Combined Structural and Computational Study of the mRubyFT Fluorescent Timer Locked in Its Blue Form
Abstract
1. Introduction
2. Results and Discussion
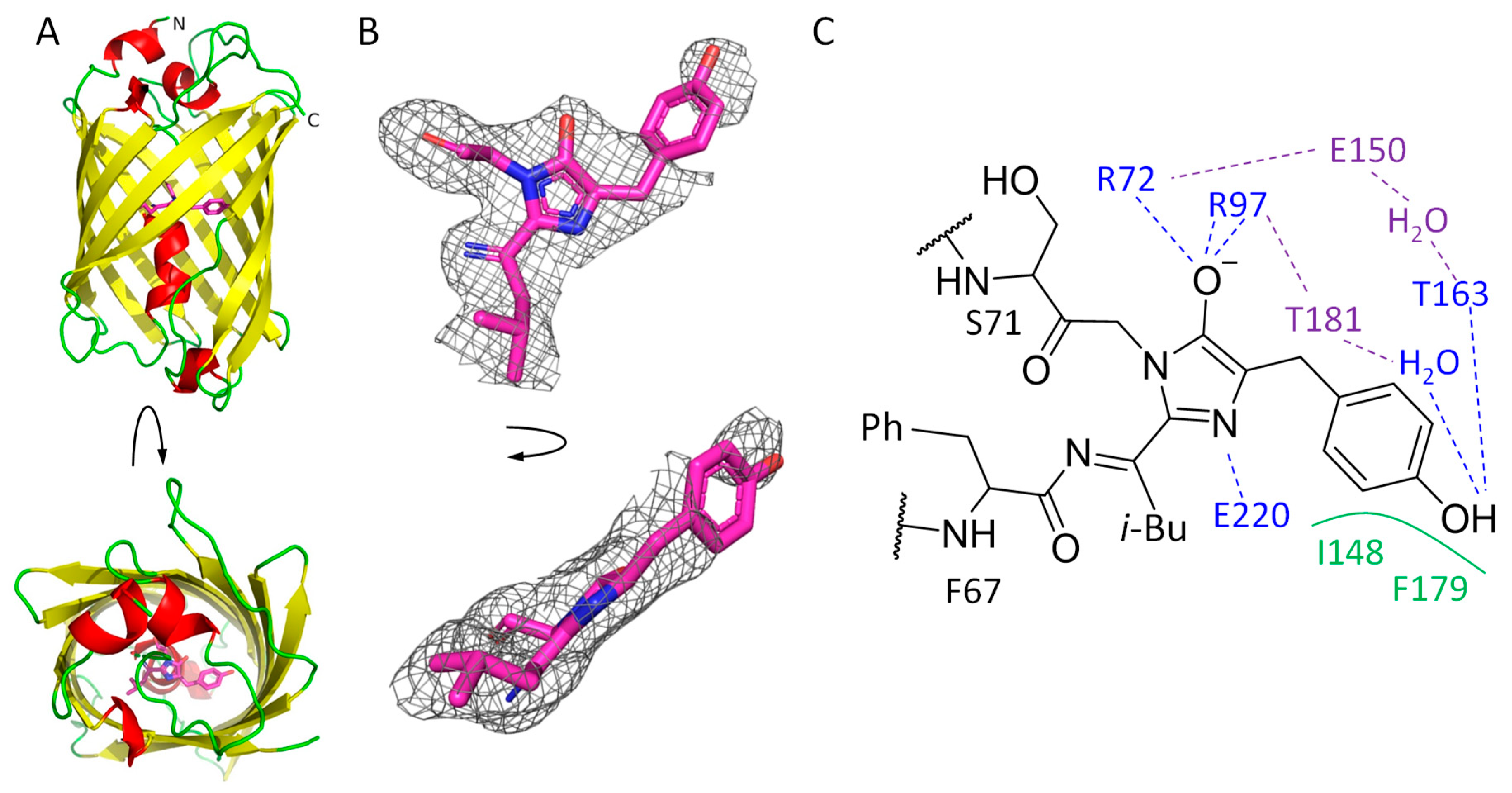
2.1. mRubyFTS148I Overall Structure and Chromophore Environment
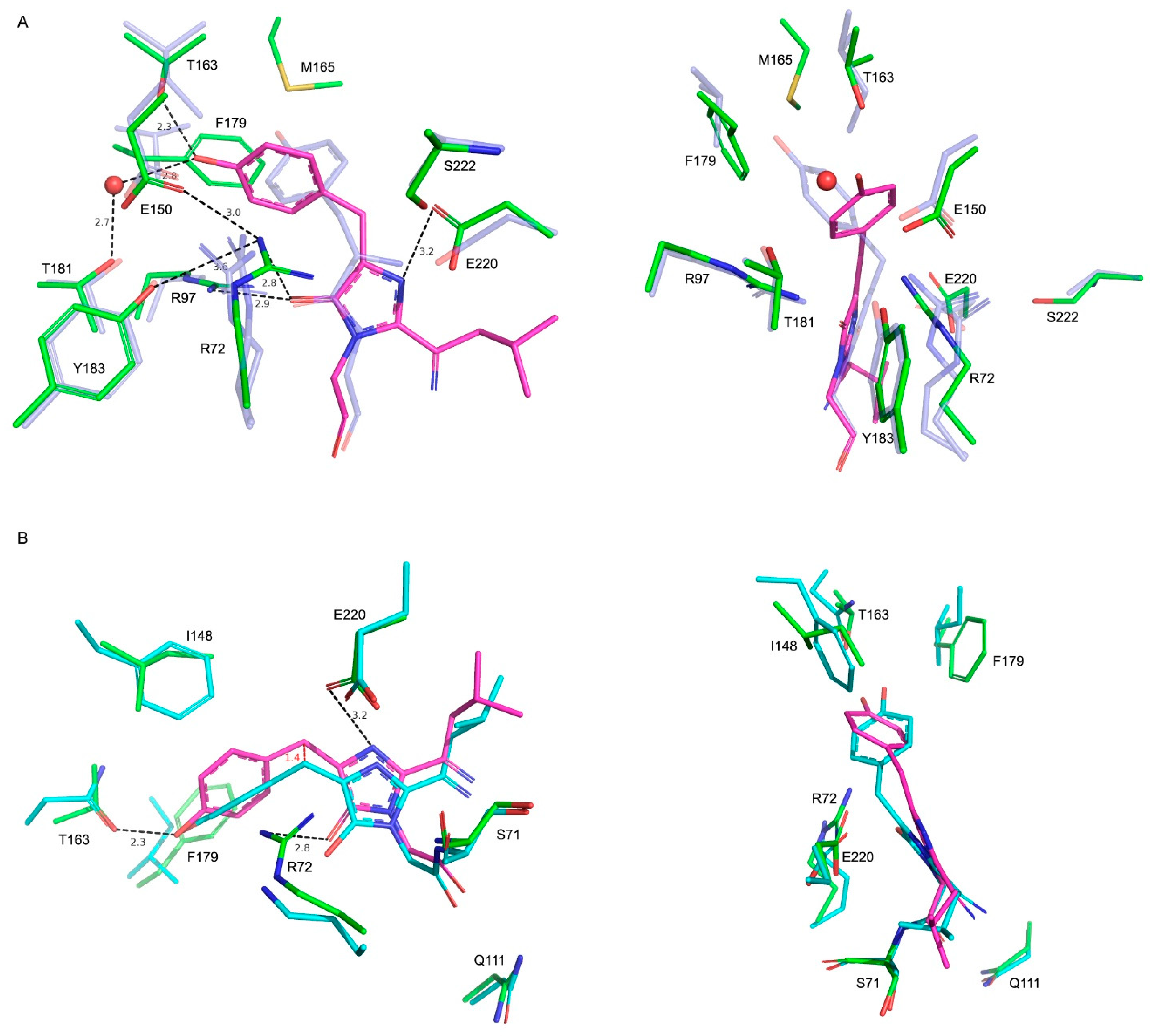
2.2. Comparison with Blue102—A Blue Form of Fluorescent Timer Fast-FT
2.3. Comparison with mTagBFP
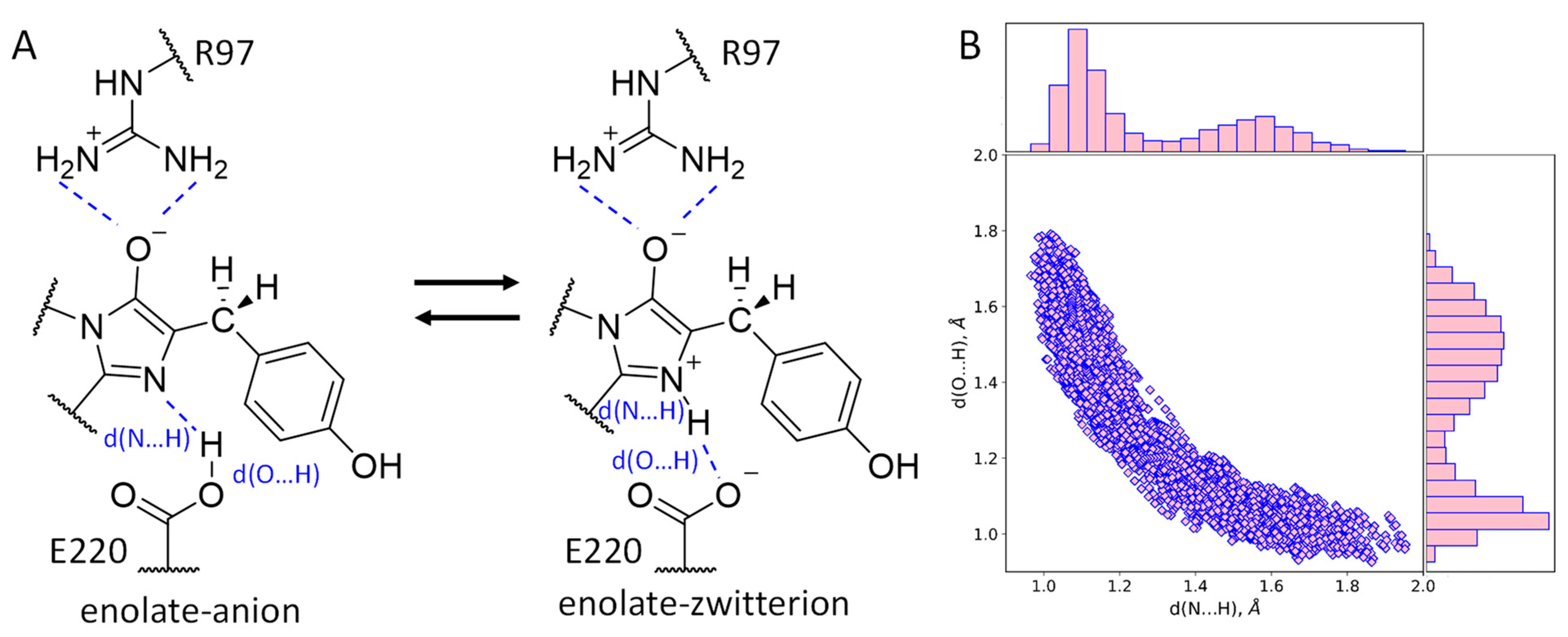
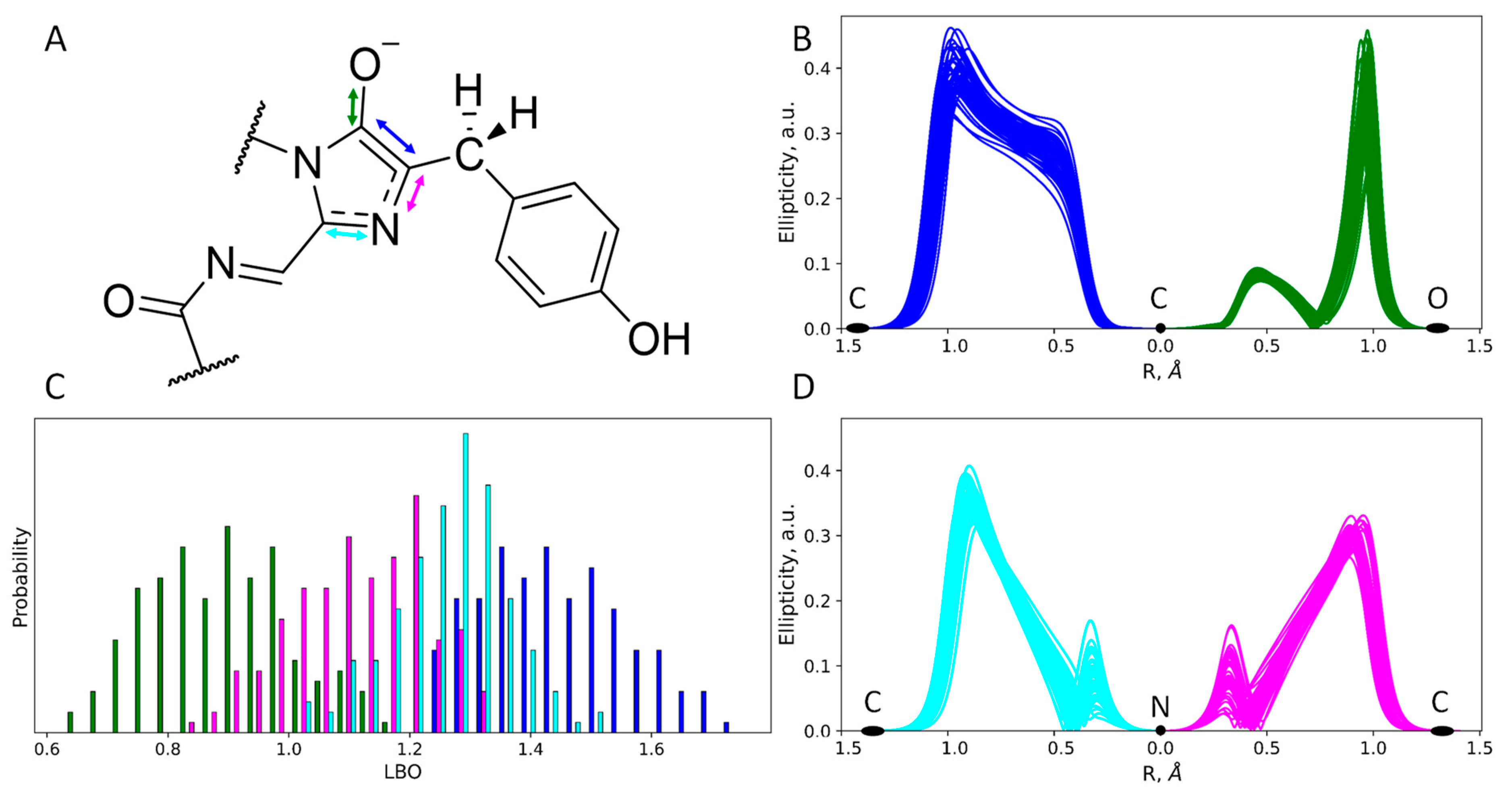
2.4. Structure and Protonation State of the Chromophore in the mRubyFTS148I
2.5. Photophysical Properties of the Chromophore: Experiment and Calculations
3. Material and Methods
3.1. mRubyFTS148I Cloning, Expression and Purification
3.2. Spectral Characterization
3.3. Crystallization and Structure Determination of mRubyFTS148I
3.4. Structural Analysis
3.5. Molecular Modeling
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Subach, O.M.; Vlaskina, A.V.; Agapova, Y.K.; Nikolaeva, A.Y.; Anokhin, K.V.; Piatkevich, K.D.; Patrushev, M.V.; Boyko, K.M.; Subach, F.V. Blue-to-Red TagFT, mTagFT, mTsFT, and Green-to-FarRed mNeptusFT2 Proteins, Genetically Encoded True and Tandem Fluorescent Timers. Int. J. Mol. Sci. 2023, 24, 3279. [Google Scholar] [CrossRef]
- Subach, F.V.; Subach, O.M.; Gundorov, I.S.; Morozova, K.S.; Piatkevich, K.D.; Cuervo, A.M.; Verkhusha, V. V Monomeric fluorescent timers that change color from blue to red report on cellular trafficking. Nat. Chem. Biol. 2009, 5, 118–126. [Google Scholar] [CrossRef]
