadic Inclusion Body Myositis
sIBM is the most recurrent sporadic muscle disease in older populations, with the slow progression of muscle impairment accompanied by muscular atrophy. It is still categorized as idiopathic inflammatory myopathies, similar to polymyositis and dermatomyositis, which are characterized by endomysial inflammation associated with various degenerative changes of the muscle fibers (Figure 2c,d) and heterogeneous muscle impairment [168]. Although the lack of response to immunosuppressive or immunomodulatory therapies clearly suggests that the disease is mainly degenerative, and the inflammatory component role is usually limited in early stages [169,170].
sIBM generally shows CD8+ T cells invasion of non-necrotic muscle fibers, incremented expression of MHC class I, rimmed vacuoles, and protein aggregates which could contain, among others, beta amyloid protein, p62 and TDP-43, like those found in Alzheimer’s disease, in amyotrophic lateral sclerosis and frontotemporal dementia [171,172].
Prof. Valerie Askanas’s group investigated the expression of proteins involved in the selective removal of aggregates in muscle biopsies of sIBM patients defined by the typical clinical features [173,174]. Their interest arose from the observation of a certain degree of phenotypic similarity between the sIBM muscle tissue and the brain of Alzheimer’s and Parkinson’s disease patients. The group showed that in sIBM but not in control samples (including polymyositis, dermatomyositis, amyotrophic lateral sclerosis, peripheral neuropathy patients and normal controls), there is increased positivity for p62, LC3 and NBR1. Specifically, in 80% of the vacuolated and 20–25% of the non-vacuolated fibers, p62 strongly accumulates within the aggregates, co-localizing with ubiquitin and p-tau [175]. Moreover, p62, even though in close proximity to, did not appear to co-localize with LC3, which was also found to be activated and accumulating in the cytoplasm of sIBM biopsies [176]. In a later work, the involvement of NBR1 was also considered [177], demonstrating that this aggrephagy adaptor is likewise present within the muscle fibers, accumulating in the form of aggregates and co-localizing with p62, LC3, ubiquitin and p-tau. Furthermore, Nicot and colleagues [178] identified, by in vitro studies, a posttranslational modification of NBR1 by the glycogen synthase kinase 3 (GSK3), which will regulate its activity and showed that a strong dephosphorilation of NBR1 is present in the muscle tissues of sIBM patients, which directly correlates with the severity of the phenotype.
Two different theories exist for the primary mechanism involved in the pathogenesis of sIBM, an autoimmune activation causing T cell-mediated cytotoxicity with consequent damage of the muscle fibers or a degenerative pathway activated by defective aggrephagy. The non-responsiveness to the immunosuppressive therapies and the observations described above seem to better support the initial involvement of a defective protein degradation in the pathogenesis of sIBM [179].
3.3. Rigid Spine Syndrome with FHL1 Mutation
Reducing body myopathy due to FHL1 mutations is an X-linked disorder, marked by the presence of intracytoplasmic inclusion bodies that contain a multitude of proteins, among which mutant FHL1 protein is the most represented.
In patients affected by rigid spine syndrome (RSS) with a mutation in the FHL1 gene, Bonaldo’s group described for the first time the activation of the pathway involved in the selective removal of aggresomes [180]. In about 20% of the muscle fibers of the proband and her brother’s biopsies, the authors identified reducing bodies (RBs) that were Congo-red positive, thus indicating an amyloid-like nature and also revealed positivity for the FHL1 antibody. Further analysis of these aggregates showed co-localization between FHL1 and p62, as well as the presence of DAPI-positive material and ubiquitin within the aggregates. Positivity for LC3 was also detected, although no co-localization with the FHL1 signal was observed. Doing Western blot on muscle protein extract, Beclin1 and BNIP3, two autophagy enhancers, were proven to be strongly induced. A slight increase in the lipidated form of LC3 (LC3-II) was also detected. The identified mutation, a cysteine to arginine conversion in position 150, fell in the second LIM domain of FHL1, which is predicted to promote misfolding of the protein. Previous reports [181,182] of patients with the same p.C150R mutation also show the presence of RBs positive for FHL1 staining. Therefore, the authors conclude that all the data presented strongly indicate the involvement of the aggresome and the autophagy machinery in the muscular pathological mechanism underlying RSS patients with p.C150R FHL1 mutation.
3.4. Oculopharyngeal Muscular Dystrophy
Oculopharingeal muscular dystrophy has mainly autosomal dominant inheritance with progressive late onset of proximal limb, facial and extraocular muscle weakness due to a poly-alanine expansion in the Poly(A) Binding Protein Nuclear 1 (PABPN1) [183,184].
It has been demonstrated that PABPN1 plays a critical role in mRNA biogenesis and myogenesis. In fact, PAPBN1 is implicated in post-transcriptional RNA processing, including long non-coding RNAs (lncRNAs) [185]. Despite being ubiquitous, its alteration results in a muscle-specific disease, with defective myoblast proliferation and differentiation correlated with shorter mRNA poly(A) tails and subsequent nuclear accumulation of poly(A) RNA and misfolded PABPN1.
Thus, it has been postulated that the pathogenetic mechanism involves a reduced bioavailability of PABPN1 due to its aggregation [184].
OPMD muscle biopsies show fiber size variability, rare internal nuclei, scattered atrophic and hypotrophic fibers and rimmed vacuoles (Figure 2e).
In a cohort of 21 Chinese patients clinically affected by OPMD and genetically characterized by five different pathological genotypes, Lin and colleagues investigated the involvement of the autophagy pathway, focusing on the physical association between PABPN1 and p62, NBR1, LC3 and the ubiquitinated protein marker FK2 [186]. In two patients’ muscle biopsies, the authors identified abnormal accumulation of PABPN1 present in 5–7% of the muscle fibers, which were also stained positive for p62, NBR1 and FK2 and, to a lesser extent, LC3. OPMD nuclear inclusions have been reported to contain ubiquitin and the chaperone HSP70 [187,188]. Therefore, the authors conclude that a pathogenic mechanism related to protein degradation involving both the UPS and the selective removal of aggregates is underlying OPMD.
3.5. PLIN4-Related Myopathy
Recently, in an Italian family affected by a late onset myopathy, with distal or less frequently limb-girdle muscle involvement and autophagic features within the muscle, we identified by multi-omic approach a potentially causative large coding expansion in the PLIN4 gene encoding for perilipin 4 [8]. Perilipin 4 is part of a family of proteins involved in the metabolism of lipids, protecting them from lipases by coating the lipid droplets (LDs). Its structure is more divergent compared to the other perilipins, containing a large amphipathic helix responsible for the interaction with the lipids, which is organized in a 3–11 alpha helix structurally similar to α-synuclein and apolipoproteins. The identified genetic mutation causes a 99 nt coding region to be repeated nine more times compared to the wild-type, thus adding 297 extra amino acids to the encoded protein.
Numerous degenerated fibers, frequent central nuclei, fibers splitting and vacuoles (Figure 2f) are observed in patients’ muscle biopsies.
The pathogenic mechanism involved is indicated to be the aggregation of the mutated protein with consequent activation of the aggrephagy pathway for the aggregates’ removal, as demonstrated by the co-localization between perilipin 4 signal and FK2, p62 and NBR1 as well as by the increased signal of WDFY3. The aggregates are present at the subsarcolemmal region of the muscle fibers and within the vacuoles, and positivity for these antibodies is directly correlated with the severity of the phenotype, thus indicating a progressive blockage and inefficiency of the aggrephagy mechanism. More recently, three unrelated families and one Chinese sporadic patient with PLIN4-related myopathy have been reported [189,190]. In these reports, the same activation of the aggrephagy pathway has been confirmed by immunostaining of the muscle biopsies. Notably, one family with a more severe and more rapidly progressing proximal phenotype was associated with a longer repeat expansion [189].

Table 2.
Clinical, laboratory and histological characteristics of muscle disorders with reported aggrephagy markers.
Table 2.
