Ubiquitination Links DNA Damage and Repair Signaling to Cancer Metabolism
Abstract
1. Introduction
2. The Effects of Ubiquitination on Cancer Metabolism
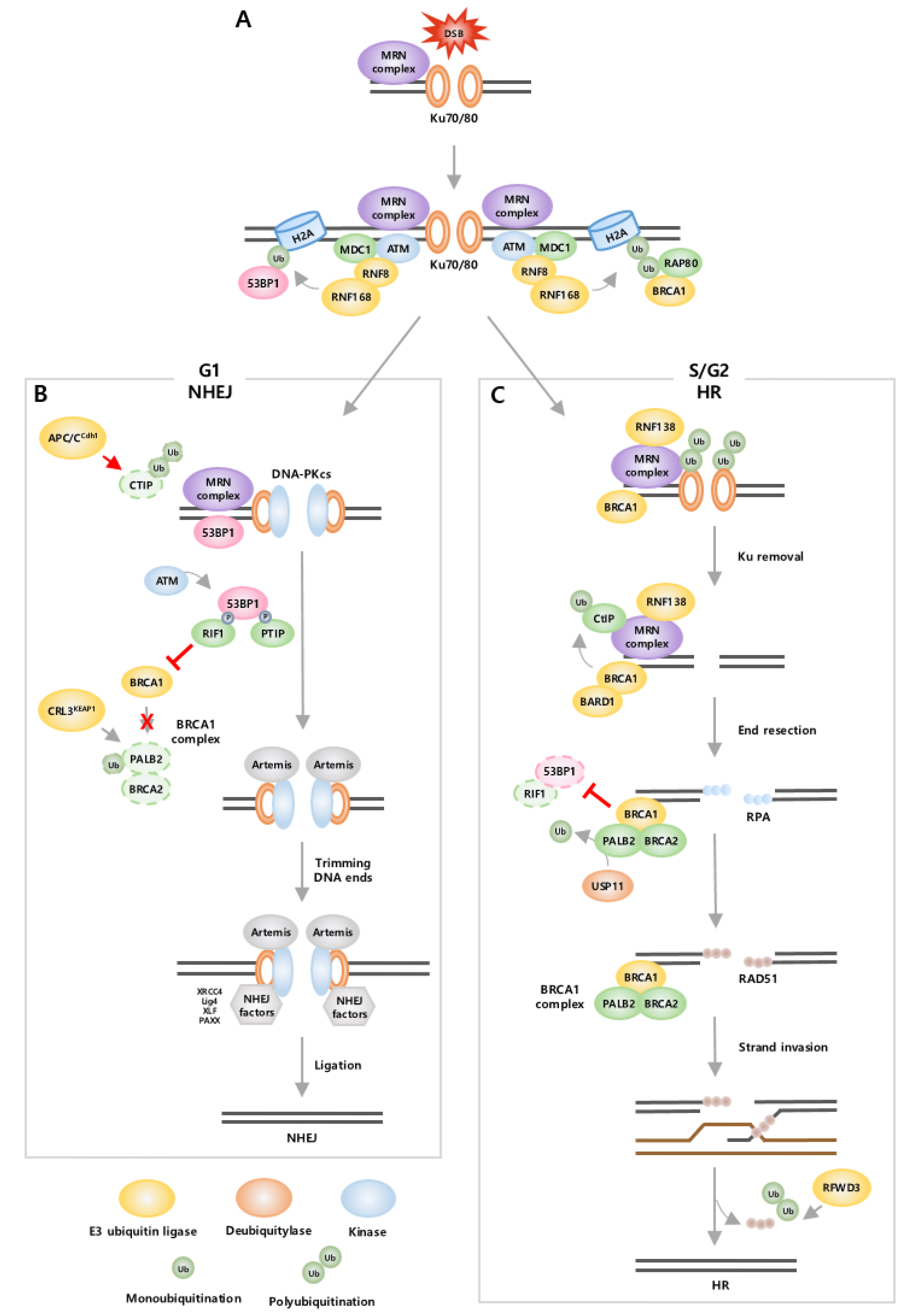
3. The Effects of Ubiquitination on DNA Damage and DNA Repair
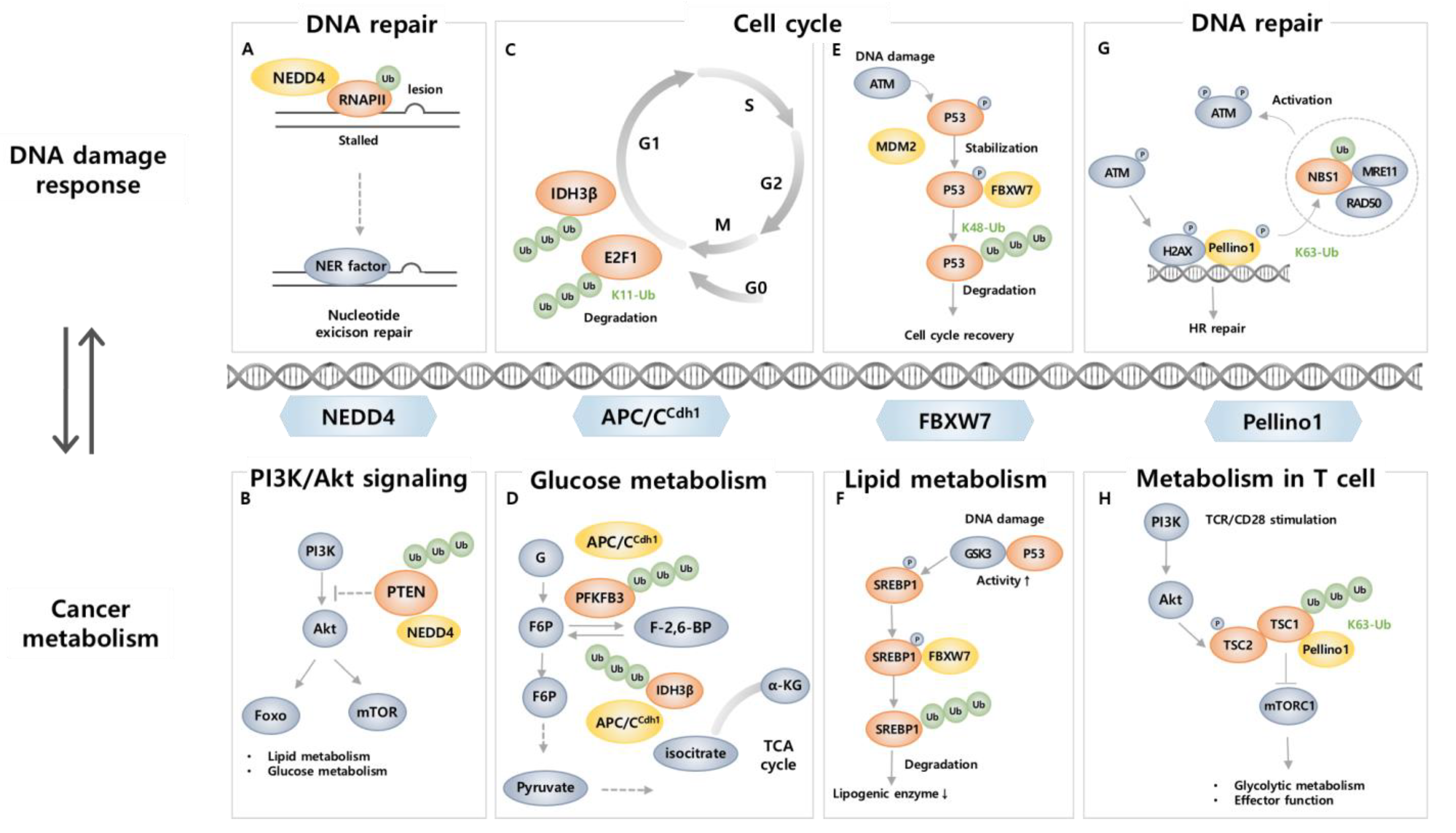
4. E3 Ligase Involved in Cancer Metabolism and DNA Damage
4.1. NEDD4
4.2. APC/CCDH1
4.3. FBXW7
4.4. Pellino1
5. Current Therapeutic Implications in Cancer Targeting Ubiquitination
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Deng, L.; Meng, T.; Chen, L.; Wei, W.; Wang, P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal. Transduct. Target. Ther. 2020, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Gu, H.; Peng, C.; Xia, F.; Cao, H.; Cui, H. Regulation of Glucose, Fatty Acid and Amino Acid Metabolism by Ubiquitination and SUMOylation for Cancer Progression. Front. Cell Dev. Biol. 2022, 10, 849625. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Liu, Z.; Yang, Q. The role of ubiquitination and deubiquitination in cancer metabolism. Mol. Cancer 2020, 19, 146. [Google Scholar] [CrossRef]
- Chan, C.H.; Li, C.F.; Yang, W.L.; Gao, Y.; Lee, S.W.; Feng, Z.; Huang, H.Y.; Tsai, K.K.C.; Flores, L.G.; Shao, Y.; et al. The Skp2-SCF E3 Ligase Regulates Akt Ubiquitination, Glycolysis, Herceptin Sensitivity, and Tumorigenesis. Cell 2012, 151, 913–914. [Google Scholar] [CrossRef] [PubMed]
- Lilley, C.E.; Chaurushiya, M.S.; Boutell, C.; Landry, S.; Suh, J.; Panier, S.; Everett, R.D.; Stewart, G.S.; Durocher, D.; Weitzman, M.D. A viral E3 ligase targets RNF8 and RNF168 to control histone ubiquitination and DNA damage responses. EMBO J. 2010, 29, 943–955. [Google Scholar] [CrossRef] [PubMed]
- Schwertman, P.; Bekker-Jensen, S.; Mailand, N. Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat. Rev. Mol. Cell Biol. 2016, 17, 379–394. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Qin, B.; Lou, Z. Ubiquitin and ubiquitin-like molecules in DNA double strand break repair. Cell Biosci. 2020, 10, 13. [Google Scholar] [CrossRef]
- Sobanski, T.; Rose, M.; Suraweera, A.; O’Byrne, K.; Richard, D.J.; Bolderson, E. Cell Metabolism and DNA Repair Pathways: Implications for Cancer Therapy. Front. Cell Dev. Biol. 2021, 9, 633305. [Google Scholar] [CrossRef]
- Senft, D.; Qi, J.; Ronai, Z.A. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat. Rev. Cancer 2018, 18, 69–88. [Google Scholar] [CrossRef]
- Liu, W.; Zheng, M.; Zhang, R.; Jiang, Q.; Du, G.; Wu, Y.; Yang, C.; Li, F.; Li, W.; Wang, L.; et al. RNF126-Mediated MRE11 Ubiquitination Activates the DNA Damage Response and Confers Resistance of Triple-Negative Breast Cancer to Radiotherapy. Adv. Sci. 2023, 10, e2203884. [Google Scholar] [CrossRef]
- Zhao, Y.; Aguilar, A.; Bernard, D.; Wang, S. Small-molecule inhibitors of the MDM2-p53 protein-protein interaction (MDM2 Inhibitors) in clinical trials for cancer treatment. J. Med. Chem. 2015, 58, 1038–1052. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Reyes, I.; Chandel, N.S. Cancer metabolism: Looking forward. Nat. Rev. Cancer 2021, 21, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, A.; Liu, J.; Zhou, Y.; Oltvai, Z.N. Catabolic efficiency of aerobic glycolysis: The Warburg effect revisited. BMC Syst. Biol. 2010, 4, 58. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.M.; Mucka, P.; Kern, J.G.; Feng, H. The emerging role and targetability of the TCA cycle in cancer metabolism. Protein Cell 2018, 9, 216–237. [Google Scholar] [CrossRef]
- Yang, Q.; Zhao, J.; Chen, D.; Wang, Y. E3 ubiquitin ligases: Styles, structures and functions. Mol. Biomed. 2021, 2, 23. [Google Scholar] [CrossRef]
- Zheng, J. Energy metabolism of cancer: Glycolysis versus oxidative phosphorylation (Review). Oncol. Lett. 2012, 4, 1151–1157. [Google Scholar] [CrossRef]
- Jiao, L.; Zhang, H.L.; Li, D.D.; Yang, K.L.; Tang, J.; Li, X.; Ji, J.; Yu, Y.; Wu, R.Y.; Ravichandran, S.; et al. Regulation of glycolytic metabolism by autophagy in liver cancer involves selective autophagic degradation of HK2 (hexokinase 2). Autophagy 2018, 14, 671–684. [Google Scholar] [CrossRef]
- Lee, H.J.; Li, C.F.; Ruan, D.; He, J.; Montal, E.D.; Lorenz, S.; Girnun, G.D.; Chan, C.H. Non-proteolytic ubiquitination of Hexokinase 2 by HectH9 controls tumor metabolism and cancer stem cell expansion. Nat. Commun. 2019, 10, 2625. [Google Scholar] [CrossRef]
- Bao, C.; Zhu, S.; Song, K.; He, C. HK2: A potential regulator of osteoarthritis via glycolytic and non-glycolytic pathways. Cell Commun. Signal. 2022, 20, 132. [Google Scholar] [CrossRef]
- Park, J.S.; Burckhardt, C.J.; Lazcano, R.; Solis, L.M.; Isogai, T.; Li, L.; Chen, C.S.; Gao, B.; Minna, J.D.; Bachoo, R.; et al. Mechanical regulation of glycolysis via cytoskeleton architecture. Nature 2020, 578, 621–626. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, Y.; Cai, Y.; Liu, R.; Lu, M.; Li, T.; Fu, Y.; Guo, M.; Huang, H.; Ou, Y.; et al. A20 targets PFKL and glycolysis to inhibit the progression of hepatocellular carcinoma. Cell Death Dis. 2020, 11, 89. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; He, D.; He, K.; Zhang, Q.; Tang, M.; Dai, J.; Lv, H.; Wang, X.; Xiang, G.; Yu, H. Downregulation of TRIM21 contributes to hepatocellular carcinoma carcinogenesis and indicates poor prognosis of cancers. Tumour Biol. 2015, 36, 8761–8772. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Liu, R.; Li, J.; Zhang, C.; Wang, Y.; Cai, Q.; Qian, X.; Xia, Y.; Zheng, Y.; Piao, Y.; et al. Stabilization of phosphofructokinase 1 platelet isoform by AKT promotes tumorigenesis. Nat. Commun. 2017, 8, 949. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Li, F.; Han, H.; Chen, Y.; Mao, Z.; Luo, J.; Zhao, Y.; Zheng, B.; Gu, W.; Zhao, W. Parkin Regulates the Activity of Pyruvate Kinase M2. J. Biol. Chem. 2016, 291, 10307–10317. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; He, J.; Wang, Y.; Feng, Q.; Zhang, Y.; Guo, J.; Li, J.; Li, S.; Wang, Y.; Yan, G.; et al. CHIP/Stub1 regulates the Warburg effect by promoting degradation of PKM2 in ovarian carcinoma. Oncogene 2017, 36, 4191–4200. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.; Zhou, Y.; Wang, R.; Chen, S.; Wang, Q.; Xu, Z.; Liu, Y.; Yang, H. TRIM58 Interacts with Pyruvate Kinase M2 to Inhibit Tumorigenicity in Human Osteosarcoma Cells. Biomed. Res. Int. 2020, 2020, 8450606. [Google Scholar] [CrossRef]
- Qiu, X.; Huang, Y.; Zhou, Y.; Zheng, F. Aberrant methylation of TRIM58 in hepatocellular carcinoma and its potential clinical implication. Oncol. Rep. 2016, 36, 811–818. [Google Scholar] [CrossRef]
- Kajiura, K.; Masuda, K.; Naruto, T.; Kohmoto, T.; Watabnabe, M.; Tsuboi, M.; Takizawa, H.; Kondo, K.; Tangoku, A.; Imoto, I. Frequent silencing of the candidate tumor suppressor TRIM58 by promoter methylation in early-stage lung adenocarcinoma. Oncotarget 2017, 8, 2890–2905. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, X.; Cai, J.; Li, Y.; Luo, Q.; Wu, H.; Yang, Z.; Wang, L.; Chen, D. Downregulation of TRIM58 expression is associated with a poor patient outcome and enhances colorectal cancer cell invasion. Oncol. Rep. 2018, 40, 1251–1260. [Google Scholar] [CrossRef]
- Chu, Z.; Huo, N.; Zhu, X.; Liu, H.; Cong, R.; Ma, L.; Kang, X.; Xue, C.; Li, J.; Li, Q.; et al. FOXO3A-induced LINC00926 suppresses breast tumor growth and metastasis through inhibition of PGK1-mediated Warburg effect. Mol. Ther. 2021, 29, 2737–2753. [Google Scholar] [CrossRef]
- Cai, Q.; Wang, S.; Jin, L.; Weng, M.; Zhou, D.; Wang, J.; Tang, Z.; Quan, Z. Long non-coding RNA GBCDRlnc1 induces chemoresistance of gallbladder cancer cells by activating autophagy. Mol. Cancer 2019, 18, 82. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Luo, Y.; Zhang, D.; Wang, X.; Zhang, P.; Li, H.; Ejaz, S.; Liang, S. PGK1-mediated cancer progression and drug resistance. Am. J. Cancer Res. 2019, 9, 2280–2302. [Google Scholar] [PubMed]
- Navale, A.M.; Paranjape, A.N. Glucose transporters: Physiological and pathological roles. Biophys. Rev. 2016, 8, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Revathidevi, S.; Munirajan, A.K. Akt in cancer: Mediator and more. Semin. Cancer Biol. 2019, 59, 80–91. [Google Scholar] [CrossRef]
- Yu, L.; Chen, X.; Sun, X.; Wang, L.; Chen, S. The Glycolytic Switch in Tumors: How Many Players Are Involved? J. Cancer 2017, 8, 3430–3440. [Google Scholar] [CrossRef]
- Eniafe, J.; Jiang, S. The functional roles of TCA cycle metabolites in cancer. Oncogene 2021, 40, 3351–3363. [Google Scholar] [CrossRef]