- Subach, O.M.; Tashkeev, A.; Vlaskina, A.V.; Petrenko, D.E.; Gaivoronskii, F.A.; Nikolaeva, A.Y.; Ivashkina, O.I.; Anokhin, K.V.; Popov, V.O.; Boyko, K.M.; et al. The mRubyFT Protein, Genetically Encoded Blue-to-Red Fluorescent Timer. Int. J. Mol. Sci. 2022, 23, 3208. [Google Scholar] [CrossRef] [PubMed]
- Okuyama, T.; Kitamura, T.; Roy, D.S.; Itohara, S.; Tonegawa, S. Ventral CA1 neurons store social memory. Science 2016, 353, 1536–1541. [Google Scholar] [CrossRef] [PubMed]
- Pletnev, S.; Subach, F.V.; Dauter, Z.; Wlodawer, A.; Verkhusha, V.V. Understanding Blue-to-Red Conversion in Monomeric Fluorescent Timers and Hydrolytic Degradation of Their Chromophores. J. Am. Chem. Soc. 2010, 132, 2243–2253. [Google Scholar] [CrossRef] [PubMed]
- Boyko, K.M.; Nikolaeva, A.Y.; Dorovatovskii, P.V.; Vlaskina, A.V.; Subach, O.M.; Subach, F.V. Three-Dimensional Structure of the S148F Mutant of Blue-to-Red Fluorescent Timer mRubyFT. Crystallogr. Reports 2022, 67, 905–908. [Google Scholar] [CrossRef]
- Barondeau, D.P.; Tainer, J.A.; Getzoff, E.D. Structural Evidence for an Enolate Intermediate in GFP Fluorophore Biosynthesis. J. Am. Chem. Soc. 2006, 128, 3166–3168. [Google Scholar] [CrossRef]
- Heim, R.; Cubitt, A.B.; Tsien, R.Y. Improved green fluorescence. Nature 1995, 373, 663–664. [Google Scholar] [CrossRef]
- Heim, R.; Prasher, D.C.; Tsien, R.Y. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl. Acad. Sci. USA 1994, 91, 12501–12504. [Google Scholar] [CrossRef]
- Ma, Y.; Sun, Q.; Zhang, H.; Peng, L.; Yu, J.-G.; Smith, S.C. The Mechanism of Cyclization in Chromophore Maturation of Green Fluorescent Protein: A Theoretical Study. J. Phys. Chem. B 2010, 114, 9698–9705. [Google Scholar] [CrossRef]