Clinical, laboratory and histological characteristics of muscle disorders with reported aggrephagy markers.
| Disease | Gene/ Inheritance | Age at Onset | Predominant Muscle Involvement | Systemic Involvement | Evolution, Loss of Ambulation | Creatin Kinase Level | Muscle Biopsy: Rimmed Vacuoles and Protein Aggregates |
|---|---|---|---|---|---|---|---|
| Myofibrillar myopathy [156,157,158,167] | MYOT/AD DES/AD | Adult Young adult | Distal Distal + limb-girdle | Rarely cardiomyopathy and peripheral neuropathy Cardiomyopathy | Slowly progressive Slowly progressive, LoA in later stages | Mild to highly elevated | Rimmed vacuoles and protein aggregates + |
| Inclusion body myopathy [174,177] | sporadic | Late adult | Quadriceps and/or finger flexors | Dysphagia, respiratory involvement | Progressive, LoA in later stages | Very variable, ranging from normal to up to 15 times UNL | Rimmed vacuoles and protein aggregates ++ |
| oculo-pharyngeal muscular dystrophy [186,191] | PABPN1 */AD | Late adult | Eyelid ptosis+ bulbar | Dysphagia, respiratory involvement, executive function deficits, rarely peripheral neuropathy | Slowly progressive | Slightly elevated in mild cases, higher severe cases | Rimmed vacuoles +/− |
| FHL1 myopathy [180] | FHL1 †/XLD | Young adult | Neck flexors and limb-girdle | Respiratory involvement, scoliosis, rigid spinel, rarely cardiomyopathy | Severe progressive course, LoA at early stages | Mildly elevated | Protein aggregates + |
| PLIN4 myopathy [8,189,190] | PLIN4 §/AD | Adult | Distal and limb-girdle | None | Slowly progressive, LoA at early stages | Normal to slightly elevated | Rimmed vacuoles and preotein aggregates + |
AD: autosomal dominant, AR: autosomal recessive, XLD: X-linked dominant LoA: loss of ambulation, UNL: upper normal limit. * Abnormal expansion of (GCN)n repeats, † p.C150R, § 99 nt coding expansion.
4. Conclusions
Aggrephagy represents an underexplored but widely-present pathological mechanism in the more general context of autophagy, with the latter already being extensively investigated in human diseases, including myopathies. To date, aggrephagy has been investigated only in a limited number of diseases, reveling a pathological role in a case of FHL1-rigid spine syndrome and in a small case series of sIBM, PLIN4-related myopathy, myotilinopathies and desminopathies and oculopharyngeal muscular dystrophy.
Notably, these diseases are extremely heterogeneous. The described myopathies are all genetically defined, apart from sIBM, which is an acquired entity, suggesting a transversal role of aggrephagy across different diseases. The clinical presentation mainly occurs in adult life, around the fifth and sixth decades, and shows a slowly progressive worsening, with the exception of RSS cases, which have an onset in early adult life and have a rapid decrease of motor functions. These observations suggest that the impairment of aggrephagy is generally a slowly ongoing progressive process that may be related to aging. On the other hand, the distribution of affected muscles is very diverse, ranging from cranial to limb-girdle to distal muscle involvement. Even cardiomyocytes can be affected, as in desminopathy, myotilinopathy [192] and FHL1-related disorders. Accumulation of protein aggregates implicating the aggrephagy pathway abnormality has been recently investigated in patients with genetic cardiomyopathy. The analysis of p62-positive aggregates in the myocardium showed a high prevalence of diffused p62-positive aggregates in phospholamban (PNL) and desmin-related (DRC) cardiomyopathy patients [193], confirming earlier findings in a mouse model of DRC desmin mutant and impaired lysosomal function [194].
Thus far, only a few players known to be directly involved in the selective removal of protein aggregates have been investigated in muscle diseases. Most likely, the fact that p62 and NBR1 appear to be more investigated in these diseases compared to the other known aggrephagy players is related to a better knowledge of their function and mechanism of action. However, the simultaneous evaluation of other proteins involved in aggrephagy is necessary for a comprehensive view of the possible pathogenic mechanism leading to protein aggregation myopathies.
Further studies are needed to better characterize the pathological role of aggrephagy in muscle diseases and to define possible therapeutic targets.
Author Contributions
Original draft preparation, S.G. and A.R.; review—editing, M.C.; review—editing and supervision, L.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Lorenzo Maggi is member of the ERN-NMD. This work was partially supported by the Italian Ministry of Health (RRC). The authors thank Rosalind Hendricks, Assistant Biomedical Librarian at “Fondazione IRCCS Istituto Neurologico Carlo Besta”, for language revision of our paper.
Conflicts of Interest
Maggi has received honoraria for speaking, advisory boards and compensation for congress participations from Sanofi Genzyme, Roche and Biogen, Amicus Theraputics, Alexion, Janssen, Lupin, outside the submitted work. The other authors declare no conflict of interest.
References
- Margeta, M. Autophagy Defects in Skeletal Myopathies. Annu. Rev. Pathol. 2020, 15, 261–285. [Google Scholar] [CrossRef]
- Franco-Romero, A.; Sandri, M. Role of autophagy in muscle disease. Mol. Aspects. Med. 2021, 82, 101041. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Petroni, G.; Amaravadi, R.K.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cadwell, K.; Cecconi, F.; Choi, A.M.K.; et al. Autophagy in major human diseases. EMBO J. 2021, 40, e108863. [Google Scholar] [CrossRef] [PubMed]
- Sugie, K. Autophagic vacuolar myopathy: Danon disease and related myopathies. Neurol. Clin. Neurosci. 2022, 10, 273–278. [Google Scholar] [CrossRef]
- Olivé, M.; Winter, L.; Fürst, D.O.; Schröder, R.; ENMC Workshop Study Group. 246th ENMC International Workshop: Protein aggregate myopathies 24-26 May 2019, Hoofddorp, The Netherlands. Neuromuscul Disord 2021, 31, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.C.; Goebel, H.H. Protein aggregate myopathies. Neurol. India 2005, 53, 273–279. [Google Scholar] [CrossRef]
- Malicdan, M.C.; Nishino, I. Autophagy in lysosomal myopathies. Brain Pathol. 2012, 22, 82–88. [Google Scholar] [CrossRef]
- Ruggieri, A.; Naumenko, S.; Smith, M.A.; Iannibelli, E.; Blasevich, F.; Bragato, C.; Gibertini, S.; Barton, K.; Vorgerd, M.; Marcus, K.; et al. Multiomic elucidation of a coding 99-mer repeat-expansion skeletal muscle disease. Acta Neuropathol. 2020, 140, 231–235. [Google Scholar] [CrossRef]
