m6Aminer: Predicting the m6Am Sites on mRNA by Fusing Multiple Sequence-Derived Features into a CatBoost-Based Classifier
Abstract
1. Introduction
2. Results
2.1. Model Performance with Different Machine Learning Algorithms and Feature Subsets
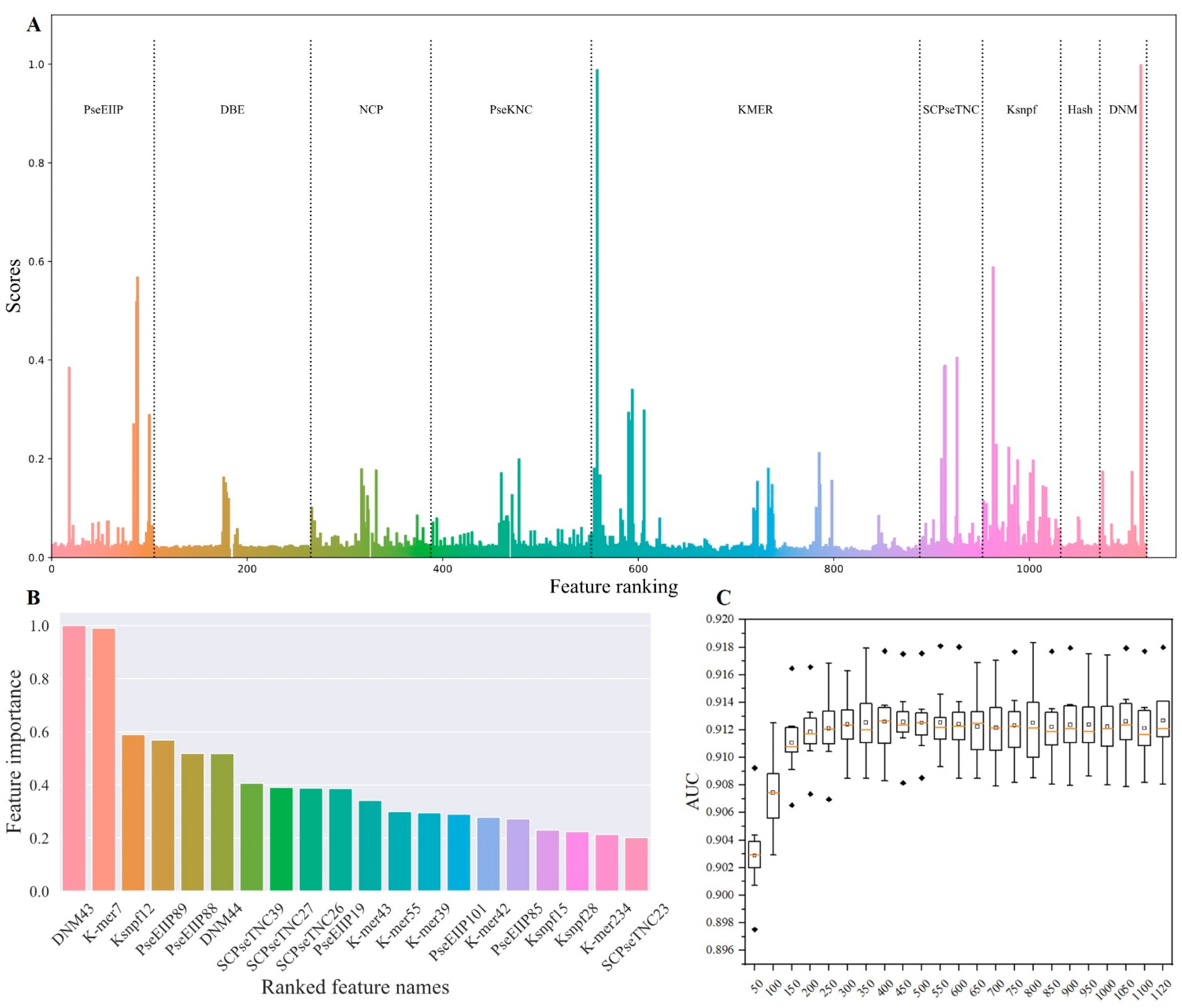
2.2. Feature Ranking and Selection
2.3. Model Optimization
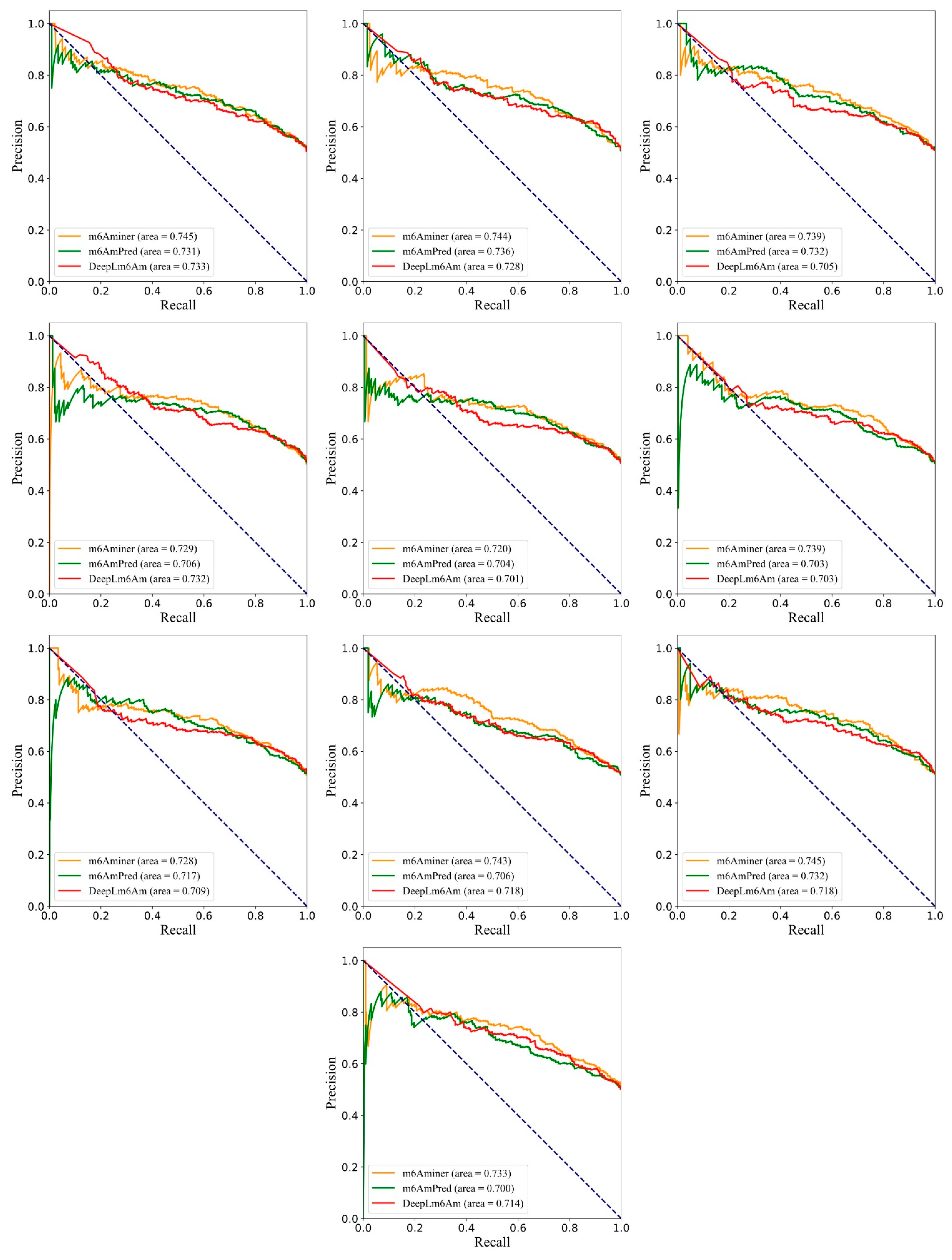
2.4. Comparison with the State-of-the-Art Predictors
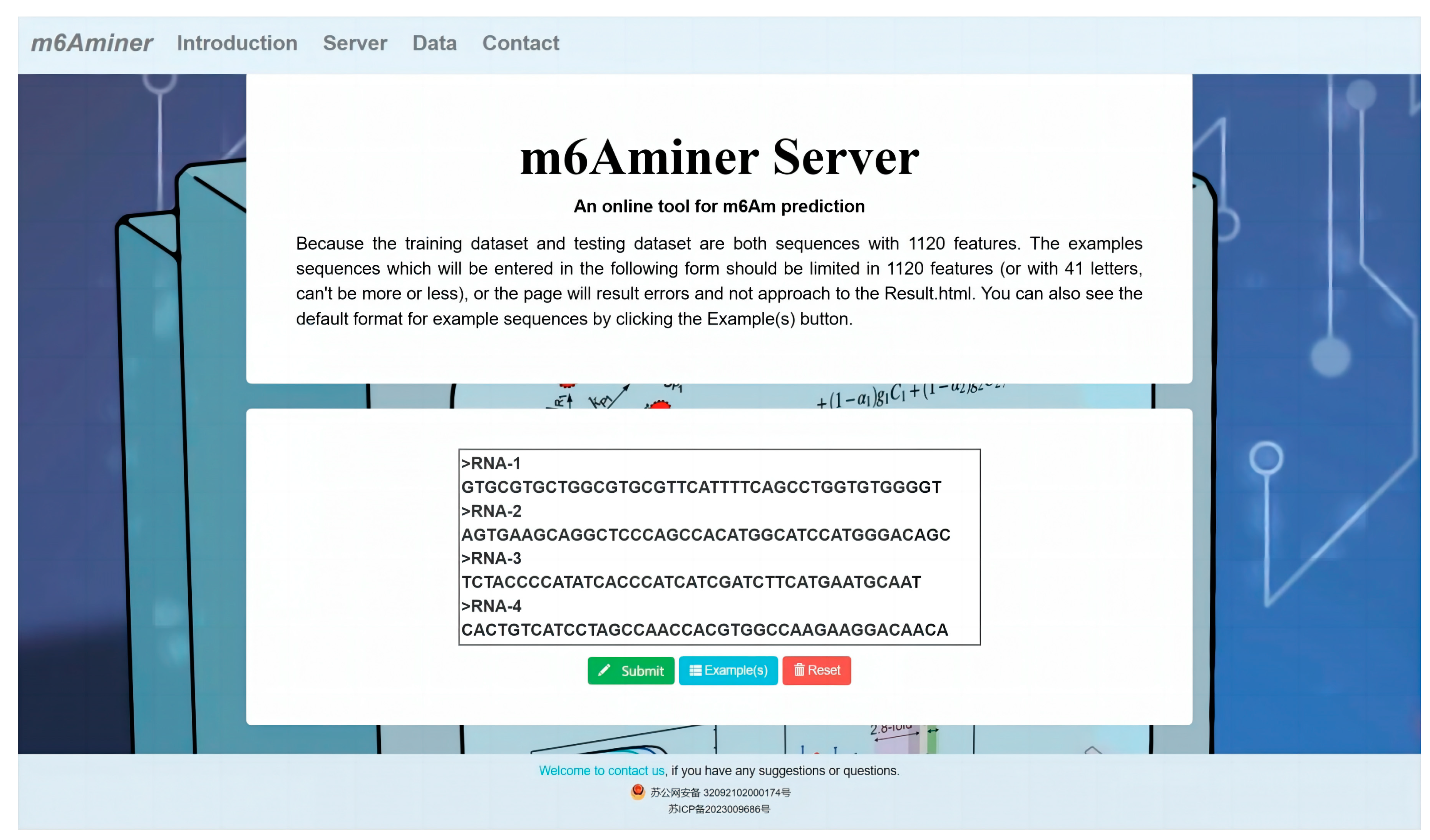
2.5. Web Server
3. Discussions
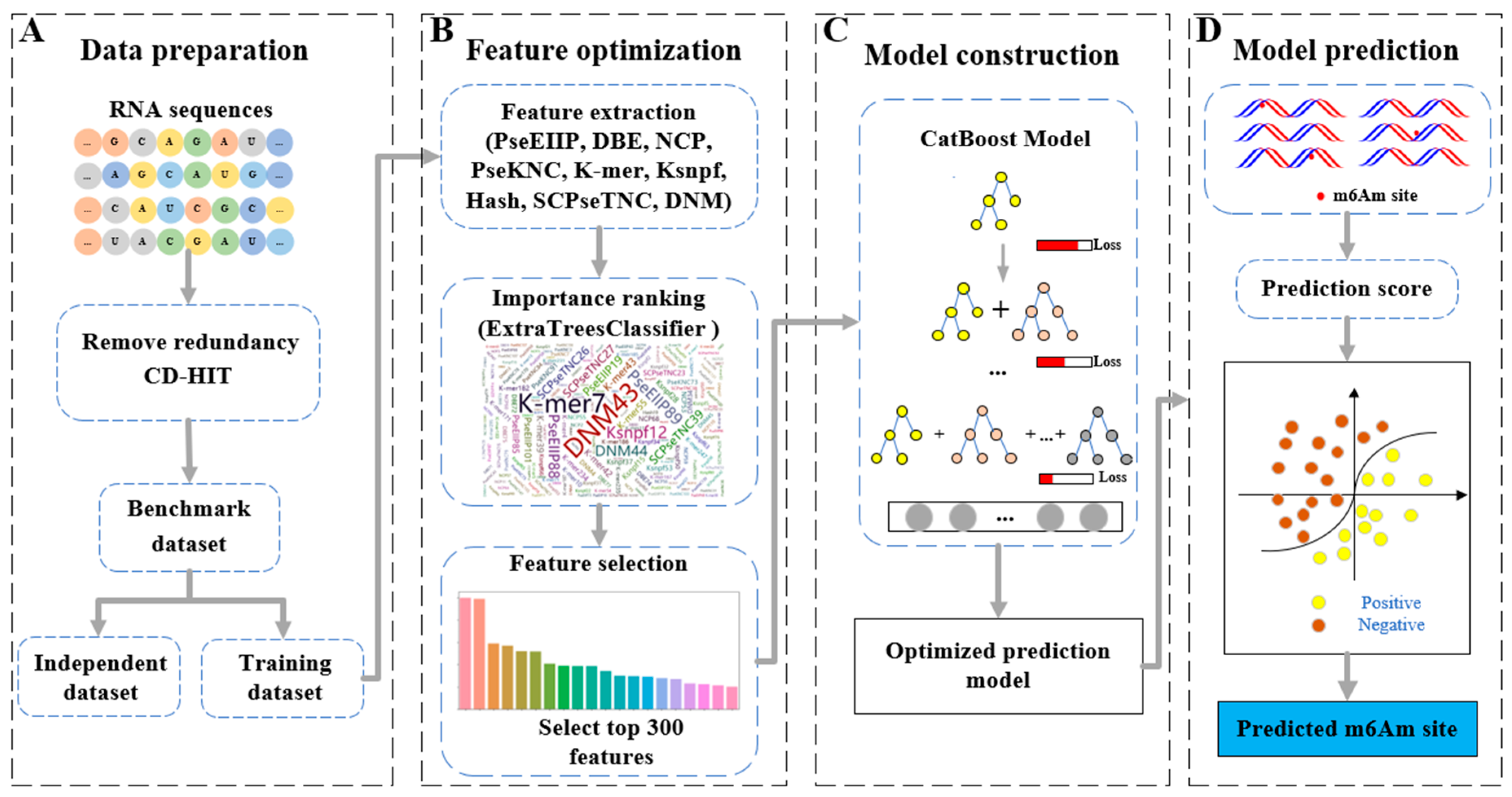
4. Materials and Methods
4.1. Benchmark Dataset
4.2. Feature Extraction
4.2.1. Pseudo Electron–Ion Interaction Potential (PseEIIP)
4.2.2. Hash Decimal Conversion Method (Hash)
4.2.3. Dinucleotide Binary Encoding (DBE)
4.2.4. Nucleotide Chemical Property (NCP)
4.2.5. Pseudo k-Tuple Composition (PseKNC)
4.2.6. Dinucleotide Numerical Mapping (DNM)
4.2.7. K Monomeric Units (K-mer)
4.2.8. Series Correlation Pseudo Trinucleotide Composition (SCPseTNC)
4.2.9. K-Spaced Nucleotide Pair Frequency (Ksnpf)
4.3. CatBoost
4.4. Performance Evaluation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cohn, E.; Volkin, W. Nucleoside-5′-Phosphates from Ribonucleic Acid. Nature 1951, 167, 483–484. [Google Scholar] [CrossRef]
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piatkowski, P.; Baginski, B.; Wirecki, T.K.; de Crecy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: A database of RNA modification pathways. Nucleic Acids Res. 2018, 46, D303–D307. [Google Scholar] [CrossRef] [PubMed]