- Miyawaki, A.; Shcherbakova, D.M.; Verkhusha, V. V Red fluorescent proteins: Chromophore formation and cellular applications. Curr. Opin. Struct. Biol. 2012, 22, 679–688. [Google Scholar] [CrossRef] [PubMed]
- Pletneva, N.V.; Pletnev, V.Z.; Lukyanov, K.A.; Gurskaya, N.G.; Goryacheva, E.A.; Martynov, V.I.; Wlodawer, A.; Dauter, Z.; Pletnev, S. Structural Evidence for a Dehydrated Intermediate in Green Fluorescent Protein Chromophore Biosynthesis. J. Biol. Chem. 2010, 285, 15978–15984. [Google Scholar] [CrossRef]
- Verkhusha, V.V.; Chudakov, D.M.; Gurskaya, N.G.; Lukyanov, S.; Lukyanov, K.A. Common Pathway for the Red Chromophore Formation in Fluorescent Proteins and Chromoproteins. Chem. Biol. 2004, 11, 845–854. [Google Scholar] [CrossRef]
- Yum, C.; Ahn, T.; Shim, W.-S. Development of a Novel Blue Fluorescent Gene-encoded Calcium Indicator Modified from GCaMP3. J. Fluoresc. 2020, 30, 1287–1293. [Google Scholar] [CrossRef] [PubMed]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkóczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.-W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. Sect. D Struct. Biol. 2019, 75, 861–877. [Google Scholar] [CrossRef] [PubMed]
- Akerboom, J.; Carreras Calderón, N.; Tian, L.; Wabnig, S.; Prigge, M.; Tolö, J.; Gordus, A.; Orger, M.B.; Severi, K.E.; Macklin, J.J.; et al. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front. Mol. Neurosci. 2013, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Kremers, G.-J.; Goedhart, J.; van den Heuvel, D.J.; Gerritsen, H.C.; Gadella, T.W.J. Improved Green and Blue Fluorescent Proteins for Expression in Bacteria and Mammalian Cells. Biochemistry 2007, 46, 3775–3783. [Google Scholar] [CrossRef]