- Goebel, H.H.; Fardeau, M.; Olivé, M.; Schröder, R. 156th ENMC International Workshop: Desmin and protein aggregate myopathies, 9-11 November 2007, Naarden, The Netherlands. Neuromuscul. Disord. 2008, 18, 583–592. [Google Scholar] [CrossRef]
- Kundra, R.; Ciryam, P.; Morimoto, R.I.; Dobson, C.M.; Vendruscolo, M. Protein homeostasis of a metastable subproteome associated with Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2017, 114, E5703–E5711. [Google Scholar] [CrossRef]
- Sandri, M.; Coletto, L.; Grumati, P.; Bonaldo, P. Misregulation of autophagy and protein degradation systems in myopathies and muscular dystrophies. J. Cell Sci. 2013, 126, 5325–5333. [Google Scholar] [CrossRef]
- Goebel, H.H.; Müller, H.D. Protein aggregate myopathies. Semin. Pediatr. Neurol. 2006, 13, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Goebel, H.H. Protein aggregate myopathies. Introduction. Brain Pathol. 2009, 19, 480–482. [Google Scholar] [CrossRef] [PubMed]
- Klinge, L.; Straub, V.; Neudorf, U.; Voit, T. Enzyme replacement therapy in classical infantile pompe disease: Results of a ten-month follow-up study. Neuropediatrics 2005, 36, 6–11. [Google Scholar] [CrossRef]
- Stevens, D.; Milani-Nejad, S.; Mozaffar, T. Pompe Disease: A Clinical, Diagnostic, and Therapeutic Overview. Curr. Treat. Options Neurol. 2022, 24, 573–588. [Google Scholar] [CrossRef]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef]
- Liutkute, M.; Samatova, E.; Rodnina, M.V. Cotranslational Folding of Proteins on the Ribosome. Biomolecules 2020, 10, 97. [Google Scholar] [CrossRef] [PubMed]
- Dobson, C.M. Protein folding and misfolding. Nature 2003, 426, 884–890. [Google Scholar] [CrossRef]
- Jahn, T.R.; Radford, S.E. The Yin and Yang of protein folding. FEBS J. 2005, 272, 5962–5970. [Google Scholar] [CrossRef]
- Dunker, A.K.; Silman, I.; Uversky, V.N.; Sussman, J.L. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 2008, 18, 756–764. [Google Scholar] [CrossRef]
- Trivedi, R.; Nagarajaram, H.A. Intrinsically Disordered Proteins: An Overview. Int. J. Mol. Sci. 2022, 23, 14050. [Google Scholar] [CrossRef] [PubMed]
- Powers, E.T.; Morimoto, R.I.; Dillin, A.; Kelly, J.W.; Balch, W.E. Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 2009, 78, 959–991. [Google Scholar] [CrossRef] [PubMed]
- Hipp, M.S.; Kasturi, P.; Hartl, F.U. The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 2019, 20, 421–435. [Google Scholar] [CrossRef] [PubMed]
- Jayaraj, G.G.; Hipp, M.S.; Hartl, F.U. Functional Modules of the Proteostasis Network. Cold Spring Harb. Perspect. Biol. 2020, 12, a033951. [Google Scholar] [CrossRef] [PubMed]
- Brehme, M.; Voisine, C.; Rolland, T.; Wachi, S.; Soper, J.H.; Zhu, Y.; Orton, K.; Villella, A.; Garza, D.; Vidal, M.; et al. A chaperome subnetwork safeguards proteostasis in aging and neurodegenerative disease. Cell Rep. 2014, 9, 1135–1150. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A.; Kwon, Y.T. Protein Quality Control by Molecular Chaperones in Neurodegeneration. Front. Neurosci. 2017, 11, 185. [Google Scholar] [CrossRef]
- Kampinga, H.H.; Hageman, J.; Vos, M.J.; Kubota, H.; Tanguay, R.M.; Bruford, E.A.; Cheetham, M.E.; Chen, B.; Hightower, L.E. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress. Chaperones. 2009, 14, 105–111. [Google Scholar] [CrossRef]
- Brocchieri, L.; Conway de Macario, E.; Macario, A.J. hsp70 genes in the human genome: Conservation and differentiation patterns predict a wide array of overlapping and specialized functions. BMC Evol. Biol. 2008, 8, 19. [Google Scholar] [CrossRef]
- Rosenzweig, R.; Nillegoda, N.B.; Mayer, M.P.; Bukau, B. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019, 20, 665–680. [Google Scholar] [CrossRef]
- Hu, C.; Yang, J.; Qi, Z.; Wu, H.; Wang, B.; Zou, F.; Mei, H.; Liu, J.; Wang, W.; Liu, Q. Heat shock proteins: Biological functions, pathological roles, and therapeutic opportunities. MedComm (2020) 2022, 3, e161. [Google Scholar] [CrossRef]
- Mayer, M.P.; Bukau, B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell Mol. Life Sci. 2005, 62, 670–684. [Google Scholar] [CrossRef]
- Piette, B.L.; Alerasool, N.; Lin, Z.Y.; Lacoste, J.; Lam, M.H.Y.; Qian, W.W.; Tran, S.; Larsen, B.; Campos, E.; Peng, J.; et al. Comprehensive interactome profiling of the human Hsp70 network highlights functional differentiation of J domains. Mol. Cell 2021, 81, 2549–2565.e2548. [Google Scholar] [CrossRef]
- Mayer, M.P. Gymnastics of molecular chaperones. Mol. Cell 2010, 39, 321–331. [Google Scholar] [CrossRef]
- Kampinga, H.H.; Craig, E.A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010, 11, 579–592. [Google Scholar] [CrossRef]
- Behnke, J.; Feige, M.J.; Hendershot, L.M. BiP and its nucleotide exchange factors Grp170 and Sil1: Mechanisms of action and biological functions. J. Mol. Biol. 2015, 427, 1589–1608. [Google Scholar] [CrossRef] [PubMed]
- Pobre, K.F.R.; Poet, G.J.; Hendershot, L.M. The endoplasmic reticulum (ER) chaperone BiP is a master regulator of ER functions: Getting by with a little help from ERdj friends. J. Biol. Chem. 2019, 294, 2098–2108. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Soroka, J.; Buchner, J. The Hsp90 chaperone machinery: Conformational dynamics and regulation by co-chaperones. Biochim. Biophys. Acta 2012, 1823, 624–635. [Google Scholar] [CrossRef] [PubMed]
- Schopf, F.H.; Biebl, M.M.; Buchner, J. The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 2017, 18, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Pezeshki, A.M.; Yu, X.; Guo, C.; Subjeck, J.R.; Wang, X.Y. The Endoplasmic Reticulum Chaperone GRP170: From Immunobiology to Cancer Therapeutics. Front. Oncol. 2014, 4, 377. [Google Scholar] [CrossRef]
- Basha, E.; O’Neill, H.; Vierling, E. Small heat shock proteins and α-crystallins: Dynamic proteins with flexible functions. Trends. Biochem. Sci. 2012, 37, 106–117. [Google Scholar] [CrossRef]
- Kim, Y.E.; Hipp, M.S.; Bracher, A.; Hayer-Hartl, M.; Hartl, F.U. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 2013, 82, 323–355. [Google Scholar] [CrossRef] [PubMed]
- Stan, G.; Lorimer, G.H.; Thirumalai, D. Friends in need: How chaperonins recognize and remodel proteins that require folding assistance. Front. Mol. Biosci. 2022, 9, 1071168. [Google Scholar] [CrossRef]
- Vijay-Kumar, S.; Bugg, C.E.; Wilkinson, K.D.; Cook, W.J. Three-dimensional structure of ubiquitin at 2.8 A resolution. Proc. Natl. Acad. Sci. USA 1985, 82, 3582–3585. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, G.; Scheid, M.; Hammerling, U.; Schlesinger, D.H.; Niall, H.D.; Boyse, E.A. Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc. Natl. Acad. Sci. USA 1975, 72, 11–15. [Google Scholar] [CrossRef]