- Nakatsukasa, K.; Nishimura, T.; Byrne, S.D.; Okamoto, M.; Takahashi-Nakaguchi, A.; Chibana, H.; Okumura, F.; Kamura, T. The Ubiquitin Ligase SCF(Ucc1) Acts as a Metabolic Switch for the Glyoxylate Cycle. Mol. Cell 2015, 59, 22–34. [Google Scholar] [CrossRef]
- Peng, M.; Yang, D.; Hou, Y.; Liu, S.; Zhao, M.; Qin, Y.; Chen, R.; Teng, Y.; Liu, M. Intracellular citrate accumulation by oxidized ATM-mediated metabolism reprogramming via PFKP and CS enhances hypoxic breast cancer cell invasion and metastasis. Cell Death Dis. 2019, 10, 228. [Google Scholar] [CrossRef]
- Lin, C.C.; Cheng, T.L.; Tsai, W.H.; Tsai, H.J.; Hu, K.H.; Chang, H.C.; Yeh, C.W.; Chen, Y.C.; Liao, C.C.; Chang, W.T. Loss of the respiratory enzyme citrate synthase directly links the Warburg effect to tumor malignancy. Sci. Rep. 2012, 2, 785. [Google Scholar] [CrossRef]
- Huang, L.; Wang, C.; Xu, H.; Peng, G. Targeting citrate as a novel therapeutic strategy in cancer treatment. Biochim. Biophys. Acta Rev. Cancer 2020, 1873, 188332. [Google Scholar] [CrossRef]
- Yang, H.; Ye, D.; Guan, K.L.; Xiong, Y. IDH1 and IDH2 mutations in tumorigenesis: Mechanistic insights and clinical perspectives. Clin. Cancer Res. 2012, 18, 5562–5571. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Hu, N.; Wang, W.; Hu, W.; Zhou, S.; Shi, J.; Li, M.; Jing, Z.; Chen, C.; Zhang, X.; et al. Loss of FBXW7 Correlates with Increased IDH1 Expression in Glioma and Enhances IDH1-Mutant Cancer Cell Sensitivity to Radiation. Cancer Res. 2022, 82, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Lambhate, S.; Bhattacharjee, D.; Jain, N. APC/C CDH1 ubiquitinates IDH2 contributing to ROS increase in mitosis. Cell. Signal. 2021, 86, 110087. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.C.; Denko, N.C. Hypoxic regulation of glutamine metabolism through HIF1 and SIAH2 supports lipid synthesis that is necessary for tumor growth. Cell. Metab. 2014, 19, 285–292. [Google Scholar] [CrossRef]
- Vatrinet, R.; Leone, G.; De Luise, M.; Girolimetti, G.; Vidone, M.; Gasparre, G.; Porcelli, A.M. The alpha-ketoglutarate dehydrogenase complex in cancer metabolic plasticity. Cancer Metab. 2017, 5, 3. [Google Scholar] [CrossRef]
- Zhao, T.; Mu, X.; You, Q. Succinate: An initiator in tumorigenesis and progression. Oncotarget 2017, 8, 53819–53828. [Google Scholar] [CrossRef]
- Ferreira, J.V.; Soares, A.R.; Ramalho, J.S.; Pereira, P.; Girao, H. K63 linked ubiquitin chain formation is a signal for HIF1A degradation by Chaperone-Mediated Autophagy. Sci. Rep. 2015, 5, 10210. [Google Scholar] [CrossRef]
- Merlo, A.; Bernardo-Castineira, C.; Saenz-de-Santa-Maria, I.; Pitiot, A.S.; Balbin, M.; Astudillo, A.; Valdes, N.; Scola, B.; Del Toro, R.; Mendez-Ferrer, S.; et al. Role of VHL, HIF1A and SDH on the expression of miR-210: Implications for tumoral pseudo-hypoxic fate. Oncotarget 2017, 8, 6700–6717. [Google Scholar] [CrossRef]
- Fu, Y.; Zou, T.; Shen, X.; Nelson, P.J.; Li, J.; Wu, C.; Yang, J.; Zheng, Y.; Bruns, C.; Zhao, Y.; et al. Lipid metabolism in cancer progression and therapeutic strategies. MedComm 2021, 2, 27–59. [Google Scholar] [CrossRef]
- Vasseur, S.; Guillaumond, F. Lipids in cancer: A global view of the contribution of lipid pathways to metastatic formation and treatment resistance. Oncogenesis 2022, 11, 46. [Google Scholar] [CrossRef]
- Ameer, F.; Scandiuzzi, L.; Hasnain, S.; Kalbacher, H.; Zaidi, N. De novo lipogenesis in health and disease. Metabolism 2014, 63, 895–902. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wang, A.; Wang, W.; Wang, Y.; Chen, H.; Liu, X.; Xia, T.; Zhang, A.; Chen, D.; Qi, H.; et al. HRD1 inhibits fatty acid oxidation and tumorigenesis by ubiquitinating CPT2 in triple-negative breast cancer. Mol. Oncol. 2021, 15, 642–656. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Temkin, S.M.; Hawkridge, A.M.; Guo, C.; Wang, W.; Wang, X.Y.; Fang, X. Fatty acid oxidation: An emerging facet of metabolic transformation in cancer. Cancer Lett. 2018, 435, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Bai, L.; Li, W.; Cui, J. The Lipid Metabolic Landscape of Cancers and New Therapeutic Perspectives. Front. Oncol. 2020, 10, 605154. [Google Scholar] [CrossRef]
- Han, C.; Yang, L.; Choi, H.H.; Baddour, J.; Achreja, A.; Liu, Y.; Li, Y.; Li, J.; Wan, G.; Huang, C.; et al. Amplification of USP13 drives ovarian cancer metabolism. Nat. Commun. 2016, 7, 13525. [Google Scholar] [CrossRef]
- Gu, L.; Zhu, Y.; Lin, X.; Lu, B.; Zhou, X.; Zhou, F.; Zhao, Q.; Prochownik, E.V.; Li, Y. The IKKbeta-USP30-ACLY Axis Controls Lipogenesis and Tumorigenesis. Hepatology 2021, 73, 160–174. [Google Scholar] [CrossRef]
- Tian, M.; Hao, F.; Jin, X.; Sun, X.; Jiang, Y.; Wang, Y.; Li, D.; Chang, T.; Zou, Y.; Peng, P.; et al. ACLY ubiquitination by CUL3-KLHL25 induces the reprogramming of fatty acid metabolism to facilitate iTreg differentiation. Elife 2021, 10, e62394. [Google Scholar] [CrossRef]
- Simeone, P.; Tacconi, S.; Longo, S.; Lanuti, P.; Bravaccini, S.; Pirini, F.; Ravaioli, S.; Dini, L.; Giudetti, A.M. Expanding Roles of De Novo Lipogenesis in Breast Cancer. Int. J. Environ. Res. Public. Health 2021, 18, 3575. [Google Scholar] [CrossRef]
- Ni, W.; Lin, S.; Bian, S.; Zheng, W.; Qu, L.; Fan, Y.; Lu, C.; Xiao, M.; Zhou, P. USP7 mediates pathological hepatic de novo lipogenesis through promoting stabilization and transcription of ZNF638. Cell Death Dis. 2020, 11, 843. [Google Scholar] [CrossRef]
- Wei, Z.; Liu, X.; Cheng, C.; Yu, W.; Yi, P. Metabolism of Amino Acids in Cancer. Front. Cell Dev. Biol. 2020, 8, 603837. [Google Scholar] [CrossRef]
- Yoo, H.C.; Yu, Y.C.; Sung, Y.; Han, J.M. Glutamine reliance in cell metabolism. Exp. Mol. Med. 2020, 52, 1496–1516. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.E.; Yoo, J.E.; Kim, J.; Kim, S.; Kim, S.; Lee, H.; Cheong, H. NEDD4L downregulates autophagy and cell growth by modulating ULK1 and a glutamine transporter. Cell Death Dis. 2020, 11, 38. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Wang, J.M.; Yan, J.; Zhang, D.L.; Liu, B.Q.; Jiang, J.Y.; Li, C.; Li, S.; Meng, X.N.; Wang, H.Q. BAG3 promotes autophagy and glutaminolysis via stabilizing glutaminase. Cell Death Dis. 2019, 10, 284. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, C.; Wu, H.; Sun, X.X.; Li, Y.; Huang, S.; Yue, X.; Lu, S.E.; Shen, Z.; Su, X.; et al. Parkin ubiquitinates phosphoglycerate dehydrogenase to suppress serine synthesis and tumor progression. J. Clin. Investig. 2020, 130, 3253–3269. [Google Scholar] [CrossRef]
- Labuschagne, C.F.; van den Broek, N.J.; Mackay, G.M.; Vousden, K.H.; Maddocks, O.D. Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 2014, 7, 1248–1258. [Google Scholar] [CrossRef] [PubMed]
- Maddocks, O.D.; Labuschagne, C.F.; Adams, P.D.; Vousden, K.H. Serine Metabolism Supports the Methionine Cycle and DNA/RNA Methylation through De Novo ATP Synthesis in Cancer Cells. Mol. Cell 2016, 61, 210–221. [Google Scholar] [CrossRef]
- Anderson, D.D.; Eom, J.Y.; Stover, P.J. Competition between sumoylation and ubiquitination of serine hydroxymethyltransferase 1 determines its nuclear localization and its accumulation in the nucleus. J. Biol. Chem. 2012, 287, 4790–4799. [Google Scholar] [CrossRef]
- Jain, M.; Nilsson, R.; Sharma, S.; Madhusudhan, N.; Kitami, T.; Souza, A.L.; Kafri, R.; Kirschner, M.W.; Clish, C.B.; Mootha, V.K. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 2012, 336, 1040–1044. [Google Scholar] [CrossRef]
- Nguyen, T.V.; Lee, J.E.; Sweredoski, M.J.; Yang, S.J.; Jeon, S.J.; Harrison, J.S.; Yim, J.H.; Lee, S.G.; Handa, H.; Kuhlman, B.; et al. Glutamine Triggers Acetylation-Dependent Degradation of Glutamine Synthetase via the Thalidomide Receptor Cereblon. Mol. Cell 2016, 61, 809–820. [Google Scholar] [CrossRef]
- Nguyen, T.V. USP15 antagonizes CRL4(CRBN)-mediated ubiquitylation of glutamine synthetase and neosubstrates. Proc. Natl. Acad. Sci. USA 2021, 118, e2111391118. [Google Scholar] [CrossRef]
- Li, Y.; Tong, Y.; Liu, J.; Lou, J. The Role of MicroRNA in DNA Damage Response. Front. Genet. 2022, 13, 850038. [Google Scholar] [CrossRef] [PubMed]
- Dantuma, N.P.; van Attikum, H. Spatiotemporal regulation of posttranslational modifications in the DNA damage response. EMBO J. 2016, 35, 6–23. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Bozzella, M.; Seluanov, A.; Gorbunova, V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle 2008, 7, 2902–2906. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; Chahwan, C.; Bailis, J.; Hunter, T.; Russell, P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell Biol. 2005, 25, 5363–5379. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, X.; Zhang, L.; Wu, C.Y.; Rezaeian, A.H.; Chan, C.H.; Li, J.M.; Wang, J.; Gao, Y.; Han, F.; et al. Skp2 E3 ligase integrates ATM activation and homologous recombination repair by ubiquitinating NBS1. Mol. Cell 2012, 46, 351–361. [Google Scholar] [CrossRef]