- Helm, M.; Alfonzo, J.D. Posttranscriptional RNA Modifications: Playing metabolic games in a cell’s chemical Legoland. Chem. Biol. 2014, 21, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef] [PubMed]
- Batista, P.J.; Molinie, B.; Wang, J.; Qu, K.; Zhang, J.; Li, L.; Bouley, D.M.; Lujan, E.; Haddad, B.; Daneshvar, K.; et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 2014, 15, 707–719. [Google Scholar] [CrossRef]
- Delaunay, S.; Frye, M. RNA modifications regulating cell fate in cancer. Nat. Cell Biol. 2019, 21, 552–559. [Google Scholar] [CrossRef]
- Jonkhout, N.; Tran, J.; Smith, M.A.; Schonrock, N.; Mattick, J.S.; Novoa, E.M. The RNA modification landscape in human disease. RNA 2017, 23, 1754–1769. [Google Scholar] [CrossRef]
- Frye, M.; Jaffrey, S.R.; Pan, T.; Rechavi, G.; Suzuki, T. RNA modifications: What have we learned and where are we headed? Nat. Rev. Genet. 2016, 17, 365–372. [Google Scholar] [CrossRef]
- Gilbert, W.V.; Bell, T.A.; Schaening, C. Messenger RNA modifications: Form, distribution, and function. Science 2016, 352, 1408–1412. [Google Scholar] [CrossRef]
- Sun, H.; Zhang, M.; Li, K.; Bai, D.; Yi, C. Cap-specific, terminal N-6-methylation by a mammalian m6Am methyltransferase. Cell Res. 2019, 29, 80–82. [Google Scholar] [CrossRef]