- Hochstrasser, M. Evolution and function of ubiquitin-like protein-conjugation systems. Nat. Cell Biol. 2000, 2, E153–E157. [Google Scholar] [CrossRef] [PubMed]
- Pickart, C.M.; Eddins, M.J. Ubiquitin: Structures, functions, mechanisms. Biochim. Biophys. Acta 2004, 1695, 55–72. [Google Scholar] [CrossRef]
- Glickman, M.H.; Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar] [CrossRef]
- Reinstein, E.; Ciechanover, A. Narrative review: Protein degradation and human diseases: The ubiquitin connection. Ann. Intern. Med. 2006, 145, 676–684. [Google Scholar] [CrossRef]
- Dikic, I.; Wakatsuki, S.; Walters, K.J. Ubiquitin-binding domains—From structures to functions. Nat. Rev. Mol. Cell Biol. 2009, 10, 659–671. [Google Scholar] [CrossRef]
- Weissman, A.M.; Shabek, N.; Ciechanover, A. The predator becomes the prey: Regulating the ubiquitin system by ubiquitylation and degradation. Nat. Rev. Mol. Cell Biol. 2011, 12, 605–620. [Google Scholar] [CrossRef]
- Ciechanover, A.; Stanhill, A. The complexity of recognition of ubiquitinated substrates by the 26S proteasome. Biochim. Biophys. Acta 2014, 1843, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef] [PubMed]
- Akutsu, M.; Dikic, I.; Bremm, A. Ubiquitin chain diversity at a glance. J. Cell Sci. 2016, 129, 875–880. [Google Scholar] [CrossRef]
- Ohtake, F.; Tsuchiya, H. The emerging complexity of ubiquitin architecture. J. Biochem. 2017, 161, 125–133. [Google Scholar] [CrossRef]
- Morris, J.R.; Garvin, A.J. SUMO in the DNA Double-Stranded Break Response: Similarities, Differences, and Cooperation with Ubiquitin. J. Mol. Biol. 2017, 429, 3376–3387. [Google Scholar] [CrossRef]
- Yin, Z.; Popelka, H.; Lei, Y.; Yang, Y.; Klionsky, D.J. The Roles of Ubiquitin in Mediating Autophagy. Cells 2020, 9, 2025. [Google Scholar] [CrossRef] [PubMed]
- Navon, A.; Ciechanover, A. The 26 S proteasome: From basic mechanisms to drug targeting. J. Biol. Chem. 2009, 284, 33713–33718. [Google Scholar] [CrossRef]
- Tanaka, K. The proteasome: Overview of structure and functions. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009, 85, 12–36. [Google Scholar] [CrossRef]
- Huang, L.; Chen, C.H. Proteasome regulators: Activators and inhibitors. Curr. Med. Chem. 2009, 16, 931–939. [Google Scholar] [CrossRef]
- Budenholzer, L.; Cheng, C.L.; Li, Y.; Hochstrasser, M. Proteasome Structure and Assembly. J. Mol. Biol. 2017, 429, 3500–3524. [Google Scholar] [CrossRef]
- Ciechanover, A.; Iwai, K. The ubiquitin system: From basic mechanisms to the patient bed. IUBMB Life 2004, 56, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Fonts, K.; Davis, C.; Tomita, T.; Elsasser, S.; Nager, A.R.; Shi, Y.; Finley, D.; Matouschek, A. The proteasome 19S cap and its ubiquitin receptors provide a versatile recognition platform for substrates. Nat. Commun. 2020, 11, 477. [Google Scholar] [CrossRef]
- Tan, J.M.; Wong, E.S.; Kirkpatrick, D.S.; Pletnikova, O.; Ko, H.S.; Tay, S.P.; Ho, M.W.; Troncoso, J.; Gygi, S.P.; Lee, M.K.; et al. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum. Mol. Genet. 2008, 17, 431–439. [Google Scholar] [CrossRef]
- Zheng, Q.; Su, H.; Tian, Z.; Wang, X. Proteasome malfunction activates macroautophagy in the heart. Am. J. Cardiovasc. Dis. 2011, 1, 214–226. [Google Scholar] [PubMed]
- Zhu, K.; Dunner, K.; McConkey, D.J. Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene 2010, 29, 451–462. [Google Scholar] [CrossRef]
- Pandey, U.B.; Nie, Z.; Batlevi, Y.; McCray, B.A.; Ritson, G.P.; Nedelsky, N.B.; Schwartz, S.L.; DiProspero, N.A.; Knight, M.A.; Schuldiner, O.; et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 2007, 447, 859–863. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Bravo-San Pedro, J.M.; Cecconi, F.; Choi, A.M.; Chu, C.T.; Codogno, P.; Colombo, M.I.; et al. Molecular definitions of autophagy and related processes. EMBO J. 2017, 36, 1811–1836. [Google Scholar] [CrossRef] [PubMed]
- Øverbye, A.; Fengsrud, M.; Seglen, P.O. Proteomic analysis of membrane-associated proteins from rat liver autophagosomes. Autophagy 2007, 3, 300–322. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Eskelinen, E.L.; Deretic, V. Autophagosomes, phagosomes, autolysosomes, phagolysosomes, autophagolysosomes... wait, I’m confused. Autophagy 2014, 10, 549–551. [Google Scholar] [CrossRef]
- Yang, Z.; Klionsky, D.J. Mammalian autophagy: Core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010, 22, 124–131. [Google Scholar] [CrossRef]
- Feng, Y.; He, D.; Yao, Z.; Klionsky, D.J. The machinery of macroautophagy. Cell Res. 2014, 24, 24–41. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Emr, S.D. Autophagy as a regulated pathway of cellular degradation. Science 2000, 290, 1717–1721. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Zhang, S.; Mizushima, N. Autophagy genes in biology and disease. Nat. Rev. Genet. 2023. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. mTOR signaling at a glance. J. Cell Sci. 2009, 122, 3589–3594. [Google Scholar] [CrossRef]
- Jung, C.H.; Ro, S.H.; Cao, J.; Otto, N.M.; Kim, D.H. mTOR regulation of autophagy. FEBS Lett. 2010, 584, 1287–1295. [Google Scholar] [CrossRef] [PubMed]
- Wong, P.M.; Puente, C.; Ganley, I.G.; Jiang, X. The ULK1 complex: Sensing nutrient signals for autophagy activation. Autophagy 2013, 9, 124–137. [Google Scholar] [CrossRef]
- Yuan, H.X.; Russell, R.C.; Guan, K.L. Regulation of PIK3C3/VPS34 complexes by MTOR in nutrient stress-induced autophagy. Autophagy 2013, 9, 1983–1995. [Google Scholar] [CrossRef]
- Jhanwar-Uniyal, M.; Amin, A.G.; Cooper, J.B.; Das, K.; Schmidt, M.H.; Murali, R. Discrete signaling mechanisms of mTORC1 and mTORC2: Connected yet apart in cellular and molecular aspects. Adv. Biol. Regul. 2017, 64, 39–48. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef]
- Dossou, A.S.; Basu, A. The Emerging Roles of mTORC1 in Macromanaging Autophagy. Cancers 2019, 11, 1422. [Google Scholar] [CrossRef]
- Deleyto-Seldas, N.; Efeyan, A. The mTOR-Autophagy Axis and the Control of Metabolism. Front. Cell Dev. Biol. 2021, 9, 655731. [Google Scholar] [CrossRef] [PubMed]
- Sandri, M. Autophagy in skeletal muscle. FEBS Lett. 2010, 584, 1411–1416. [Google Scholar] [CrossRef] [PubMed]