- Kim, H.; Kim, D.; Choi, H.; Shin, G.; Lee, J.K. Deubiquitinase USP2 stabilizes the MRE11-RAD50-NBS1 complex at DNA double-strand break sites by counteracting the ubiquitination of NBS1. J. Biol. Chem. 2023, 299, 102752. [Google Scholar] [CrossRef] [PubMed]
- Ha, G.H.; Ji, J.H.; Chae, S.; Park, J.; Kim, S.; Lee, J.K.; Kim, Y.; Min, S.; Park, J.M.; Kang, T.H.; et al. Pellino1 regulates reversible ATM activation via NBS1 ubiquitination at DNA double-strand breaks. Nat. Commun. 2019, 10, 1577. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Huang, J. MRN complex is an essential effector of DNA damage repair. J. Zhejiang Univ. Sci. B 2021, 22, 31–37. [Google Scholar] [CrossRef]
- Stucki, M.; Clapperton, J.A.; Mohammad, D.; Yaffe, M.B.; Smerdon, S.J.; Jackson, S.P. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 2005, 123, 1213–1226. [Google Scholar] [CrossRef]
- Hodge, C.D.; Ismail, I.H.; Edwards, R.A.; Hura, G.L.; Xiao, A.T.; Tainer, J.A.; Hendzel, M.J.; Glover, J.N. RNF8 E3 Ubiquitin Ligase Stimulates Ubc13 E2 Conjugating Activity That Is Essential for DNA Double Strand Break Signaling and BRCA1 Tumor Suppressor Recruitment. J. Biol. Chem. 2016, 291, 9396–9410. [Google Scholar] [CrossRef]
- Nowsheen, S.; Aziz, K.; Aziz, A.; Deng, M.; Qin, B.; Luo, K.; Jeganathan, K.B.; Zhang, H.; Liu, T.; Yu, J.; et al. L3MBTL2 orchestrates ubiquitin signalling by dictating the sequential recruitment of RNF8 and RNF168 after DNA damage. Nat. Cell Biol. 2018, 20, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Borsos, B.N.; Majoros, H.; Pankotai, T. Ubiquitylation-Mediated Fine-Tuning of DNA Double-Strand Break Repair. Cancers 2020, 12, 1617. [Google Scholar] [CrossRef] [PubMed]
- Uckelmann, M.; Sixma, T.K. Histone ubiquitination in the DNA damage response. DNA Repair. 2017, 56, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Gudjonsson, T.; Altmeyer, M.; Savic, V.; Toledo, L.; Dinant, C.; Grofte, M.; Bartkova, J.; Poulsen, M.; Oka, Y.; Bekker-Jensen, S.; et al. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell 2012, 150, 697–709. [Google Scholar] [CrossRef]
- Mosbech, A.; Lukas, C.; Bekker-Jensen, S.; Mailand, N. The deubiquitylating enzyme USP44 counteracts the DNA double-strand break response mediated by the RNF8 and RNF168 ubiquitin ligases. J. Biol. Chem. 2013, 288, 16579–16587. [Google Scholar] [CrossRef]
- Juang, Y.C.; Landry, M.C.; Sanches, M.; Vittal, V.; Leung, C.C.; Ceccarelli, D.F.; Mateo, A.R.; Pruneda, J.N.; Mao, D.Y.; Szilard, R.K.; et al. OTUB1 co-opts Lys48-linked ubiquitin recognition to suppress E2 enzyme function. Mol. Cell 2012, 45, 384–397. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Yin, N.; Xie, T.; Zheng, Y.; Xia, N.; Shang, J.; Chen, F.; Zhang, H.; Yu, J.; Liu, F. Quantitative assessment of HR and NHEJ activities via CRISPR/Cas9-induced oligodeoxynucleotide-mediated DSB repair. DNA Repair 2018, 70, 67–71. [Google Scholar] [CrossRef]
- Escribano-Diaz, C.; Orthwein, A.; Fradet-Turcotte, A.; Xing, M.; Young, J.T.; Tkac, J.; Cook, M.A.; Rosebrock, A.P.; Munro, M.; Canny, M.D.; et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell 2013, 49, 872–883. [Google Scholar] [CrossRef]
- Brandsma, I.; Gent, D.C. Pathway choice in DNA double strand break repair: Observations of a balancing act. Genome Integr. 2012, 3, 9. [Google Scholar] [CrossRef]
- Lei, T.; Du, S.; Peng, Z.; Chen, L. Multifaceted regulation and functions of 53BP1 in NHEJ-mediated DSB repair (Review). Int. J. Mol. Med. 2022, 50, 90. [Google Scholar] [CrossRef]
- Callen, E.; Di Virgilio, M.; Kruhlak, M.J.; Nieto-Soler, M.; Wong, N.; Chen, H.T.; Faryabi, R.B.; Polato, F.; Santos, M.; Starnes, L.M.; et al. 53BP1 mediates productive and mutagenic DNA repair through distinct phosphoprotein interactions. Cell 2013, 153, 1266–1280. [Google Scholar] [CrossRef] [PubMed]
- Orthwein, A.; Noordermeer, S.M.; Wilson, M.D.; Landry, S.; Enchev, R.I.; Sherker, A.; Munro, M.; Pinder, J.; Salsman, J.; Dellaire, G.; et al. A mechanism for the suppression of homologous recombination in G1 cells. Nature 2015, 528, 422–426. [Google Scholar] [CrossRef]
- Wu, S.; Zhou, J.; Zhang, K.; Chen, H.; Luo, M.; Lu, Y.; Sun, Y.; Chen, Y. Molecular Mechanisms of PALB2 Function and Its Role in Breast Cancer Management. Front. Oncol. 2020, 10, 301. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, D.; Furst, A.; Meaburn, K.; Lezaja, A.; Wen, Y.; Altmeyer, M.; Reina-San-Martin, B.; Soutoglou, E. Activation of homologous recombination in G1 preserves centromeric integrity. Nature 2021, 600, 748–753. [Google Scholar] [CrossRef]
- Germani, A.; Prabel, A.; Mourah, S.; Podgorniak, M.P.; Di Carlo, A.; Ehrlich, R.; Gisselbrecht, S.; Varin-Blank, N.; Calvo, F.; Bruzzoni-Giovanelli, H. SIAH-1 interacts with CtIP and promotes its degradation by the proteasome pathway. Oncogene 2003, 22, 8845–8851. [Google Scholar] [CrossRef]
- Zhao, F.; Kim, W.; Kloeber, J.A.; Lou, Z. DNA end resection and its role in DNA replication and DSB repair choice in mammalian cells. Exp. Mol. Med. 2020, 52, 1705–1714. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Fu, S.; Lai, M.; Baer, R.; Chen, J. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes. Dev. 2006, 20, 1721–1726. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.R.; Taylor, M.R.; Boulton, S.J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 2012, 47, 497–510. [Google Scholar] [CrossRef]
- Harris, J.L.; Khanna, K.K. BRCA1 A-complex fine tunes repair functions of BRCA1. Aging 2011, 3, 461–463. [Google Scholar] [CrossRef]
- Savage, K.I.; Harkin, D.P. BRCA1, a ’complex’ protein involved in the maintenance of genomic stability. FEBS J. 2015, 282, 630–646. [Google Scholar] [CrossRef]
- You, Z.; Bailis, J.M. DNA damage and decisions: CtIP coordinates DNA repair and cell cycle checkpoints. Trends Cell Biol. 2010, 20, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Carreira, A.; Kowalczykowski, S.C. Two classes of BRC repeats in BRCA2 promote RAD51 nucleoprotein filament function by distinct mechanisms. Proc. Natl. Acad. Sci. USA 2011, 108, 10448–10453. [Google Scholar] [CrossRef] [PubMed]
- Aquila, L.; Atanassov, B.S. Regulation of Histone Ubiquitination in Response to DNA Double Strand Breaks. Cells 2020, 9, 1699. [Google Scholar] [CrossRef] [PubMed]
- Helleday, T.; Petermann, E.; Lundin, C.; Hodgson, B.; Sharma, R.A. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer 2008, 8, 193–204. [Google Scholar] [CrossRef]
- Li, L.Y.; Guan, Y.D.; Chen, X.S.; Yang, J.M.; Cheng, Y. DNA Repair Pathways in Cancer Therapy and Resistance. Front. Pharmacol. 2020, 11, 629266. [Google Scholar] [CrossRef]
- Popovic, D.; Vucic, D.; Dikic, I. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 2014, 20, 1242–1253. [Google Scholar] [CrossRef]
- Moretton, A.; Loizou, J.I. Interplay between Cellular Metabolism and the DNA Damage Response in Cancer. Cancers 2020, 12, 2051. [Google Scholar] [CrossRef]
- Turgeon, M.O.; Perry, N.J.S.; Poulogiannis, G. DNA Damage, Repair, and Cancer Metabolism. Front. Oncol. 2018, 8, 15. [Google Scholar] [CrossRef]
- Jiang, P.; Du, W.; Wu, M. Regulation of the pentose phosphate pathway in cancer. Protein Cell 2014, 5, 592–602. [Google Scholar] [CrossRef]
- Rotin, D.; Kumar, S. Physiological functions of the HECT family of ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2009, 10, 398–409. [Google Scholar] [CrossRef]
- Dove, K.K.; Stieglitz, B.; Duncan, E.D.; Rittinger, K.; Klevit, R.E. Molecular insights into RBR E3 ligase ubiquitin transfer mechanisms. EMBO Rep. 2016, 17, 1221–1235. [Google Scholar] [CrossRef] [PubMed]
- Sherpa, D.; Chrustowicz, J.; Schulman, B.A. How the ends signal the end: Regulation by E3 ubiquitin ligases recognizing protein termini. Mol. Cell 2022, 82, 1424–1438. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Fang, S.; Gu, W. The Molecular Mechanism of Metabolic Remodeling in Lung Cancer. J. Cancer 2020, 11, 1403–1411. [Google Scholar] [CrossRef] [PubMed]
- Migita, T.; Narita, T.; Nomura, K.; Miyagi, E.; Inazuka, F.; Matsuura, M.; Ushijima, M.; Mashima, T.; Seimiya, H.; Satoh, Y.; et al. ATP citrate lyase: Activation and therapeutic implications in non-small cell lung cancer. Cancer Res. 2008, 68, 8547–8554. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Ai, W.; Zhang, Y.; Hu, Q.; Gan, M.; Wang, J.B.; Han, T. ARHGEF3 regulates the stability of ACLY to promote the proliferation of lung cancer. Cell Death Dis. 2022, 13, 870. [Google Scholar] [CrossRef] [PubMed]