- Keith, J.M.; Ensinger, M.J.; Moss, B. HeLa cell RNA (2′-O-methyladenosine-N6-)-methyltransferase specific for the capped 5′-end of messenger RNA. J. Biol. Chem. 1978, 253, 5033–5039. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Gershowitz, A.; Moss, B. N6, O2′-dimethyladenosine a novel methylated ribonucleoside next to the 5′ terminal of animal cell and virus mRNAs. Nature 1975, 257, 251–253. [Google Scholar] [CrossRef] [PubMed]
- Akichika, S.; Hirano, S.; Shichino, Y.; Suzuki, T.; Nishimasu, H.; Ishitani, R.; Sugita, A.; Hirose, Y.; Iwasaki, S.; Nureki, O.; et al. Cap-specific terminal N-6-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 2019, 363, eaav0080. [Google Scholar] [CrossRef]
- Sendinc, E.; Valle-Garcia, D.; Dhall, A.; Chen, H.; Henriques, T.; Navarrete-Perea, J.; Sheng, W.; Gygi, S.P.; Adelman, K.; Shi, Y. PCIF1 Catalyzes m6Am mRNA Methylation to Regulate Gene Expression. Mol. Cell 2019, 75, 620–630.e9. [Google Scholar] [CrossRef] [PubMed]
- Mauer, J.; Luo, X.; Blanjoie, A.; Jiao, X.; Grozhik, A.V.; Patil, D.P.; Linder, B.; Pickering, B.F.; Vasseur, J.J.; Chen, Q.; et al. Reversible methylation of m(6)A(m) in the 5′ cap controls mRNA stability. Nature 2017, 541, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Relier, S.; Ripoll, J.; Guillorit, H.; Amalric, A.; Achour, C.; Boissiere, F.; Vialaret, J.; Attina, A.; Debart, F.; Choquet, A.; et al. FTO-mediated cytoplasmic m6Am demethylation adjusts stem-like properties in colorectal cancer cell. Nat. Commun. 2021, 12, 1716. [Google Scholar] [CrossRef]
- Li, X.; Xiong, X.; Yi, C. Epitranscriptome sequencing technologies: Decoding RNA modifications. Nat. Methods 2016, 14, 23–31. [Google Scholar] [CrossRef]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef]