- Bonaldo, P.; Sandri, M. Cellular and molecular mechanisms of muscle atrophy. Dis. Model. Mech. 2013, 6, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Leduc-Gaudet, J.P.; Franco-Romero, A.; Cefis, M.; Moamer, A.; Broering, F.E.; Milan, G.; Sartori, R.; Chaffer, T.J.; Dulac, M.; Marcangeli, V.; et al. MYTHO is a novel regulator of skeletal muscle autophagy and integrity. Nat. Commun. 2023, 14, 1199. [Google Scholar] [CrossRef] [PubMed]
- Onishi, M.; Yamano, K.; Sato, M.; Matsuda, N.; Okamoto, K. Molecular mechanisms and physiological functions of mitophagy. EMBO J. 2021, 40, e104705. [Google Scholar] [CrossRef]
- Papandreou, M.E.; Tavernarakis, N. Nucleophagy: From homeostasis to disease. Cell Death. Differ. 2019, 26, 630–639. [Google Scholar] [CrossRef]
- Beese, C.J.; Brynjólfsdóttir, S.H.; Frankel, L.B. Selective Autophagy of the Protein Homeostasis Machinery: Ribophagy, Proteaphagy and ER-Phagy. Front. Cell Dev. Biol. 2019, 7, 373. [Google Scholar] [CrossRef]
- Shin, D.W. Lipophagy: Molecular Mechanisms and Implications in Metabolic Disorders. Mol. Cells 2020, 43, 686–693. [Google Scholar] [CrossRef]
- Papadopoulos, C.; Kravic, B.; Meyer, H. Repair or Lysophagy: Dealing with Damaged Lysosomes. J. Mol. Biol. 2020, 432, 231–239. [Google Scholar] [CrossRef]
- Lamark, T.; Johansen, T. Aggrephagy: Selective disposal of protein aggregates by macroautophagy. Int. J. Cell Biol. 2012, 2012, 736905. [Google Scholar] [CrossRef]
- Fink, A.L. Protein aggregation: Folding aggregates, inclusion bodies and amyloid. Fold. Des. 1998, 3, R9–R23. [Google Scholar] [CrossRef]
- Kopito, R.R. Aggresomes, inclusion bodies and protein aggregation. Trends. Cell Biol. 2000, 10, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Johnston, J.A.; Ward, C.L.; Kopito, R.R. Aggresomes: A cellular response to misfolded proteins. J. Cell. Biol. 1998, 143, 1883–1898. [Google Scholar] [CrossRef]
- García-Mata, R.; Bebök, Z.; Sorscher, E.J.; Sztul, E.S. Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J. Cell Biol. 1999, 146, 1239–1254. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, Y.; Kovacs, J.J.; McLaurin, A.; Vance, J.M.; Ito, A.; Yao, T.P. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 2003, 115, 727–738. [Google Scholar] [CrossRef]
- Yan, J. Interplay between HDAC6 and its interacting partners: Essential roles in the aggresome-autophagy pathway and neurodegenerative diseases. DNA Cell Biol. 2014, 33, 567–580. [Google Scholar] [CrossRef]
- Hubbert, C.; Guardiola, A.; Shao, R.; Kawaguchi, Y.; Ito, A.; Nixon, A.; Yoshida, M.; Wang, X.F.; Yao, T.P. HDAC6 is a microtubule-associated deacetylase. Nature 2002, 417, 455–458. [Google Scholar] [CrossRef] [PubMed]
- Iwata, A.; Riley, B.E.; Johnston, J.A.; Kopito, R.R. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J. Biol. Chem. 2005, 280, 40282–40292. [Google Scholar] [CrossRef]
- Lee, J.Y.; Koga, H.; Kawaguchi, Y.; Tang, W.; Wong, E.; Gao, Y.S.; Pandey, U.B.; Kaushik, S.; Tresse, E.; Lu, J.; et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010, 29, 969–980. [Google Scholar] [CrossRef]
- Bjørkøy, G.; Lamark, T.; Brech, A.; Outzen, H.; Perander, M.; Overvatn, A.; Stenmark, H.; Johansen, T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005, 171, 603–614. [Google Scholar] [CrossRef]
- Pankiv, S.; Clausen, T.H.; Lamark, T.; Brech, A.; Bruun, J.A.; Outzen, H.; Øvervatn, A.; Bjørkøy, G.; Johansen, T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007, 282, 24131–24145. [Google Scholar] [CrossRef] [PubMed]
- Fecto, F.; Yan, J.; Vemula, S.P.; Liu, E.; Yang, Y.; Chen, W.; Zheng, J.G.; Shi, Y.; Siddique, N.; Arrat, H.; et al. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch. Neurol. 2011, 68, 1440–1446. [Google Scholar] [CrossRef] [PubMed]
- Goode, A.; Butler, K.; Long, J.; Cavey, J.; Scott, D.; Shaw, B.; Sollenberger, J.; Gell, C.; Johansen, T.; Oldham, N.J.; et al. Defective recognition of LC3B by mutant SQSTM1/p62 implicates impairment of autophagy as a pathogenic mechanism in ALS-FTLD. Autophagy 2016, 12, 1094–1104. [Google Scholar] [CrossRef]
- Haack, T.B.; Ignatius, E.; Calvo-Garrido, J.; Iuso, A.; Isohanni, P.; Maffezzini, C.; Lönnqvist, T.; Suomalainen, A.; Gorza, M.; Kremer, L.S.; et al. Absence of the Autophagy Adaptor SQSTM1/p62 Causes Childhood-Onset Neurodegeneration with Ataxia, Dystonia, and Gaze Palsy. Am. J. Hum. Genet. 2016, 99, 735–743. [Google Scholar] [CrossRef]
- Bucelli, R.C.; Arhzaouy, K.; Pestronk, A.; Pittman, S.K.; Rojas, L.; Sue, C.M.; Evilä, A.; Hackman, P.; Udd, B.; Harms, M.B.; et al. SQSTM1 splice site mutation in distal myopathy with rimmed vacuoles. Neurology 2015, 85, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Hocking, L.J.; Lucas, G.J.; Daroszewska, A.; Mangion, J.; Olavesen, M.; Cundy, T.; Nicholson, G.C.; Ward, L.; Bennett, S.T.; Wuyts, W.; et al. Domain-specific mutations in sequestosome 1 (SQSTM1) cause familial and sporadic Paget’s disease. Hum. Mol. Genet. 2002, 11, 2735–2739. [Google Scholar] [CrossRef]
- Pankiv, S.; Lamark, T.; Bruun, J.A.; Øvervatn, A.; Bjørkøy, G.; Johansen, T. Nucleocytoplasmic shuttling of p62/SQSTM1 and its role in recruitment of nuclear polyubiquitinated proteins to promyelocytic leukemia bodies. J. Biol. Chem. 2010, 285, 5941–5953. [Google Scholar] [CrossRef] [PubMed]
- Isogai, S.; Morimoto, D.; Arita, K.; Unzai, S.; Tenno, T.; Hasegawa, J.; Sou, Y.S.; Komatsu, M.; Tanaka, K.; Shirakawa, M.; et al. Crystal structure of the ubiquitin-associated (UBA) domain of p62 and its interaction with ubiquitin. J. Biol. Chem. 2011, 286, 31864–31874. [Google Scholar] [CrossRef]
- Lim, J.; Lachenmayer, M.L.; Wu, S.; Liu, W.; Kundu, M.; Wang, R.; Komatsu, M.; Oh, Y.J.; Zhao, Y.; Yue, Z. Proteotoxic stress induces phosphorylation of p62/SQSTM1 by ULK1 to regulate selective autophagic clearance of protein aggregates. PLoS Genet. 2015, 11, e1004987. [Google Scholar] [CrossRef]
- Ciuffa, R.; Lamark, T.; Tarafder, A.K.; Guesdon, A.; Rybina, S.; Hagen, W.J.; Johansen, T.; Sachse, C. The selective autophagy receptor p62 forms a flexible filamentous helical scaffold. Cell Rep. 2015, 11, 748–758. [Google Scholar] [CrossRef]
- Ichimura, Y.; Kumanomidou, T.; Sou, Y.S.; Mizushima, T.; Ezaki, J.; Ueno, T.; Kominami, E.; Yamane, T.; Tanaka, K.; Komatsu, M. Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 2008, 283, 22847–22857. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Wu, R.; Zheng, J.; Li, P.; Yu, L. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 2018, 28, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Zaffagnini, G.; Savova, A.; Danieli, A.; Romanov, J.; Tremel, S.; Ebner, M.; Peterbauer, T.; Sztacho, M.; Trapannone, R.; Tarafder, A.K.; et al. p62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J. 2018, 37, e98308. [Google Scholar] [CrossRef] [PubMed]