- Mena, M.A.; Treynor, T.P.; Mayo, S.L.; Daugherty, P.S. Blue fluorescent proteins with enhanced brightness and photostability from a structurally targeted library. Nat. Biotechnol. 2006, 24, 1569–1571. [Google Scholar] [CrossRef] [PubMed]
- Subach, O.M.; Cranfill, P.J.; Davidson, M.W.; Verkhusha, V.V. An Enhanced Monomeric Blue Fluorescent Protein with the High Chemical Stability of the Chromophore. PLoS ONE 2011, 6, e28674. [Google Scholar] [CrossRef]
- Subach, O.M.; Malashkevich, V.N.; Zencheck, W.D.; Morozova, K.S.; Piatkevich, K.D.; Almo, S.C.; Verkhusha, V.V. Structural Characterization of Acylimine-Containing Blue and Red Chromophores in mTagBFP and TagRFP Fluorescent Proteins. Chem. Biol. 2010, 17, 333–341. [Google Scholar] [CrossRef]
- Acharya, A.; Bogdanov, A.M.; Grigorenko, B.L.; Bravaya, K.B.; Nemukhin, A.V.; Lukyanov, K.A.; Krylov, A.I. Photoinduced Chemistry in Fluorescent Proteins: Curse or Blessing? Chem. Rev. 2017, 117, 758–795. [Google Scholar] [CrossRef]
- Bravaya, K.B.; Grigorenko, B.L.; Nemukhin, A.V.; Krylov, A.I. Quantum Chemistry Behind Bioimaging: Insights from Ab Initio Studies of Fluorescent Proteins and Their Chromophores. Acc. Chem. Res. 2012, 45, 265–275. [Google Scholar] [CrossRef]
- Khrenova, M.G.; Mulashkin, F.D.; Bulavko, E.S.; Zakharova, T.M.; Nemukhin, A.V. Dipole Moment Variation Clears Up Electronic Excitations in the π-Stacked Complexes of Fluorescent Protein Chromophores. J. Chem. Inf. Model. 2020, 60, 6288–6297. [Google Scholar] [CrossRef]