- Sun, H.; Li, K.; Zhang, X.; Liu, J.; Zhang, M.; Meng, H.; Yi, C. m6Am-seq reveals the dynamic m6Am methylation in the human transcriptome. Nat. Commun. 2021, 12, 4778. [Google Scholar] [CrossRef]
- Boulias, K.; Toczydlowska-Socha, D.; Hawley, B.R.; Liberman, N.; Takashima, K.; Zaccara, S.; Guez, T.; Vasseur, J.J.; Debart, F.; Aravind, L.; et al. Identification of the m6Am Methyltransferase PCIF1 Reveals the Location and Functions of m6Am in the Transcriptome. Mol. Cell 2019, 75, 631–643.e8. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, Z.; Mao, X.; Li, Q. m7GPredictor: An improved machine learning-based model for predicting internal m7G modifications using sequence properties. Anal. Biochem. 2020, 609, 113905. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Song, B.; Tang, Y.; Chen, K.; Wei, Z.; Meng, J. m5UPred: A Web Server for the Prediction of RNA 5-Methyluridine Sites from Sequences. Mol. Ther. Nucleic Acids 2020, 22, 742–747. [Google Scholar] [CrossRef] [PubMed]
- Zou, Q.; Xing, P.; Wei, L.; Liu, B. Gene2vec: Gene subsequence embedding for prediction of mammalian N-6-methyladenosine sites from mRNA. RNA 2019, 25, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Song, B.; Chen, K.; Lu, Z.; Rong, R.; Zhong, Y.; Meng, J. m6AmPred: Identifying RNA N6, 2′-O-dimethyladenosine m6Am sites based on sequence-derived information. Methods 2022, 203, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Su, W.; Lou, L.; Qiu, W.; Xiao, X.; Xu, Z. DLm6Am: A Deep-Learning-Based Tool for Identifying N6,2′-O-Dimethyladenosine Sites in RNA Sequences. Int. J. Mol. Sci. 2022, 23, 11026. [Google Scholar] [CrossRef]