- Kirkin, V.; Lamark, T.; Sou, Y.S.; Bjørkøy, G.; Nunn, J.L.; Bruun, J.A.; Shvets, E.; McEwan, D.G.; Clausen, T.H.; Wild, P.; et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 2009, 33, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Waters, S.; Marchbank, K.; Solomon, E.; Whitehouse, C.; Gautel, M. Interactions with LC3 and polyubiquitin chains link nbr1 to autophagic protein turnover. FEBS Lett. 2009, 583, 1846–1852. [Google Scholar] [CrossRef]
- Rasmussen, N.L.; Kournoutis, A.; Lamark, T.; Johansen, T. NBR1: The archetypal selective autophagy receptor. J. Cell Biol. 2022, 221, e202208092. [Google Scholar] [CrossRef]
- Johansen, T.; Lamark, T. Selective Autophagy: ATG8 Family Proteins, LIR Motifs and Cargo Receptors. J. Mol. Biol. 2020, 432, 80–103. [Google Scholar] [CrossRef]
- Marchbank, K.; Waters, S.; Roberts, R.G.; Solomon, E.; Whitehouse, C.A. MAP1B Interaction with the FW Domain of the Autophagic Receptor Nbr1 Facilitates Its Association to the Microtubule Network. Int. J. Cell Biol. 2012, 2012, 208014. [Google Scholar] [CrossRef]
- Mardakheh, F.K.; Auciello, G.; Dafforn, T.R.; Rappoport, J.Z.; Heath, J.K. Nbr1 is a novel inhibitor of ligand-mediated receptor tyrosine kinase degradation. Mol. Cell Biol. 2010, 30, 5672–5685. [Google Scholar] [CrossRef]
- Turco, E.; Savova, A.; Gere, F.; Ferrari, L.; Romanov, J.; Schuschnig, M.; Martens, S. Reconstitution defines the roles of p62, NBR1 and TAX1BP1 in ubiquitin condensate formation and autophagy initiation. Nat. Commun. 2021, 12, 5212. [Google Scholar] [CrossRef]
- Chauveau, C.; Bonnemann, C.G.; Julien, C.; Kho, A.L.; Marks, H.; Talim, B.; Maury, P.; Arne-Bes, M.C.; Uro-Coste, E.; Alexandrovich, A.; et al. Recessive TTN truncating mutations define novel forms of core myopathy with heart disease. Hum. Mol. Genet. 2014, 23, 980–991. [Google Scholar] [CrossRef] [PubMed]
- Lange, S.; Xiang, F.; Yakovenko, A.; Vihola, A.; Hackman, P.; Rostkova, E.; Kristensen, J.; Brandmeier, B.; Franzen, G.; Hedberg, B.; et al. The kinase domain of titin controls muscle gene expression and protein turnover. Science 2005, 308, 1599–1603. [Google Scholar] [CrossRef] [PubMed]
- Topaloglu, H. Core myopathies—A short review. Acta Myol. 2020, 39, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Amburgey, K.; McNamara, N.; Bennett, L.R.; McCormick, M.E.; Acsadi, G.; Dowling, J.J. Prevalence of congenital myopathies in a representative pediatric united states population. Ann. Neurol. 2011, 70, 662–665. [Google Scholar] [CrossRef] [PubMed]
- Simonsen, A.; Birkeland, H.C.; Gillooly, D.J.; Mizushima, N.; Kuma, A.; Yoshimori, T.; Slagsvold, T.; Brech, A.; Stenmark, H. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J. Cell Sci. 2004, 117, 4239–4251. [Google Scholar] [CrossRef] [PubMed]
- Filimonenko, M.; Isakson, P.; Finley, K.D.; Anderson, M.; Jeong, H.; Melia, T.J.; Bartlett, B.J.; Myers, K.M.; Birkeland, H.C.; Lamark, T.; et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol. Cell 2010, 38, 265–279. [Google Scholar] [CrossRef]
- Clausen, T.H.; Lamark, T.; Isakson, P.; Finley, K.; Larsen, K.B.; Brech, A.; Øvervatn, A.; Stenmark, H.; Bjørkøy, G.; Simonsen, A.; et al. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy 2010, 6, 330–344. [Google Scholar] [CrossRef]
- Isakson, P.; Holland, P.; Simonsen, A. The role of ALFY in selective autophagy. Cell Death. Differ. 2013, 20, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Psakhye, I.; Jentsch, S. Autophagic clearance of polyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell 2014, 158, 549–563. [Google Scholar] [CrossRef]
- Maruyama, H.; Morino, H.; Ito, H.; Izumi, Y.; Kato, H.; Watanabe, Y.; Kinoshita, Y.; Kamada, M.; Nodera, H.; Suzuki, H.; et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature 2010, 465, 223–226. [Google Scholar] [CrossRef] [PubMed]
- Del Bo, R.; Tiloca, C.; Pensato, V.; Corrado, L.; Ratti, A.; Ticozzi, N.; Corti, S.; Castellotti, B.; Mazzini, L.; Sorarù, G.; et al. Novel optineurin mutations in patients with familial and sporadic amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry 2011, 82, 1239–1243. [Google Scholar] [CrossRef]
- Osawa, T.; Mizuno, Y.; Fujita, Y.; Takatama, M.; Nakazato, Y.; Okamoto, K. Optineurin in neurodegenerative diseases. Neuropathology 2011, 31, 569–574. [Google Scholar] [CrossRef]
- Iida, A.; Hosono, N.; Sano, M.; Kamei, T.; Oshima, S.; Tokuda, T.; Kubo, M.; Nakamura, Y.; Ikegawa, S. Optineurin mutations in Japanese amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry 2012, 83, 233–235. [Google Scholar] [CrossRef]
- Rezaie, T.; Child, A.; Hitchings, R.; Brice, G.; Miller, L.; Coca-Prados, M.; Héon, E.; Krupin, T.; Ritch, R.; Kreutzer, D.; et al. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science 2002, 295, 1077–1079. [Google Scholar] [CrossRef]
- Korac, J.; Schaeffer, V.; Kovacevic, I.; Clement, A.M.; Jungblut, B.; Behl, C.; Terzic, J.; Dikic, I. Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. J. Cell Sci. 2013, 126, 580–592. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.C.; Li, H.Y.; Chen, G.C.; Chern, Y.; Tu, P.H. Mutations in the ubiquitin-binding domain of OPTN/optineurin interfere with autophagy-mediated degradation of misfolded proteins by a dominant-negative mechanism. Autophagy 2015, 11, 685–700. [Google Scholar] [CrossRef]
- Newman, A.C.; Scholefield, C.L.; Kemp, A.J.; Newman, M.; McIver, E.G.; Kamal, A.; Wilkinson, S. TBK1 kinase addiction in lung cancer cells is mediated via autophagy of Tax1bp1/Ndp52 and non-canonical NF-κB signalling. PLoS ONE 2012, 7, e50672. [Google Scholar] [CrossRef] [PubMed]
- Sarraf, S.A.; Shah, H.V.; Kanfer, G.; Pickrell, A.M.; Holtzclaw, L.A.; Ward, M.E.; Youle, R.J. Loss of TAX1BP1-Directed Autophagy Results in Protein Aggregate Accumulation in the Brain. Mol. Cell 2020, 80, 779–795.e710. [Google Scholar] [CrossRef] [PubMed]
- White, J.; Suklabaidya, S.; Vo, M.T.; Choi, Y.B.; Harhaj, E.W. Multifaceted roles of TAX1BP1 in autophagy. Autophagy 2023, 19, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Lu, C.; Chen, Y.; Li, S.; Ma, N.; Tao, X.; Li, Y.; Wang, J.; Zhou, M.; Yan, Y.B.; et al. CCT2 is an aggrephagy receptor for clearance of solid protein aggregates. Cell 2022, 185, 1325–1345.e1322. [Google Scholar] [CrossRef] [PubMed]
- Conway, O.; Akpinar, H.A.; Rogov, V.V.; Kirkin, V. Selective Autophagy Receptors in Neuronal Health and Disease. J. Mol. Biol. 2020, 432, 2483–2509. [Google Scholar] [CrossRef] [PubMed]