- Sivanand, S.; Rhoades, S.; Jiang, Q.; Lee, J.V.; Benci, J.; Zhang, J.; Yuan, S.; Viney, I.; Zhao, S.; Carrer, A.; et al. Nuclear Acetyl-CoA Production by ACLY Promotes Homologous Recombination. Mol. Cell 2017, 67, 252–265.e6. [Google Scholar] [CrossRef]
- Densham, R.M.; Garvin, A.J.; Stone, H.R.; Strachan, J.; Baldock, R.A.; Daza-Martin, M.; Fletcher, A.; Blair-Reid, S.; Beesley, J.; Johal, B.; et al. Human BRCA1-BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nat. Struct. Mol. Biol. 2016, 23, 647–655. [Google Scholar] [CrossRef]
- Gou, Y.; Jin, D.; He, S.; Han, S.; Bai, Q. RNF168 is highly expressed in esophageal squamous cell carcinoma and contributes to the malignant behaviors in association with the Wnt/beta-catenin signaling pathway. Aging 2021, 13, 5403–5414. [Google Scholar] [CrossRef]
- Xie, T.; Qin, H.; Yuan, Z.; Zhang, Y.; Li, X.; Zheng, L. Emerging Roles of RNF168 in Tumor Progression. Molecules 2023, 28, 1417. [Google Scholar] [CrossRef]
- Chroma, K.; Mistrik, M.; Moudry, P.; Gursky, J.; Liptay, M.; Strauss, R.; Skrott, Z.; Vrtel, R.; Bartkova, J.; Kramara, J.; et al. Tumors overexpressing RNF168 show altered DNA repair and responses to genotoxic treatments, genomic instability and resistance to proteotoxic stress. Oncogene 2017, 36, 2405–2422. [Google Scholar] [CrossRef]
- Ingham, R.J.; Gish, G.; Pawson, T. The Nedd4 family of E3 ubiquitin ligases: Functional diversity within a common modular architecture. Oncogene 2004, 23, 1972–1984. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.W.; Hu, X.; Ye, M.; Lin, M.; Chu, M.; Shen, X. NEDD4 E3 ligase: Functions and mechanism in human cancer. Semin. Cancer Biol. 2020, 67, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.D.; Harreman, M.; Svejstrup, J.Q. Ubiquitylation and degradation of elongating RNA polymerase II: The last resort. Biochim. Biophys. Acta 2013, 1829, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Anindya, R.; Aygun, O.; Svejstrup, J.Q. Damage-induced ubiquitylation of human RNA polymerase II by the ubiquitin ligase Nedd4, but not Cockayne syndrome proteins or BRCA1. Mol. Cell 2007, 28, 386–397. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Fan, C.D.; Wang, X. Regulation of Mdm2 protein stability and the p53 response by NEDD4-1 E3 ligase. Oncogene 2015, 34, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Harris, S.L.; Levine, A.J. The p53 pathway: Positive and negative feedback loops. Oncogene 2005, 24, 2899–2908. [Google Scholar] [CrossRef]
- Trotman, L.C.; Wang, X.; Alimonti, A.; Chen, Z.; Teruya-Feldstein, J.; Yang, H.; Pavletich, N.P.; Carver, B.S.; Cordon-Cardo, C.; Erdjument-Bromage, H.; et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell 2007, 128, 141–156. [Google Scholar] [CrossRef]
- Wang, X.; Trotman, L.C.; Koppie, T.; Alimonti, A.; Chen, Z.; Gao, Z.; Wang, J.; Erdjument-Bromage, H.; Tempst, P.; Cordon-Cardo, C.; et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell 2007, 128, 129–139. [Google Scholar] [CrossRef]
- Chen, C.Y.; Chen, J.; He, L.; Stiles, B.L. PTEN: Tumor Suppressor and Metabolic Regulator. Front. Endocrinol. 2018, 9, 338. [Google Scholar] [CrossRef]
- Yang, J.; Nie, J.; Ma, X.; Wei, Y.; Peng, Y.; Wei, X. Targeting PI3K in cancer: Mechanisms and advances in clinical trials. Mol. Cancer 2019, 18, 26. [Google Scholar] [CrossRef]
- Pal, A.; Barber, T.M.; Van de Bunt, M.; Rudge, S.A.; Zhang, Q.; Lachlan, K.L.; Cooper, N.S.; Linden, H.; Levy, J.C.; Wakelam, M.J.; et al. PTEN mutations as a cause of constitutive insulin sensitivity and obesity. N. Engl. J. Med. 2012, 367, 1002–1011. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.; Federico, L.; Naples, M.; Avramoglu, R.K.; Meshkani, R.; Zhang, J.; Tsai, J.; Hussain, M.; Dai, K.; Iqbal, J.; et al. Phosphatase and tensin homolog (PTEN) regulates hepatic lipogenesis, microsomal triglyceride transfer protein, and the secretion of apolipoprotein B-containing lipoproteins. Hepatology 2008, 48, 1799–1809. [Google Scholar] [CrossRef] [PubMed]
- Koundouros, N.; Poulogiannis, G. Reprogramming of fatty acid metabolism in cancer. Br. J. Cancer 2020, 122, 4–22. [Google Scholar] [CrossRef]
- Hosios, A.M.; Manning, B.D. Cancer Signaling Drives Cancer Metabolism: AKT and the Warburg Effect. Cancer Res. 2021, 81, 4896–4898. [Google Scholar] [CrossRef] [PubMed]
- Lien, E.C.; Lyssiotis, C.A.; Cantley, L.C. Metabolic Reprogramming by the PI3K-Akt-mTOR Pathway in Cancer. Recent. Results Cancer Res. 2016, 207, 39–72. [Google Scholar] [PubMed]
- Fan, C.D.; Lum, M.A.; Xu, C.; Black, J.D.; Wang, X. Ubiquitin-dependent regulation of phospho-AKT dynamics by the ubiquitin E3 ligase, NEDD4-1, in the insulin-like growth factor-1 response. J. Biol. Chem. 2013, 288, 1674–1684. [Google Scholar] [CrossRef]
- Huang, X.; Gu, H.; Zhang, E.; Chen, Q.; Cao, W.; Yan, H.; Chen, J.; Yang, L.; Lv, N.; He, J.; et al. The NEDD4-1 E3 ubiquitin ligase: A potential molecular target for bortezomib sensitivity in multiple myeloma. Int. J. Cancer 2020, 146, 1963–1978. [Google Scholar] [CrossRef] [PubMed]
- Platta, H.W.; Abrahamsen, H.; Thoresen, S.B.; Stenmark, H. Nedd4-dependent lysine-11-linked polyubiquitination of the tumour suppressor Beclin 1. Biochem. J. 2012, 441, 399–406. [Google Scholar] [CrossRef]
- Almeida, A. Regulation of APC/C-Cdh1 and its function in neuronal survival. Mol. Neurobiol. 2012, 46, 547–554. [Google Scholar] [CrossRef]
- Kalucka, J.; Missiaen, R.; Georgiadou, M.; Schoors, S.; Lange, C.; De Bock, K.; Dewerchin, M.; Carmeliet, P. Metabolic control of the cell cycle. Cell Cycle 2015, 14, 3379–3388. [Google Scholar] [CrossRef]
- Campos, A.; Clemente-Blanco, A. Cell Cycle and DNA Repair Regulation in the Damage Response: Protein Phosphatases Take Over the Reins. Int. J. Mol. Sci. 2020, 21, 446. [Google Scholar] [CrossRef] [PubMed]
- De Boer, H.R.; Guerrero Llobet, S.; van Vugt, M.A. Controlling the response to DNA damage by the APC/C-Cdh1. Cell Mol. Life Sci. 2016, 73, 949–960. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.; Pagano, M. Linking metabolism and cell cycle progression via the APC/CCdh1 and SCFbetaTrCP ubiquitin ligases. Proc. Natl. Acad. Sci. USA 2011, 108, 20857–20858. [Google Scholar] [CrossRef]
- Alhmoud, J.F.; Woolley, J.F.; Al Moustafa, A.E.; Malki, M.I. DNA Damage/Repair Management in Cancers. Cancers 2020, 12, 1050. [Google Scholar] [CrossRef]
- Budhavarapu, V.N.; White, E.D.; Mahanic, C.S.; Chen, L.; Lin, F.T.; Lin, W.C. Regulation of E2F1 by APC/C Cdh1 via K11 linkage-specific ubiquitin chain formation. Cell Cycle 2012, 11, 2030–2038. [Google Scholar] [CrossRef] [PubMed]
- Lafranchi, L.; de Boer, H.R.; de Vries, E.G.; Ong, S.E.; Sartori, A.A.; van Vugt, M.A. APC/C(Cdh1) controls CtIP stability during the cell cycle and in response to DNA damage. EMBO J. 2014, 33, 2860–2879. [Google Scholar] [CrossRef]
- Makharashvili, N.; Paull, T.T. CtIP: A DNA damage response protein at the intersection of DNA metabolism. DNA Repair 2015, 32, 75–81. [Google Scholar] [CrossRef]
- Pascale, R.M.; Calvisi, D.F.; Simile, M.M.; Feo, C.F.; Feo, F. The Warburg Effect 97 Years after Its Discovery. Cancers 2020, 12, 2819. [Google Scholar] [CrossRef]
- Kotowski, K.; Rosik, J.; Machaj, F.; Supplitt, S.; Wiczew, D.; Jablonska, K.; Wiechec, E.; Ghavami, S.; Dziegiel, P. Role of PFKFB3 and PFKFB4 in Cancer: Genetic Basis, Impact on Disease Development/Progression, and Potential as Therapeutic Targets. Cancers 2021, 13, 909. [Google Scholar] [CrossRef]
- Almeida, A.; Bolanos, J.P.; Moncada, S. E3 ubiquitin ligase APC/C-Cdh1 accounts for the Warburg effect by linking glycolysis to cell proliferation. Proc. Natl. Acad. Sci. USA 2010, 107, 738–741. [Google Scholar] [CrossRef]
- Tudzarova, S.; Colombo, S.L.; Stoeber, K.; Carcamo, S.; Williams, G.H.; Moncada, S. Two ubiquitin ligases, APC/C-Cdh1 and SKP1-CUL1-F (SCF)-beta-TrCP, sequentially regulate glycolysis during the cell cycle. Proc. Natl. Acad. Sci. USA 2011, 108, 5278–5283. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Zhang, W.; Xue, L.; Wang, Y.; Fu, M.; Ma, L.; Song, Y.; Zhan, Q.M. APC/C-CDH1-Regulated IDH3beta Coordinates with the Cell Cycle to Promote Cell Proliferation. Cancer Res. 2019, 79, 3281–3293. [Google Scholar] [CrossRef]
- Tyagi, A.; Sarodaya, N.; Kaushal, K.; Chandrasekaran, A.P.; Antao, A.M.; Suresh, B.; Rhie, B.H.; Kim, K.S.; Ramakrishna, S. E3 Ubiquitin Ligase APC/C(Cdh1) Regulation of Phenylalanine Hydroxylase Stability and Function. Int. J. Mol. Sci. 2020, 21, 9076. [Google Scholar] [CrossRef]