- Khrenova, M.G.; Mulashkin, F.D.; Nemukhin, A.V. Modeling Spectral Tuning in Red Fluorescent Proteins Using the Dipole Moment Variation upon Excitation. J. Chem. Inf. Model. 2021, 61, 5125–5132. [Google Scholar] [CrossRef]
- Winter, G.; Waterman, D.G.; Parkhurst, J.M.; Brewster, A.S.; Gildea, R.J.; Gerstel, M.; Fuentes-Montero, L.; Vollmar, M.; Michels-Clark, T.; Young, I.D.; et al. DIALS: Implementation and evaluation of a new integration package. Acta Crystallogr. Sect. D Struct. Biol. 2018, 74, 85–97. [Google Scholar] [CrossRef]
- Evans, P. Scaling and assessment of data quality. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006, 62, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Vagin, A.; Teplyakov, A. MOLREP: An Automated Program for Molecular Replacement. J. Appl. Crystallogr. 1997, 30, 1022–1025. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Skubák, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC 5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Smart, O.S.; Womack, T.O.; Flensburg, C.; Keller, P.; Paciorek, W.; Sharff, A.; Vonrhein, C.; Bricogne, G. Exploiting structure similarity in refinement: Automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 368–380. [Google Scholar] [CrossRef]
- Collaborative Computational Project, N. 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994, 50, 760–763. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Krissinel, E.; Henrick, K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2256–2268. [Google Scholar] [CrossRef]