- Sciarretta, S.; Maejima, Y.; Zablocki, D.; Sadoshima, J. The Role of Autophagy in the Heart. Annu. Rev. Physiol. 2018, 80, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Kitada, M.; Koya, D. Autophagy in metabolic disease and ageing. Nat. Rev. Endocrinol. 2021, 17, 647–661. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Wu, H.; Chen, Y.; Zhang, J.; Zheng, M.; Chen, G.; Li, L.; Lu, Q. The Therapeutic and Pathogenic Role of Autophagy in Autoimmune Diseases. Front. Immunol. 2018, 9, 1512. [Google Scholar] [CrossRef] [PubMed]
- Amaravadi, R.K.; Kimmelman, A.C.; Debnath, J. Targeting Autophagy in Cancer: Recent Advances and Future Directions. Cancer Discov 2019, 9, 1167–1181. [Google Scholar] [CrossRef] [PubMed]
- Papandreou, M.E.; Tavernarakis, N. Selective Autophagy as a Potential Therapeutic Target in Age-Associated Pathologies. Metabolites 2021, 11, 588. [Google Scholar] [CrossRef] [PubMed]
- Kocak, M.; Ezazi Erdi, S.; Jorba, G.; Maestro, I.; Farrés, J.; Kirkin, V.; Martinez, A.; Pless, O. Targeting autophagy in disease: Established and new strategies. Autophagy 2022, 18, 473–495. [Google Scholar] [CrossRef]
- Lu, G.; Wang, Y.; Shi, Y.; Zhang, Z.; Huang, C.; He, W.; Wang, C.; Shen, H.M. Autophagy in health and disease: From molecular mechanisms to therapeutic target. MedComm (2020) 2022, 3, e150. [Google Scholar] [CrossRef]
- Saunders, R.N.; Metcalfe, M.S.; Nicholson, M.L. Rapamycin in transplantation: A review of the evidence. Kidney Int. 2001, 59, 3–16. [Google Scholar] [CrossRef]
- Bhaoighill, M.N.; Dunlop, E.A. Mechanistic target of rapamycin inhibitors: Successes and challenges as cancer therapeutics. Cancer Drug Resist 2019, 2, 1069–1085. [Google Scholar] [CrossRef]
- Liu, Q.; Chang, J.W.; Wang, J.; Kang, S.A.; Thoreen, C.C.; Markhard, A.; Hur, W.; Zhang, J.; Sim, T.; Sabatini, D.M.; et al. Discovery of 1-(4-(4-propionylpiperazin-1-yl)-3-(trifluoromethyl)phenyl)-9-(quinolin-3-yl)benzo[h][1,6]naphthyridin-2(1H)-one as a highly potent, selective mammalian target of rapamycin (mTOR) inhibitor for the treatment of cancer. J. Med. Chem. 2010, 53, 7146–7155. [Google Scholar] [CrossRef]
- Wang, C.; Wang, X.; Su, Z.; Fei, H.; Liu, X.; Pan, Q. The novel mTOR inhibitor Torin-2 induces autophagy and downregulates the expression of UHRF1 to suppress hepatocarcinoma cell growth. Oncol. Rep. 2015, 34, 1708–1716. [Google Scholar] [CrossRef] [PubMed]
- Verbaanderd, C.; Maes, H.; Schaaf, M.B.; Sukhatme, V.P.; Pantziarka, P.; Sukhatme, V.; Agostinis, P.; Bouche, G. Repurposing Drugs in Oncology (ReDO)-chloroquine and hydroxychloroquine as anti-cancer agents. Ecancermedicalscience 2017, 11, 781. [Google Scholar] [CrossRef] [PubMed]
- Kalid, O.; Gotliv, I.; Levy-Apter, E.; Beker, D.F.; Cherniavsky-Lev, M.; Rotem, E.; Miron, N. PTX80, a novel compound targeting the autophagy receptor p62/SQSTM1 for treatment of cancer. Chem. Biol. Drug. Des. 2022, 100, 623–638. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Cho, W.J.; Najy, A.J.; Saliganan, A.D.; Pham, T.; Rakowski, J.; Loughery, B.; Ji, C.H.; Sakr, W.; Kim, S.; et al. p62/SQSTM1-induced caspase-8 aggresomes are essential for ionizing radiation-mediated apoptosis. Cell Death. Dis. 2021, 12, 997. [Google Scholar] [CrossRef]
- Dagvadorj, A.; Olivé, M.; Urtizberea, J.A.; Halle, M.; Shatunov, A.; Bönnemann, C.; Park, K.Y.; Goebel, H.H.; Ferrer, I.; Vicart, P.; et al. A series of West European patients with severe cardiac and skeletal myopathy associated with a de novo R406W mutation in desmin. J. Neurol. 2004, 251, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Selcen, D.; Ohno, K.; Engel, A.G. Myofibrillar myopathy: Clinical, morphological and genetic studies in 63 patients. Brain 2004, 127, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Selcen, D. Myofibrillar myopathies. Neuromuscul. Disord. 2011, 21, 161–171. [Google Scholar] [CrossRef]
- Hedberg-Oldfors, C.; Meyer, R.; Nolte, K.; Abdul Rahim, Y.; Lindberg, C.; Karason, K.; Thuestad, I.J.; Visuttijai, K.; Geijer, M.; Begemann, M.; et al. Loss of supervillin causes myopathy with myofibrillar disorganization and autophagic vacuoles. Brain 2020, 143, 2406–2420. [Google Scholar] [CrossRef]
- Dafsari, H.S.; Kocaturk, N.M.; Daimagüler, H.S.; Brunn, A.; Dötsch, J.; Weis, J.; Deckert, M.; Cirak, S. Bi-allelic mutations in uncoordinated mutant number-45 myosin chaperone B are a cause for congenital myopathy. Acta Neuropathol. Commun. 2019, 7, 211. [Google Scholar] [CrossRef]
- Winter, L.; Goldmann, W.H. Biomechanical characterization of myofibrillar myopathies. Cell Biol. Int. 2015, 39, 361–363. [Google Scholar] [CrossRef] [PubMed]
- Straussberg, R.; Schottmann, G.; Sadeh, M.; Gill, E.; Seifert, F.; Halevy, A.; Qassem, K.; Rendu, J.; van der Ven, P.F.; Stenzel, W.; et al. Kyphoscoliosis peptidase (KY) mutation causes a novel congenital myopathy with core targetoid defects. Acta Neuropathol. 2016, 132, 475–478. [Google Scholar] [CrossRef] [PubMed]
- Gundesli, H.; Talim, B.; Korkusuz, P.; Balci-Hayta, B.; Cirak, S.; Akarsu, N.A.; Topaloglu, H.; Dincer, P. Mutation in exon 1f of PLEC, leading to disruption of plectin isoform 1f, causes autosomal-recessive limb-girdle muscular dystrophy. Am. J. Hum. Genet. 2010, 87, 834–841. [Google Scholar] [CrossRef]
- O’Grady, G.L.; Best, H.A.; Sztal, T.E.; Schartner, V.; Sanjuan-Vazquez, M.; Donkervoort, S.; Abath Neto, O.; Sutton, R.B.; Ilkovski, B.; Romero, N.B.; et al. Variants in the Oxidoreductase PYROXD1 Cause Early-Onset Myopathy with Internalized Nuclei and Myofibrillar Disorganization. Am. J. Hum. Genet. 2016, 99, 1086–1105. [Google Scholar] [CrossRef] [PubMed]
- De Bleecker, J.L.; Engel, A.G.; Ertl, B.B. Myofibrillar myopathy with abnormal foci of desmin positivity. II. Immunocytochemical analysis reveals accumulation of multiple other proteins. J. Neuropathol. Exp. Neurol. 1996, 55, 563–577. [Google Scholar] [CrossRef]
- Ferrer, I.; Martín, B.; Castaño, J.G.; Lucas, J.J.; Moreno, D.; Olivé, M. Proteasomal expression, induction of immunoproteasome subunits, and local MHC class I presentation in myofibrillar myopathy and inclusion body myositis. J. Neuropathol. Exp. Neurol. 2004, 63, 484–498. [Google Scholar] [CrossRef] [PubMed]
- Olivé, M.; van Leeuwen, F.W.; Janué, A.; Moreno, D.; Torrejón-Escribano, B.; Ferrer, I. Expression of mutant ubiquitin (UBB+1) and p62 in myotilinopathies and desminopathies. Neuropathol. Appl. Neurobiol. 2008, 34, 76–87. [Google Scholar] [CrossRef]