- Zhao, J.; Zhou, R.; Hui, K.; Yang, Y.; Zhang, Q.; Ci, Y.; Shi, L.; Xu, C.; Huang, F.; Hu, Y. Selenite inhibits glutamine metabolism and induces apoptosis by regulating GLS1 protein degradation via APC/C-CDH1 pathway in colorectal cancer cells. Oncotarget 2017, 8, 18832–18847. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Bellon, M.; Ju, M.; Zhao, L.; Wei, M.; Fu, L.; Nicot, C. Clinical significance of FBXW7 loss of function in human cancers. Mol. Cancer 2022, 21, 87. [Google Scholar] [CrossRef]
- Cui, D.; Xiong, X.; Shu, J.; Dai, X.; Sun, Y.; Zhao, Y. FBXW7 Confers Radiation Survival by Targeting p53 for Degradation. Cell Rep. 2020, 30, 497–509.e4. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.; Kaur, E.; Kharat, S.S.; Hussain, M.; Damodaran, A.P.; Kulshrestha, S.; Sengupta, S. Abrogation of FBW7alpha-dependent p53 degradation enhances p53’s function as a tumor suppressor. J. Biol. Chem. 2019, 294, 13224–13232. [Google Scholar] [CrossRef]
- Galindo-Moreno, M.; Giraldez, S.; Limon-Mortes, M.C.; Belmonte-Fernandez, A.; Reed, S.I.; Saez, C.; Japon, M.A.; Tortolero, M.; Romero, F. SCF(FBXW7)-mediated degradation of p53 promotes cell recovery after UV-induced DNA damage. FASEB J. 2019, 33, 11420–11430. [Google Scholar] [CrossRef]
- Galli, F.; Rossi, M.; D’Alessandra, Y.; De Simone, M.; Lopardo, T.; Haupt, Y.; Alsheich-Bartok, O.; Anzi, S.; Shaulian, E.; Calabro, V.; et al. MDM2 and Fbw7 cooperate to induce p63 protein degradation following DNA damage and cell differentiation. J. Cell. Sci. 2010, 123, 2423–2433. [Google Scholar] [CrossRef]
- Giraldez, S.; Herrero-Ruiz, J.; Mora-Santos, M.; Japon, M.A.; Tortolero, M.; Romero, F. SCF(FBXW7alpha) modulates the intra-S-phase DNA-damage checkpoint by regulating Polo like kinase-1 stability. Oncotarget 2014, 5, 4370–4383. [Google Scholar] [CrossRef]
- Kharat, S.S.; Tripathi, V.; Damodaran, A.P.; Priyadarshini, R.; Chandra, S.; Tikoo, S.; Nandhakumar, R.; Srivastava, V.; Priya, S.; Hussain, M.; et al. Mitotic phosphorylation of Bloom helicase at Thr182 is required for its proteasomal degradation and maintenance of chromosomal stability. Oncogene 2016, 35, 1025–1038. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.; Liu, W.; Song, R.; Shah, J.J.; Feng, X.; Tsang, C.K.; Morgan, K.M.; Bunting, S.F.; Inuzuka, H.; Zheng, X.F.; et al. SOX9 is targeted for proteasomal degradation by the E3 ligase FBW7 in response to DNA damage. Nucleic Acids Res. 2016, 44, 8855–8869. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Karnak, D.; Tan, M.; Lawrence, T.S.; Morgan, M.A.; Sun, Y. FBXW7 Facilitates Nonhomologous End-Joining via K63-Linked Polyubiquitylation of XRCC4. Mol. Cell 2016, 61, 419–433. [Google Scholar] [CrossRef] [PubMed]
- Kaur, E.; Agrawal, R.; Sengupta, S. Functions of BLM Helicase in Cells: Is It Acting Like a Double-Edged Sword? Front. Genet. 2021, 12, 634789. [Google Scholar] [CrossRef]
- Stracker, T.H.; Usui, T.; Petrini, J.H. Taking the time to make important decisions: The checkpoint effector kinases Chk1 and Chk2 and the DNA damage response. DNA Repair 2009, 8, 1047–1054. [Google Scholar] [CrossRef]
- Bengoechea-Alonso, M.T.; Aldaalis, A.; Ericsson, J. Loss of the Fbw7 tumor suppressor rewires cholesterol metabolism in cancer cells leading to activation of the PI3K-AKT signalling axis. Front. Oncol. 2022, 12, 990672. [Google Scholar] [CrossRef]
- Mao, J.H.; Kim, I.J.; Wu, D.; Climent, J.; Kang, H.C.; DelRosario, R.; Balmain, A. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science 2008, 321, 1499–1502. [Google Scholar] [CrossRef]
- Popov, N.; Schulein, C.; Jaenicke, L.A.; Eilers, M. Ubiquitylation of the amino terminus of Myc by SCF(beta-TrCP) antagonizes SCF(Fbw7)-mediated turnover. Nat. Cell Biol. 2010, 12, 973–981. [Google Scholar] [CrossRef]
- Chandra, S.; Priyadarshini, R.; Madhavan, V.; Tikoo, S.; Hussain, M.; Mudgal, R.; Modi, P.; Srivastava, V.; Sengupta, S. Enhancement of c-Myc degradation by BLM helicase leads to delayed tumor initiation. J. Cell Sci. 2013, 126, 3782–3795. [Google Scholar] [CrossRef]
- Flugel, D.; Gorlach, A.; Kietzmann, T. GSK-3beta regulates cell growth, migration, and angiogenesis via Fbw7 and USP28-dependent degradation of HIF-1alpha. Blood 2012, 119, 1292–1301. [Google Scholar] [CrossRef]
- Cassavaugh, J.M.; Hale, S.A.; Wellman, T.L.; Howe, A.K.; Wong, C.; Lounsbury, K.M. Negative regulation of HIF-1alpha by an FBW7-mediated degradation pathway during hypoxia. J. Cell. Biochem. 2011, 112, 3882–3890. [Google Scholar] [CrossRef] [PubMed]
- Floter, J.; Kaymak, I.; Schulze, A. Regulation of Metabolic Activity by p53. Metabolites 2017, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Schauvliege, R.; Janssens, S.; Beyaert, R. Pellino proteins: Novel players in TLR and IL-1R signalling. J. Cell. Mol. Med. 2007, 11, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.W.; Park, H.H.; Kim, S.; Chung, J.M.; Noh, H.J.; Kim, S.K.; Song, H.K.; Lee, C.W.; Morgan, M.J.; Kang, H.C.; et al. PELI1 Selectively Targets Kinase-Active RIP3 for Ubiquitylation-Dependent Proteasomal Degradation. Mol. Cell 2018, 70, 920–935.e7. [Google Scholar] [CrossRef]
- Park, H.Y.; Go, H.; Song, H.R.; Kim, S.; Ha, G.H.; Jeon, Y.K.; Kim, J.E.; Lee, H.; Cho, H.; Kang, H.C.; et al. Pellino 1 promotes lymphomagenesis by deregulating BCL6 polyubiquitination. J. Clin. Investig. 2014, 124, 4976–4988. [Google Scholar] [CrossRef]
- Chang, M.; Jin, W.; Sun, S.C. Peli1 facilitates TRIF-dependent Toll-like receptor signaling and proinflammatory cytokine production. Nat. Immunol. 2009, 10, 1089–1095. [Google Scholar] [CrossRef]
- Moynagh, P.N. The roles of Pellino E3 ubiquitin ligases in immunity. Nat. Rev. Immunol. 2014, 14, 122–131. [Google Scholar] [CrossRef]
- Schauvliege, R.; Janssens, S.; Beyaert, R. Pellino proteins are more than scaffold proteins in TLR/IL-1R signalling: A role as novel RING E3-ubiquitin-ligases. FEBS Lett. 2006, 580, 4697–4702. [Google Scholar] [CrossRef]
- Park, J.; Park, H.Y.; Kim, S.; Kim, H.S.; Park, J.Y.; Go, H.; Lee, C.W. Pellino 1 inactivates mitotic spindle checkpoint by targeting BubR1 for ubiquitinational degradation. Oncotarget 2017, 8, 32055–32067. [Google Scholar] [CrossRef]
- Ko, C.J.; Zhang, L.; Jie, Z.; Zhu, L.; Zhou, X.; Xie, X.; Gao, T.; Yang, J.Y.; Cheng, X.; Sun, S.C. The E3 ubiquitin ligase Peli1 regulates the metabolic actions of mTORC1 to suppress antitumor T cell responses. EMBO J. 2021, 40, e104532. [Google Scholar] [CrossRef]
- Selvaraju, V.; Thirunavukkarasu, M.; Joshi, M.; Oriowo, B.; Shaikh, I.A.; Rishi, M.T.; Tapias, L.; Coca-Soliz, V.; Saad, I.; Campbell, J.; et al. Deletion of newly described pro-survival molecule Pellino-1 increases oxidative stress, downregulates cIAP2/NF-kappaB cell survival pathway, reduces angiogenic response, and thereby aggravates tissue function in mouse ischemic models. Basic. Res. Cardiol. 2020, 115, 45. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Yang, J.; Chen, H.; Li, J.; Que, L.; Zhu, G.; Liu, L.; Ha, T.; Chen, Q.; Li, C.; et al. Peli1 induction impairs cardiac microvascular endothelium through Hsp90 dissociation from IRE1alpha. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 2606–2617. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, S.Y.; Bae, S.; Lee, J.K.; Hwang, K.; Go, H.; Lee, C.W. Pellino1 promotes chronic inflammatory skin disease via keratinocyte hyperproliferation and induction of the T helper 17 response. Exp. Mol. Med. 2020, 52, 1537–1549. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lee, S.Y.; Jeon, Y.; Kim, K.M.; Lee, J.K.; Ko, J.; Park, E.J.; Yoon, J.S.; Kang, B.E.; Ryu, D.; et al. The Pellino1-PKCtheta Signaling Axis Is an Essential Target for Improving Antitumor CD8+ T-lymphocyte Function. Cancer Immunol. Res. 2022, 10, 327–342. [Google Scholar] [CrossRef]
- Zheng, T.; Zhou, Y.; Xu, X.; Qi, X.; Liu, J.; Pu, Y.; Zhang, S.; Gao, X.; Luo, X.; Li, M.; et al. MiR-30c-5p loss-induced PELI1 accumulation regulates cell proliferation and migration via activating PI3K/AKT pathway in papillary thyroid carcinoma. J. Transl. Med. 2022, 20, 20. [Google Scholar] [CrossRef]
- White, R.R.; Vijg, J. Do DNA Double-Strand Breaks Drive Aging? Mol. Cell 2016, 63, 729–738. [Google Scholar] [CrossRef]
- Blundred, R.M.; Stewart, G.S. DNA double-strand break repair, immunodeficiency and the RIDDLE syndrome. Expert. Rev. Clin. Immunol. 2011, 7, 169–185. [Google Scholar] [CrossRef]
- Gennery, A.R.; Cant, A.J.; Jeggo, P.A. Immunodeficiency associated with DNA repair defects. Clin. Exp. Immunol. 2000, 121, 1–7. [Google Scholar] [CrossRef]