- Denning, E.J.; Priyakumar, U.D.; Nilsson, L.; Mackerell, A.D. Impact of 2′-hydroxyl sampling on the conformational properties of RNA: Update of the CHARMM all-atom additive force field for RNA. J. Comput. Chem. 2011, 32, 1929–1943. [Google Scholar] [CrossRef]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.M.; Mittal, J.; Feig, M.; MacKerell, A.D. Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of the Backbone ϕ, ψ and Side-Chain χ1 and χ2 Dihedral Angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM general force field (CGenFF): A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Phillips, J.C.; Hardy, D.J.; Maia, J.D.C.; Stone, J.E.; Ribeiro, J.V.; Bernardi, R.C.; Buch, R.; Fiorin, G.; Hénin, J.; Jiang, W.; et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [Google Scholar] [CrossRef]
- Adamo, C.; Barone, V. Toward reliable density functional methods without adjustable parameters: The PBE0 model. J. Chem. Phys. 1999, 110, 6158. [Google Scholar] [CrossRef]
- Grimme, S.; Antony, J.; Ehrlich, S.; Krieg, H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010, 132, 154104. [Google Scholar] [CrossRef] [PubMed]
- Melo, M.C.R.; Bernardi, R.C.; Rudack, T.; Scheurer, M.; Riplinger, C.; Phillips, J.C.; Maia, J.D.C.; Rocha, G.B.; Ribeiro, J.V.; Stone, J.E.; et al. NAMD goes quantum: An integrative suite for hybrid simulations. Nat. Methods 2018, 15, 351–354. [Google Scholar] [CrossRef] [PubMed]
- TeraChem v 1.9, PetaChem, LLC. Available online: www.petachem.com (accessed on 25 April 2023).