- Dubowitz, V.; Sewry, C.; Oldfors, A. Muscle Biopsy A Practical Approach, 5th ed.; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Machado, P.M.; Dimachkie, M.M.; Barohn, R.J. Sporadic inclusion body myositis: New insights and potential therapy. Curr. Opin. Neurol. 2014, 27, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Dimachkie, M.M. Idiopathic inflammatory myopathies. J. Neuroimmunol. 2011, 231, 32–42. [Google Scholar] [CrossRef]
- Dalakas, M.C. Mechanisms of disease: Signaling pathways and immunobiology of inflammatory myopathies. Nat. Clin. Pract. Rheumatol. 2006, 2, 219–227. [Google Scholar] [CrossRef]
- Brady, S.; Squier, W.; Sewry, C.; Hanna, M.; Hilton-Jones, D.; Holton, J.L. A retrospective cohort study identifying the principal pathological features useful in the diagnosis of inclusion body myositis. BMJ Open 2014, 4, e004552. [Google Scholar] [CrossRef] [PubMed]
- Rose, M.R.; Group, E.I.W. 188th ENMC International Workshop: Inclusion Body Myositis, 2-4 December 2011, Naarden, The Netherlands. Neuromuscul. Disord. 2013, 23, 1044–1055. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, S.A. Inclusion body myositis: Clinical features and pathogenesis. Nat. Rev. Rheumatol. 2019, 15, 257–272. [Google Scholar] [CrossRef]
- Nogalska, A.; Terracciano, C.; D’Agostino, C.; King Engel, W.; Askanas, V. p62/SQSTM1 is overexpressed and prominently accumulated in inclusions of sporadic inclusion-body myositis muscle fibers, and can help differentiating it from polymyositis and dermatomyositis. Acta Neuropathol. 2009, 118, 407–413. [Google Scholar] [CrossRef]
- Nogalska, A.; D’Agostino, C.; Terracciano, C.; Engel, W.K.; Askanas, V. Impaired autophagy in sporadic inclusion-body myositis and in endoplasmic reticulum stress-provoked cultured human muscle fibers. Am. J. Pathol. 2010, 177, 1377–1387. [Google Scholar] [CrossRef] [PubMed]
- D’Agostino, C.; Nogalska, A.; Cacciottolo, M.; Engel, W.K.; Askanas, V. Abnormalities of NBR1, a novel autophagy-associated protein, in muscle fibers of sporadic inclusion-body myositis. Acta Neuropathol. 2011, 122, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Nicot, A.S.; Lo Verso, F.; Ratti, F.; Pilot-Storck, F.; Streichenberger, N.; Sandri, M.; Schaeffer, L.; Goillot, E. Phosphorylation of NBR1 by GSK3 modulates protein aggregation. Autophagy 2014, 10, 1036–1053. [Google Scholar] [CrossRef] [PubMed]
- Britson, K.A.; Yang, S.Y.; Lloyd, T.E. New Developments in the Genetics of Inclusion Body Myositis. Curr. Rheumatol. Rep. 2018, 20, 26. [Google Scholar] [CrossRef]
- Sabatelli, P.; Castagnaro, S.; Tagliavini, F.; Chrisam, M.; Sardone, F.; Demay, L.; Richard, P.; Santi, S.; Maraldi, N.M.; Merlini, L.; et al. Aggresome-Autophagy Involvement in a Sarcopenic Patient with Rigid Spine Syndrome and a p.C150R Mutation in FHL1 Gene. Front. Aging Neurosci. 2014, 6, 215. [Google Scholar] [CrossRef]
- Schessl, J.; Columbus, A.; Hu, Y.; Zou, Y.; Voit, T.; Goebel, H.H.; Bönnemann, C.G. Familial reducing body myopathy with cytoplasmic bodies and rigid spine revisited: Identification of a second LIM domain mutation in FHL1. Neuropediatrics 2010, 41, 43–46. [Google Scholar] [CrossRef]
- Selcen, D.; Bromberg, M.B.; Chin, S.S.; Engel, A.G. Reducing bodies and myofibrillar myopathy features in FHL1 muscular dystrophy. Neurology 2011, 77, 1951–1959. [Google Scholar] [CrossRef] [PubMed]
- Victor, M.; Hayes, R.; Adams, R.D. Oculopharyngeal muscular dystrophy. A familial disease of late life characterized by dysphagia and progressive ptosis of the evelids. N. Engl. J. Med. 1962, 267, 1267–1272. [Google Scholar] [CrossRef] [PubMed]
- Anvar, S.Y.; Raz, Y.; Verway, N.; van der Sluijs, B.; Venema, A.; Goeman, J.J.; Vissing, J.; van der Maarel, S.M.; ‘t Hoen, P.A.; van Engelen, B.G.; et al. A decline in PABPN1 induces progressive muscle weakness in oculopharyngeal muscle dystrophy and in muscle aging. Aging 2013, 5, 412–426. [Google Scholar] [CrossRef]
- Beaulieu, Y.B.; Kleinman, C.L.; Landry-Voyer, A.M.; Majewski, J.; Bachand, F. Polyadenylation-dependent control of long noncoding RNA expression by the poly(A)-binding protein nuclear 1. PLoS Genet. 2012, 8, e1003078. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Yang, K.; Lin, M.T.; Zheng, F.Z.; Chen, L.; Ding, Y.L.; Ye, Z.X.; Lin, X.; Wang, N.; Wang, Z.Q. The phenotypic and genotypic features of Chinese patients with oculopharyngeal muscular dystrophy. Ann. Clin. Transl. Neurol. 2023, 10, 426–439. [Google Scholar] [CrossRef]
- Askanas, V.; Serdaroglu, P.; Engel, W.K.; Alvarez, R.B. Immunolocalization of ubiquitin in muscle biopsies of patients with inclusion body myositis and oculopharyngeal muscular dystrophy. Neurosci. Lett. 1991, 130, 73–76. [Google Scholar] [CrossRef]
- Abu-Baker, A.; Messaed, C.; Laganiere, J.; Gaspar, C.; Brais, B.; Rouleau, G.A. Involvement of the ubiquitin-proteasome pathway and molecular chaperones in oculopharyngeal muscular dystrophy. Hum. Mol. Genet. 2003, 12, 2609–2623. [Google Scholar] [CrossRef]
- Yang, K.; Zeng, Y.H.; Qiu, Y.S.; Lin, F.; Chen, H.Z.; Jin, M.; Chen, L.; Zheng, F.Z.; Ding, Y.L.; Cao, C.Y.; et al. Expanding the phenotype and genotype spectra of PLIN4-associated myopathy with rimmed ubiquitin-positive autophagic vacuolation. Acta Neuropathol. 2022, 143, 733–735. [Google Scholar] [CrossRef]
- Wang, Q.; Yu, M.; Zhang, W.; Gang, Q.; Xie, Z.; Xu, J.; Zhou, C.; Wang, D.; Meng, L.; Lv, H.; et al. Subsarcolemmal and cytoplasmic p62 positivity and rimmed vacuoles are distinctive for PLIN4-myopathy. Ann. Clin. Transl. Neurol. 2022, 9, 1813–1819. [Google Scholar] [CrossRef]
- Tankink, M.; Horlings, C.G.C.; Voermans, N.; van der Sluijs, B.; Kessels, R.P.C.; van Engelen, B.; Raaphorst, J. Behavioural Impairment and Frontotemporal Dementia in Oculopharyngeal Muscular Dystrophy. J. Neuromuscul. Dis. 2022, 9, 129–135. [Google Scholar] [CrossRef]
- Olivé, M.; Goldfarb, L.G.; Shatunov, A.; Fischer, D.; Ferrer, I. Myotilinopathy: Refining the clinical and myopathological phenotype. Brain 2005, 128, 2315–2326. [Google Scholar] [CrossRef] [PubMed]
- van der Klooster, Z.J.; Sepehrkhouy, S.; Dooijes, D.; Te Rijdt, W.P.; Schuiringa, F.S.A.M.; Lingeman, J.; van Tintelen, J.P.; Harakalova, M.; Goldschmeding, R.; Suurmeijer, A.J.H.; et al. P62-positive aggregates are homogenously distributed in the myocardium and associated with the type of mutation in genetic cardiomyopathy. J. Cell Mol. Med. 2021, 25, 3160–3166. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Su, H.; Ranek, M.J.; Wang, X. Autophagy and p62 in cardiac proteinopathy. Circ. Res. 2011, 109, 296–308. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).