- Asada-Utsugi, M.; Uemura, K.; Ayaki, T.; M, T.U.; Minamiyama, S.; Hikiami, R.; Morimura, T.; Shodai, A.; Ueki, T.; Takahashi, R.; et al. Failure of DNA double-strand break repair by tau mediates Alzheimer’s disease pathology in vitro. Commun. Biol. 2022, 5, 358. [Google Scholar] [CrossRef]
- Aparicio, T.; Baer, R.; Gautier, J. DNA double-strand break repair pathway choice and cancer. DNA Repair 2014, 19, 169–175. [Google Scholar] [CrossRef]
- Li, J.; Williams, B.L.; Haire, L.F.; Goldberg, M.; Wilker, E.; Durocher, D.; Yaffe, M.B.; Jackson, S.P.; Smerdon, S.J. Structural and functional versatility of the FHA domain in DNA-damage signaling by the tumor suppressor kinase Chk2. Mol. Cell 2002, 9, 1045–1054. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Lu, L.Y.; Yang, C.Y.; Wang, S.; Yu, X. The FHA and BRCT domains recognize ADP-ribosylation during DNA damage response. Genes. Dev. 2013, 27, 1752–1768. [Google Scholar] [CrossRef]
- Xiao, Y.; Jin, J.; Zou, Q.; Hu, H.; Cheng, X.; Sun, S.C. Peli1 negatively regulates type I interferon induction and antiviral immunity in the CNS. Cell Biosci. 2015, 5, 34. [Google Scholar] [CrossRef]
- Hartlova, A.; Erttmann, S.F.; Raffi, F.A.; Schmalz, A.M.; Resch, U.; Anugula, S.; Lienenklaus, S.; Nilsson, L.M.; Kroger, A.; Nilsson, J.A.; et al. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity 2015, 42, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Fabre, M.S.; Stanton, N.M.; Slatter, T.L.; Lee, S.; Senanayake, D.; Gordon, R.M.A.; Castro, M.L.; Rowe, M.R.; Taha, A.; Royds, J.A.; et al. The oncogene BCL6 is up-regulated in glioblastoma in response to DNA damage, and drives survival after therapy. PLoS ONE 2020, 15, e0231470. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Feng, J.; Yuan, K.; Wu, Z.; Hu, L.; Lu, Y.; Li, K.; Guo, J.; Chen, J.; Ma, C.; et al. The oncoprotein BCL6 enables solid tumor cells to evade genotoxic stress. Elife 2022, 11, e69255. [Google Scholar] [CrossRef]
- Liu, B.; Liu, X.; Han, L.; Chen, X.; Wu, X.; Wu, J.; Yan, D.; Wang, Y.; Liu, S.; Shan, L.; et al. BRD4-directed super-enhancer organization of transcription repression programs links to chemotherapeutic efficacy in breast cancer. Proc. Natl. Acad. Sci. USA 2022, 119, e2109133119. [Google Scholar] [CrossRef]
- Jeon, Y.K.; Kim, C.K.; Hwang, K.R.; Park, H.Y.; Koh, J.; Chung, D.H.; Lee, C.W.; Ha, G.H. Pellino-1 promotes lung carcinogenesis via the stabilization of Slug and Snail through K63-mediated polyubiquitination. Cell Death Differ. 2017, 24, 469–480. [Google Scholar] [CrossRef]
- Hoxhaj, G.; Manning, B.D. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer 2020, 20, 74–88. [Google Scholar] [CrossRef]
- Shi, R.; Tang, Y.Q.; Miao, H. Metabolism in tumor microenvironment: Implications for cancer immunotherapy. MedComm 2020, 1, 47–68. [Google Scholar] [CrossRef]
- Maimela, N.R.; Liu, S.; Zhang, Y. Fates of CD8+ T cells in Tumor Microenvironment. Comput. Struct. Biotechnol. J. 2019, 17, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Reina-Campos, M.; Scharping, N.E.; Goldrath, A.W. CD8(+) T cell metabolism in infection and cancer. Nat. Rev. Immunol. 2021, 21, 718–738. [Google Scholar] [CrossRef] [PubMed]
- Biau, J.; Chautard, E.; Verrelle, P.; Dutreix, M. Altering DNA Repair to Improve Radiation Therapy: Specific and Multiple Pathway Targeting. Front. Oncol. 2019, 9, 1009. [Google Scholar] [CrossRef]
- Huang, R.X.; Zhou, P.K. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal. Transduct. Target. Ther. 2020, 5, 60. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Sun, S.C. Targeting ubiquitin signaling for cancer immunotherapy. Signal. Transduct. Target. Ther. 2021, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Miller, Z.; Jun, Y.; Lee, W.; Kim, K.B. Next-generation proteasome inhibitors for cancer therapy. Transl. Res. 2018, 198, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.; Tonon, G.; Bashari, M.H.; Vallet, S.; Antonini, E.; Goldschmidt, H.; Schulze-Bergkamen, H.; Opferman, J.T.; Sattler, M.; Anderson, K.C.; et al. Targeting Mcl-1 for multiple myeloma (MM) therapy: Drug-induced generation of Mcl-1 fragment Mcl-1(128-350) triggers MM cell death via c-Jun upregulation. Cancer Lett. 2014, 343, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Hambley, B.; Caimi, P.F.; William, B.M. Bortezomib for the treatment of mantle cell lymphoma: An update. Ther. Adv. Hematol. 2016, 7, 196–208. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Egashira, N. Pathological Mechanisms of Bortezomib-Induced Peripheral Neuropathy. Int. J. Mol. Sci. 2021, 22, 888. [Google Scholar] [CrossRef]
- Hyer, M.L.; Milhollen, M.A.; Ciavarri, J.; Fleming, P.; Traore, T.; Sappal, D.; Huck, J.; Shi, J.; Gavin, J.; Brownell, J.; et al. A small-molecule inhibitor of the ubiquitin activating enzyme for cancer treatment. Nat. Med. 2018, 24, 186–193. [Google Scholar] [CrossRef]
- Du, X.; Song, H.; Shen, N.; Hua, R.; Yang, G. The Molecular Basis of Ubiquitin-Conjugating Enzymes (E2s) as a Potential Target for Cancer Therapy. Int. J. Mol. Sci. 2021, 22, 3440. [Google Scholar] [CrossRef] [PubMed]
- Ceccarelli, D.F.; Tang, X.; Pelletier, B.; Orlicky, S.; Xie, W.; Plantevin, V.; Neculai, D.; Chou, Y.C.; Ogunjimi, A.; Al-Hakim, A.; et al. An allosteric inhibitor of the human Cdc34 ubiquitin-conjugating enzyme. Cell 2011, 145, 1075–1087. [Google Scholar] [CrossRef] [PubMed]
- Gombodorj, N.; Yokobori, T.; Yoshiyama, S.; Kawabata-Iwakawa, R.; Rokudai, S.; Horikoshi, I.; Nishiyama, M.; Nakano, T. Inhibition of Ubiquitin-conjugating Enzyme E2 May Activate the Degradation of Hypoxia-inducible Factors and, thus, Overcome Cellular Resistance to Radiation in Colorectal Cancer. Anticancer. Res. 2017, 37, 2425–2436. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Fan, Y.H.; Xu, X.; Zhang, H.; Dou, J.; Tang, Y.; Zhong, X.; Rojas, Y.; Yu, Y.; Zhao, Y.; et al. A small-molecule inhibitor of UBE2N induces neuroblastoma cell death via activation of p53 and JNK pathways. Cell Death Dis. 2014, 5, e1079. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, S.; Takeuchi, T.; Rotinsulu, H.; Mangindaan, R.E.; van Soest, R.W.; Ukai, K.; Kobayashi, H.; Namikoshi, M.; Ohta, T.; Yokosawa, H. Leucettamol A: A new inhibitor of Ubc13-Uev1A interaction isolated from a marine sponge, Leucetta aff. microrhaphis. Bioorg Med. Chem. Lett. 2008, 18, 6319–6320. [Google Scholar] [CrossRef] [PubMed]
- Ushiyama, S.; Umaoka, H.; Kato, H.; Suwa, Y.; Morioka, H.; Rotinsulu, H.; Losung, F.; Mangindaan, R.E.; de Voogd, N.J.; Yokosawa, H.; et al. Manadosterols A and B, sulfonated sterol dimers inhibiting the Ubc13-Uev1A interaction, isolated from the marine sponge Lissodendryx fibrosa. J. Nat. Prod. 2012, 75, 1495–1499. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Gu, X.; Liu, F.; Rui, Z.; Liu, M.; Zhao, L. Antitumor effects of hsa-miR661-3p on non-small cell lung cancer in vivo and in vitro. Oncol. Rep. 2019, 41, 2987–2996. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wu, X.; Yang, C.; Rashid, K.; Ma, C.; Hu, M.; Ding, Q.; Jiang, H. Anticancer effect of icaritin on prostate cancer via regulating miR-381-3p and its target gene UBE2C. Cancer Med. 2019, 8, 7833–7845. [Google Scholar] [CrossRef]
- Tung, C.W.; Huang, P.Y.; Chan, S.C.; Cheng, P.H.; Yang, S.H. The regulatory roles of microRNAs toward pathogenesis and treatments in Huntington’s disease. J. Biomed. Sci. 2021, 28, 59. [Google Scholar] [CrossRef]
- Sun, Y. E3 ubiquitin ligases as cancer targets and biomarkers. Neoplasia 2006, 8, 645–654. [Google Scholar] [CrossRef]
- Secchiero, P.; di Iasio, M.G.; Gonelli, A.; Zauli, G. The MDM2 inhibitor Nutlins as an innovative therapeutic tool for the treatment of haematological malignancies. Curr. Pharm. Des. 2008, 14, 2100–2110. [Google Scholar] [CrossRef] [PubMed]
- Trino, S.; Iacobucci, I.; Erriquez, D.; Laurenzana, I.; De Luca, L.; Ferrari, A.; Ghelli Luserna Di Rora, A.; Papayannidis, C.; Derenzini, E.; Simonetti, G.; et al. Targeting the p53-MDM2 interaction by the small-molecule MDM2 antagonist Nutlin-3a: A new challenged target therapy in adult Philadelphia positive acute lymphoblastic leukemia patients. Oncotarget 2016, 7, 12951–12961. [Google Scholar] [CrossRef] [PubMed]
- Saleh, M.N.; Patel, M.R.; Bauer, T.M.; Goel, S.; Falchook, G.S.; Shapiro, G.I.; Chung, K.Y.; Infante, J.R.; Conry, R.M.; Rabinowits, G.; et al. Phase 1 Trial of ALRN-6924, a Dual Inhibitor of MDMX and MDM2, in Patients with Solid Tumors and Lymphomas Bearing Wild-type TP53. Clin. Cancer Res. 2021, 27, 5236–5247. [Google Scholar] [CrossRef] [PubMed]