- Lu, T.; Chen, F. Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef]
- Lu, T.; Chen, F. Bond Order Analysis Based on the Laplacian of Electron Density in Fuzzy Overlap Space. J. Phys. Chem. A 2013, 117, 3100–3108. [Google Scholar] [CrossRef]
- Bader, R.F.W.; Slee, T.S.; Cremer, D.; Kraka, E. Description of conjugation and hyperconjugation in terms of electron distributions. J. Am. Chem. Soc. 1983, 105, 5061–5068. [Google Scholar] [CrossRef]
- Mardirossian, N.; Head-Gordon, M. Mapping the genome of meta-generalized gradient approximation density functionals: The search for B97M-V. J. Chem. Phys. 2015, 142, 074111. [Google Scholar] [CrossRef] [PubMed]
- Neese, F. The ORCA program system. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2012, 2, 73–78. [Google Scholar] [CrossRef]


| Data Collection | |
|---|---|
| Diffraction source | ”Belok-RSA“ beamline, NRC “Kurchatov Institute” |
| Wavelength (Å) | 0.79 |
| Temperature (K) | 100 |
| Detector | CCD |
| Crystal-to-detector distance (mm) | 120.00 |
| Rotation range per image (°) | 1.0 |
| Total rotation range (°) | 130 |
| Space group | P212121 |
| a, b, c (Å) | 31.79; 66.83; 97.76 |
| α, β, γ (°) | 90.0; 90.0; 90.0 |
| Unique reflections | 19640 (1116) |
| Resolution range (Å) | 48.9–1.80 (1.84–1.80) |
| Completeness (%) | 98.2 (96.7) |
| Average redundancy | 4.9 (5.2) |
| 〈I/σ(I)〉 | 6.1 (0.2) |
| Rmeas (%) | 9.0 (133.0) |
| CC1/2 | 100.0 (51.0) |
| Refinement | |
| Rfact (%) | 22.7 |
| Rfree.(%) | 26.8 |
| Bonds (Å) | 0.01 |
| Angles (°) | 2.04 |
| Ramachandran plot | |
| Most favored (%) | 97.2 |
| Allowed (%) | 2.8 |
| No. atoms | |
| Protein | 1741 |
| Water | 76 |
| Chromophore | 23 |
| Other ligands | 0 |
| B-factors (Å2) | |
| Protein | 29.5 |
| Water | 37.1 |
| Chromophore | 38.8 |
| PDB ID | 7Q6B |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boyko, K.M.; Khrenova, M.G.; Nikolaeva, A.Y.; Dorovatovskii, P.V.; Vlaskina, A.V.; Subach, O.M.; Popov, V.O.; Subach, F.V. Combined Structural and Computational Study of the mRubyFT Fluorescent Timer Locked in Its Blue Form. Int. J. Mol. Sci. 2023, 24, 7906. https://doi.org/10.3390/ijms24097906
Boyko KM, Khrenova MG, Nikolaeva AY, Dorovatovskii PV, Vlaskina AV, Subach OM, Popov VO, Subach FV. Combined Structural and Computational Study of the mRubyFT Fluorescent Timer Locked in Its Blue Form. International Journal of Molecular Sciences. 2023; 24(9):7906. https://doi.org/10.3390/ijms24097906
Chicago/Turabian StyleBoyko, Konstantin M., Maria G. Khrenova, Alena Y. Nikolaeva, Pavel V. Dorovatovskii, Anna V. Vlaskina, Oksana M. Subach, Vladimir O. Popov, and Fedor V. Subach. 2023. "Combined Structural and Computational Study of the mRubyFT Fluorescent Timer Locked in Its Blue Form" International Journal of Molecular Sciences 24, no. 9: 7906. https://doi.org/10.3390/ijms24097906
APA StyleBoyko, K. M., Khrenova, M. G., Nikolaeva, A. Y., Dorovatovskii, P. V., Vlaskina, A. V., Subach, O. M., Popov, V. O., & Subach, F. V. (2023). Combined Structural and Computational Study of the mRubyFT Fluorescent Timer Locked in Its Blue Form. International Journal of Molecular Sciences, 24(9), 7906. https://doi.org/10.3390/ijms24097906