- Breja, M.; Jain, S.K. Analyzing Linguistic Features for Answer Re-Ranking of Why-Questions. J. Cases Inf. Technol. 2021, 24, 1–16. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Nair, A.S.; Sreenadhan, S.P. A coding measure scheme employing electron-ion interaction pseudopotential (EIIP). Bioinformation 2006, 1, 197–202. [Google Scholar]
- Han, S.; Liang, Y.; Ma, Q.; Xu, Y.; Zhang, Y.; Du, W.; Wang, C.; Li, Y. LncFinder: An integrated platform for long non-coding RNA identification utilizing sequence intrinsic composition, structural information and physicochemical property. Brief. Bioinform. 2019, 20, 2009–2027. [Google Scholar] [CrossRef]
- Bonidia, R.P.; Sampaio, L.D.H.; Domingues, D.S.; Paschoal, A.R.; Lopes, F.M.; de Carvalho, A.; Sanches, D.S. Feature extraction approaches for biological sequences: A comparative study of mathematical features. Brief. Bioinform. 2021, 22, bbab011. [Google Scholar] [CrossRef] [PubMed]
- Dou, L.; Li, X.; Ding, H.; Xu, L.; Xiang, H. Prediction of m5C Modifications in RNA Sequences by Combining Multiple Sequence Features. Mol. Ther. Nucleic Acids 2020, 21, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, J.; Chen, H.; Zhang, D. Research on Counting Algorithm of k-mer Occurrence in DNA Sequence. Comput. Eng. 2007, 33, 40–42. [Google Scholar]