- Cossu, F.; Milani, M.; Mastrangelo, E.; Lecis, D. Targeting the BIR Domains of Inhibitor of Apoptosis (IAP) Proteins in Cancer Treatment. Comput. Struct. Biotechnol. J. 2019, 17, 142–150. [Google Scholar] [CrossRef]
- Greil, C.; Engelhardt, M.; Wasch, R. The Role of the APC/C and Its Coactivators Cdh1 and Cdc20 in Cancer Development and Therapy. Front. Genet. 2022, 13, 941565. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, B.; Wang, Y.; Shang, G. Cdc20 inhibitor apcin inhibits the growth and invasion of osteosarcoma cells. Oncol. Rep. 2018, 40, 841–848. [Google Scholar] [CrossRef]
- Marashiyan, M.; Kalhor, H.; Ganji, M.; Rahimi, H. Effects of tosyl-l-arginine methyl ester (TAME) on the APC/c subunits: An in silico investigation for inhibiting cell cycle. J. Mol. Graph. Model. 2020, 97, 107563. [Google Scholar] [CrossRef]
- De, K.; Grubb, T.M.; Zalenski, A.A.; Pfaff, K.E.; Pal, D.; Majumder, S.; Summers, M.K.; Venere, M. Hyperphosphorylation of CDH1 in Glioblastoma Cancer Stem Cells Attenuates APC/C(CDH1) Activity and Pharmacologic Inhibition of APC/C(CDH1/CDC20) Compromises Viability. Mol. Cancer Res. 2019, 17, 1519–1530. [Google Scholar] [CrossRef]
- Huang, H.L.; Weng, H.Y.; Wang, L.Q.; Yu, C.H.; Huang, Q.J.; Zhao, P.P.; Wen, J.Z.; Zhou, H.; Qu, L.H. Triggering Fbw7-mediated proteasomal degradation of c-Myc by oridonin induces cell growth inhibition and apoptosis. Mol. Cancer Ther. 2012, 11, 1155–1165. [Google Scholar] [CrossRef]
- Chan, C.H.; Morrow, J.K.; Li, C.F.; Gao, Y.; Jin, G.; Moten, A.; Stagg, L.J.; Ladbury, J.E.; Cai, Z.; Xu, D.; et al. Pharmacological inactivation of Skp2 SCF ubiquitin ligase restricts cancer stem cell traits and cancer progression. Cell 2013, 154, 556–568. [Google Scholar] [CrossRef]
- Liao, Y.J.; Bai, H.Y.; Li, Z.H.; Zou, J.; Chen, J.W.; Zheng, F.; Zhang, J.X.; Mai, S.J.; Zeng, M.S.; Sun, H.D.; et al. Longikaurin A, a natural ent-kaurane, induces G2/M phase arrest via downregulation of Skp2 and apoptosis induction through ROS/JNK/c-Jun pathway in hepatocellular carcinoma cells. Cell Death Dis. 2014, 5, e1137. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Zhou, X.; Wang, L.; Yin, X.; Wang, Z. Curcumin inhibits cell growth and invasion and induces apoptosis through down-regulation of Skp2 in pancreatic cancer cells. Am. J. Cancer Res. 2016, 6, 1949–1962. [Google Scholar]
- Zhou, L.; Yu, X.; Li, M.; Gong, G.; Liu, W.; Li, T.; Zuo, H.; Li, W.; Gao, F.; Liu, H. Cdh1-mediated Skp2 degradation by dioscin reprogrammes aerobic glycolysis and inhibits colorectal cancer cells growth. EBioMedicine 2020, 51, 102570. [Google Scholar] [CrossRef]
- Nakajima, H.; Fujiwara, H.; Furuichi, Y.; Tanaka, K.; Shimbara, N. A novel small-molecule inhibitor of NF-kappaB signaling. Biochem. Biophys. Res. Commun. 2008, 368, 1007–1013. [Google Scholar] [CrossRef] [PubMed]
- Blees, J.S.; Bokesch, H.R.; Rubsamen, D.; Schulz, K.; Milke, L.; Bajer, M.M.; Gustafson, K.R.; Henrich, C.J.; McMahon, J.B.; Colburn, N.H.; et al. Erioflorin stabilizes the tumor suppressor Pdcd4 by inhibiting its interaction with the E3-ligase beta-TrCP1. PLoS ONE 2012, 7, e46567. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, Y.; Ding, S.; Li, J.; Song, N.; Ren, Y.; Hong, D.; Wu, C.; Li, B.; Wang, F.; et al. Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade. Cell Res. 2018, 28, 1186–1194. [Google Scholar] [CrossRef] [PubMed]
- Han, K.H.; Kwak, M.; Lee, T.H.; Park, M.S.; Jeong, I.H.; Kim, M.J.; Jin, J.O.; Lee, P.C. USP14 Inhibition Regulates Tumorigenesis by Inducing Autophagy in Lung Cancer In Vitro. Int. J. Mol. Sci. 2019, 20, 5300. [Google Scholar] [CrossRef]
- Kapuria, V.; Peterson, L.F.; Fang, D.; Bornmann, W.G.; Talpaz, M.; Donato, N.J. Deubiquitinase inhibition by small-molecule WP1130 triggers aggresome formation and tumor cell apoptosis. Cancer Res. 2010, 70, 9265–9276. [Google Scholar] [CrossRef]
- Kim, S.; Woo, S.M.; Min, K.J.; Seo, S.U.; Lee, T.J.; Kubatka, P.; Kim, D.E.; Kwon, T.K. WP1130 Enhances TRAIL-Induced Apoptosis through USP9X-Dependent miR-708-Mediated Downregulation of c-FLIP. Cancers 2019, 11, 344. [Google Scholar] [CrossRef]
- Colland, F.; Formstecher, E.; Jacq, X.; Reverdy, C.; Planquette, C.; Conrath, S.; Trouplin, V.; Bianchi, J.; Aushev, V.N.; Camonis, J.; et al. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol. Cancer Ther. 2009, 8, 2286–2295. [Google Scholar] [CrossRef]
- Li, P.; Liu, H.M. Recent advances in the development of ubiquitin-specific-processing protease 7 (USP7) inhibitors. Eur. J. Med. Chem. 2020, 191, 112107. [Google Scholar] [CrossRef] [PubMed]
| Name | Type | Substrate | Ubiquitination Residue | Cancer Type | Ref. (PMID Number) |
|---|---|---|---|---|---|
| E4F1 | atypical | p53 | K48 | osteosarcoma, hepatocellular carcinoma | 17110336, 24163401 |
| HectH9 | HECT | c-Myc | K63 | retinoblastoma | 31677785 |
| HUWE1 | HECT | HAUSP | K63 | lung cancer, prostatic adenocarcinoma | 30176860, 27934968 |
| c-Myc | K48, K63 | breast cancer | 30176860 | ||
| p53 | K48 | B cell lymphoma | 30176860 | ||
| WIPI2 | unknown | pancreatic cancer | 30340022, 34502089 | ||
| HK2 | K63 | prostate cancer | 31201299 | ||
| NEDD4 | HECT | AKT | K63, K48 | breast cancer, hepatocellular carcinoma | 23195959, 31390487 |
| PTEN | K63 | prostate cancer | 17218261, 17218260 | ||
| Beclin-1 | K11 | adenocarcinoma | 21936852 | ||
| NEDD4L | HECT | ULK1 | K27, K29 | adenocarcinoma | 27932573 |
| UBR5 | HECT | citrate synthase (CS) | unknown | breast cancer | 30850587 |
| WWP1 | HECT | PTEN | K27 | prostate cancer | 31097636 |
| WWP2 | HECT | PTEN | unknown | prostate cancer | 21532586 |
| PARKIN | RBR | mTOR | unknown | ganglioglioma | 28803490 |
| HIF-1α | unknown | breast cancer | 29180628 | ||
| PKM2 | unknown | lung cancer, glioblastoma | 26975375 | ||
| PHGDH | unknown | breast cancer, lung cancer | 32478681 | ||
| APC/CCDH1 | multimeric RING | PFKFB3 | unknown | neuroblastoma, adenocarcinoma | 20080744, 24604252, 21402913 |
| PAH | K48, K63 (no direct evidence) | hepatocellular carcinoma | 33260674 | ||
| GLS1 | unknown | colorectal cancer | 27902968 | ||
| IDH3β | unknown | esophageal squamous cell carcinoma | 31053633 | ||
| BRCA1 | RING | AKT | K48 | breast cancer | 19074868 |
| CARP1/2 | RING | p53 | K48 | lung cancer, colorectal cancer | 17121812 |
| COP1 | RING | p53 | K48 | osteosarcoma | 15103385 |
| FASN | unknown | adenocarcinoma | 23269672 | ||
| CRL4A | RING | AMPKα | K29, K63 | ovarian cancer | 30807229 |
| CUL3-KLHL20 | RING | ULK1 | K48 | adenocarcinoma | 26687681 |
| CUL3-KLHL22 | RING | DEPDC5 | K48 | breast cancer | 29769719 |
| CUL3-KLHL25 | RING | ACLY | unknown | lung cancer | 27664236 |
| CUL3-SPOP | RING | PrLZ | K48 | prostate cancer | 35194188 |
| CUL4-DDB1 | RING | Raptor | K63 | lung cancer | 18235224 |
| CUL4-DDB1 (AMBRA1) | RING | Beclin-1 | K63 | colon cancer, pancreatic ductal adenocacinoma | 27308402, 23974797 |
| CUL5 | RING | p53 | K48 | B-cell chronic lymphocytic leukemia | 17449237, 32194910 |
| CUL7 | RING | p53 | unknown | lung cancer, breast cancer | 17942889, 25003318 |
| GID | RING | AMPK | K48 | adenocarcinoma, osteosarcoma | 31795790, 25679763 |
| Gp78 | RING | HMGCR | unknown | breast cancer, prostate cancer | 22143767, 33718197 |
| MAGE-TRIM28 | RING | FBP1 | unknown | hepatocellular carcinoma | 28394358 |
| MDM2 | RING | p53 | K48 | breast cancer | 9153396, 34650049 |
| MDM2 | RING | PGAM | unknown | lung cancer | 24567357 |
| MKRN1 | RING | p53 | K48 | lung cancer, colon cancer | 19536131, 20930521 |
| MULAN | RING | AKT | K48 | adenocarcinoma | 22410793 |
| Pirh2 | RING | p53 | K48 | osteosarcoma | 12654245 |
| RFP | RING | PTEN | K48 | plasmacytoma, myeloma, endometrial cancer, colon carcinoma | 23419514 |
| RING1 | RING | p53 | K48 | liver cancer | 29187402 |
| RNF2 | RING | AMBRA1 | K48 | thymus lymphoma | 24980959, 28789945 |
| RNF5 | RING | SLC1A5/ SLC38A2 | unknown | breast cancer | 25759021 |
| RNF8 | RING | histone H3 | K48 | breast cancer, epidermoid carcinoma | 28507061 |
| RNF115 | RING | p53 | unknown | lung adenocarcinoma | 32553631 |
| RNF126 | RING | mTOR | K48 | acute myeloid leukemia | 32131492 |
| PDK | unknown | breast cancer | 27462466 | ||
| RNF139 | RING | SREBP1 precursor | unknown | renal cell carcinoma | 20068067 |
| RNF145 | RING | HMGCR | unknown | adenocarcinoma, hepatocellular carcinoma | 29374057, 30543180 |
| RNF152 | RING | RagA | K63 | non-small cell lung cancer | 25936802, 32486221 |
| Rheb | unknown | colon cancer | 30514904, 32486221 | ||
| RNF216 | RING | Beclin-1 | K48 | adenocarcinoma | 29361549 |
| SCFFBXL6 | RING | HSP90AA1 | K63 | hepatocellular carcinoma | 32576198 |
| SCFFBXL8 | RING | mTOR | K48 | colorectal carcinoma | 27916606 |
| SCFFBXL14 | RING | c-Myc | K48? | glioma | 27923907 |
| SCFFBXL18 | RING | AKT | K63 | glioma | 27926990 |
| SCFFBXL20 | RING | VPS34 | unknown | adenocarcinoma, glioma | 25593308 |
| SCFFBXO32 | RING | c-Myc | K48 | ovary cancer | 25944903 |
| SCFFBXW7 | RING | SREBP1 | unknown | glioma | 34737211, 36176395 |
| mTOR | K48 | breast cancer | 30086763, 18787170 | ||
| HIF-1α | K48 | ovarian cancer | 21964756 | ||
| c-Myc | K48 | T-acute lymphoblastic leukemia, uterine cancer, colorectal cancer, bladder cancer, lung cancer | 30086763, 33260160, 23750012, 20852628, 28665315 | ||
| p53 | K48 | adenocarcinoma, colorectal cancer | 31905981, 25314076 | ||
| SCFSKP2 | RING | c-Myc | K48 | B cell lymphoma | 12769844, 12769843, 28665315 |
| AKT | K63 | breast cancer | 22632973 | ||
| RagA | K63 | adenocarcinoma | 26051179 | ||
| SCFβ-TrCP | RING | DEPTOR | unknown | breast cancer, ovarian cancer | 22017876, 22454292 |
| Myc | K33, K48, K63 | osteosarcoma | 20852628 | ||
| PFKFB3 | unknown | adenocarcinoma | 21402913 | ||
| NRF2 | unknown | endometrioid carcinoma | 25937177 | ||
| SIAH2 | RING | α-KGDHC | unknown | adenocarcinoma, tongue cancer, renal cancer | 15466852, 24506869 |
| Synoviolin | RING | p53 | K48 | colon cancer | 25128494, 20930521, 17170702 |
| TOPORS | RING | HIF-1α | K63 | colon cancer | 23722539 |
| TRAF2 | RING | mLST8 | K63 | ovarian cancer, melanoma, adenocarcinoma | 28489822 |
| TRAF4 | RING | AKT | K63 | lung cancer | 24154876 |
| TRAF6 | RING | p62 | K63 | prostate cancer, lung cancer | 23911927 |
| AKT | K63 | prostate cancer | 19713527 | ||
| HIF-1α | K63 | colon cancer, adenocarcinoma | 23722539 | ||
| ULK1 | K63 | chronic myeloid leukemia | 30929559 | ||
| Beclin-1 | K63 | leukemia | 20501938 | ||
| HK2 | K63 | liver cancer | 28980855 | ||
| TRC8/RNF139 | RING | HMGCR | unknown | renal cell carcinoma | 20068067 |
| TRIM16 | RING | NRF2 | K63 | adenocarcinoma | 30525100 |
| ULK1 | K63 | acute monocytic leukemia | 27693506 | ||
| TRIM21 | RING | PFKP | K48 | glioma | 29038421 |
| TRIM25 | RING | PTEN | K63 | lung cancer | 33931764 |
| TRIM31 | RING | TSC1-TSC2 | K48 | hepatocellular carcinoma | 28967907 |
| p53 | K48 | breast cancer | 34650049 | ||
| TRIM32 | RING | ULK1 | K63 | colon cancer, lung cancer, hepatocellular carcinoma | 31123703 |
| TRIM45 | RING | p53 | K48 | glioma | 28542145 |
| TRIM50 | RING | Beclin-1 | K63 | adenocarcinoma | 29604308 |
| TTC3 | RING | AKT | K48 | B Cell Lymphoma | 20059950 |
| VHL | RING | HIF-1α | K48 | adenocarcinoma | 25958982, 12086861 |
| XIAP | RING | HIF-1α | K63 | osteosarcoma, renal cell carcinoma | 28666324 |
| ZNRF1 | RING | AKT | K48 | neuroblastoma | 22057101 |
| Peli1 | RING-like | PKC theta | K48 | T cell lymphoma, colon adenocarcinoma | 35058288 |
| TSC1 | K63 | melanoma | 33215753 | ||
| CHIP | U-box | AKT | K48 | breast cancer, adenocarcinoma | 21767636 |
| p53 | K48 | lung cancer, colon cancer | 29953728 |
| Name | Type | Substrate | Ubiquitination Residue | Ref. (PMID Number) |
|---|---|---|---|---|
| ARF-BP1/Mule | HECT | p53 | unknown | 15989956 |
| MCL-1 | unknown | 15989957 | ||
| E6-AP | HECT | p53 | unknown | 31749782 |
| HUWE1 | HECT | histone H1 | unknown | 29127375 |
| ITCH | HECT | p73 | unknown | 15678106 |
| p63 | unknown | 16908849 | ||
| WWOX | K63 | 25331887 | ||
| H1.2 | K48, K63 | 30517763 | ||
| NEDD4 | HECT | RNA PolⅡ | unknown | 17996703 |
| Mdm2 | K63 | 24413081 | ||
| NEDD4L | HECT | OGG1 | unknown | 33282879 |
| Rsp5 | HECT | RNA PolⅡ | unknown | 9108033 |
| Smurf1 | HECT | RhoB | unknown | 25249323 |
| Smurf2 | HECT | H2AX | unknown | 31533041 |
| RNF20 | unknown | 33097595 | ||
| TRIP12 | HECT | USP7/HAUSP | K48 | 27800609 |
| UBR5 | HECT | ATMIN | unknown | 25092319 |
| WWP2 | HECT | SOX2 | unknown | 25042802, 34193614 |
| APC/CCdc20 | multimeric RING | MCL-1 | unknown | 29987118 |
| BIM | unknown | 24871945 | ||
| APC/CCDH1 | multimeric RING | CtIP | unknown | 25349192 |
| E2F1 | K11 | 22580462 | ||
| BRCA1 | RING | CtIP | unknown | 16818604 |
| BRCA1-BARD1 | RING | histone H2A | mono | 33589814 |
| RNA PolⅡ | K6 | 15886201, 15905410 | ||
| CHFR | RING | PARP1 | K48, K63 | 23268447 |
| COP1 | RING | p53 | unknown | 16931761 |
| CRL4ADDB1 | RING | p53 | unknown | 17967871 |
| p73 | unknown | 23085759 | ||
| CRL4Cdt2 | RING | p21 | unknown | 18794347 |
| Cul4B | RING | p53 | K48 (no direct evidence) | 33524014 |
| HUWE1 | unknown | 25883150 | ||
| CUL4-DDB-ROC1 | RING | histone H3, H4 | unknown | 16678110 |
| FANCL | RING | FANCD2 | unknown | 17352736 |
| MARCH7 | RING | Mdm2 | K63 | 29295817 |
| Mdm2 | RING | p53 | K48 | 12507556 |
| p73 | unknown | 34716260 | ||
| Pirh2 | RING | p53 | K48 | 12654245 |
| CHK2 | unknown | 23449389 | ||
| p73 | K11, K29, K48, K63 | 21994467 | ||
| Rad5 | RING | PCNA | K63 | 18757916 |
| Rad6 | RING | PCNA | K63 | 12226657 |
| Rad18 | RING | PCNA | K63 | 19851286 |
| RFWD3-Mdm2 | RING | p53 | unknown | 20173098 |
| RNF2 | RING | H2AX | unknown | 21676867 |
| RNF8 | RING | MDC1 | K63 | 18006705, 31182912 |
| histone H1 | K63 | 29127375 | ||
| histone H2A, H2AX | K63 | 22980979 | ||
| JMJD2A | K48 | 22373579 | ||
| Ku80 | K48 | 22266820 | ||
| PCNA | unknown | 18948756 | ||
| NBS1 | unknown | 23115235 | ||
| RecQL4 | K6, K27, K29 | 33674555 | ||
| RNF19A | RING | BARD1 | K63 | 34789768 |
| RNF20/RNF40 | RING | histone H2B | unknown | 30692271 |
| RNF111 | RING | XPC | unknown | 23751493 |
| RNF168 | RING | histone H2A | K27, K63 | 25578731, 22980979 |
| JMJD2A | unknown | 22373579 | ||
| H2AX | unknown | 31533041 | ||
| SCFFBXW7 | RING | p53 | K48 | 31346036, 31337255 |
| p63 | unknown | 20571051 | ||
| XRCC4 | K63 | 26774286 | ||
| PLK1 | K48 | 24970797 | ||
| SOX9 | unknown | 27566146 | ||
| BLM | K48 | 26028025 | ||
| SCFSKP2 | RING | NBS1 | K63 | 22464731 |
| SCFβ-TrCP | RING | BIM | unknown | 19150432 |
| CDC25A | unknown | 14681206 | ||
| CLASPIN | unknown | 16885022 | ||
| SCFβ-TrCP1 | RING | Mdm2 | K48 | 33676897 |
| SCFβ-TrCP2 | RING | Mdm2 | K63 | 33676897 |
| TRIM17 | RING | MCL-1 | unknown | 22976837 |
| TRIM24 | RING | p53 | unknown | 24820418 |
| UHRF1 | RING | RIF1 | K63 | 26727879 |
| Peli1 | RING-like | NBS1 | K63 | 30952868 |
| PRP19 | U-box | RPA | K63 | 24332808 |
| Target | Compounds | Chemical Formula |
|---|---|---|
| 20S Proteasome | Bortezomib | C19H25BN4O4 |
| Carfilzomib | C40H57N5O7 | |
| Oprozomib | C25H32N4O7S | |
| Ixazomib | C14H19BCl2N2O4 | |
| E1 enzyme | TAK-243 (MLN7243) | C19H20F3N5O5S2 |
| E2 enzyme | CC0651 | C20H21Cl2NO6 |
| NSC697923 | C11H9NO5S | |
| Leucettamol A | C30H52N2O2 | |
| Manadosterol A | C54H83Na5O21S5 | |
| Manadosterol B | C54H84Na4O18S4 | |
| E3 ligase | Nutlin-3a | C30H30Cl2N4O4 |
| KRT-232 (AMG 232) | C28H35Cl2NO5S | |
| Milademetan (DS-3032) | C30H34Cl2FN5O4 | |
| HDM201 | C26H24Cl2N6O4 | |
| ALRN-6924 | C95H140N20O23 | |
| GDC-0917 | C29H36N6O4S | |
| Debio1143 (Xevinapant) | C32H43N5O4 | |
| Apcin | C13H14Cl3N7O4 | |
| TAME | C14H22N4O4S | |
| Oridonin | C20H28O6 | |
| SZL-P1-41 | C24H24N2O3S | |
| Longikaurin A | C20H28O5 | |
| Curcumin | C21H20O6 | |
| Dioscin | C45H72O16 | |
| GS143 | C28H19FN2O4 | |
| Erioflorin | C19H24O6 | |
| Deubiquitinase (DUB) | IU1 | C18H21FN2O |
| IU1-47 | C19H23ClN2O | |
| WP1130 | C19H18BrN3O | |
| HBX 41,108 | C13H3ClN4O |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koo, S.-Y.; Park, E.-J.; Noh, H.-J.; Jo, S.-M.; Ko, B.-K.; Shin, H.-J.; Lee, C.-W. Ubiquitination Links DNA Damage and Repair Signaling to Cancer Metabolism. Int. J. Mol. Sci. 2023, 24, 8441. https://doi.org/10.3390/ijms24098441
Koo S-Y, Park E-J, Noh H-J, Jo S-M, Ko B-K, Shin H-J, Lee C-W. Ubiquitination Links DNA Damage and Repair Signaling to Cancer Metabolism. International Journal of Molecular Sciences. 2023; 24(9):8441. https://doi.org/10.3390/ijms24098441
Chicago/Turabian StyleKoo, Seo-Young, Eun-Ji Park, Hyun-Ji Noh, Su-Mi Jo, Bo-Kyoung Ko, Hyun-Jin Shin, and Chang-Woo Lee. 2023. "Ubiquitination Links DNA Damage and Repair Signaling to Cancer Metabolism" International Journal of Molecular Sciences 24, no. 9: 8441. https://doi.org/10.3390/ijms24098441
APA StyleKoo, S.-Y., Park, E.-J., Noh, H.-J., Jo, S.-M., Ko, B.-K., Shin, H.-J., & Lee, C.-W. (2023). Ubiquitination Links DNA Damage and Repair Signaling to Cancer Metabolism. International Journal of Molecular Sciences, 24(9), 8441. https://doi.org/10.3390/ijms24098441