- Hasan, M.M.; Manavalan, B.; Shoombuatong, W.; Khatun, M.S.; Kurata, H. i4mC-Mouse: Improved identification of DNA N4-methylcytosine sites in the mouse genome using multiple encoding schemes. Comput. Struct. Biotechnol. J. 2020, 18, 906–912. [Google Scholar] [CrossRef] [PubMed]
- Qiang, X.; Chen, H.; Ye, X.; Su, R.; Wei, L. M6AMRFS: Robust Prediction of N6-Methyladenosine Sites with Sequence-Based Features in Multiple Species. Front. Genet. 2018, 9, 495. [Google Scholar] [CrossRef]
- Liu, K.; Chen, W. iMRM: A platform for simultaneously identifying multiple kinds of RNA modifications. Bioinformatics 2020, 36, 3336–3342. [Google Scholar] [CrossRef]
- Bari, A.; Reaz, M.R.; Jeong, B.S. Effective DNA Encoding for Splice Site Prediction Using SVM. Match-Commun. Math. Comput. Chem. 2014, 71, 241–258. [Google Scholar]
- Chen, W.; Lei, T.Y.; Jin, D.C.; Lin, H.; Chou, K.C. PseKNC: A flexible web server for generating pseudo K-tuple nucleotide composition. Anal. Biochem. 2014, 456, 53–60. [Google Scholar] [CrossRef]
- Musleh, S.; Islam, M.T.; Qureshi, R.; Alajez, N.M.; Alam, T. MSLP: mRNA subcellular localization predictor based on machine learning techniques. BMC Bioinform. 2023, 24, 109. [Google Scholar]
- Fan, Y.; Wang, W.; Zhu, Q. iterb-PPse: Identification of transcriptional terminators in bacterial by incorporating nucleotide properties into PseKNC. PLoS ONE 2020, 15, e0228479. [Google Scholar] [CrossRef]
- Tang, Q.; Nie, F.; Kang, J.; Chen, W. mRNALocater: Enhance the prediction accuracy of eukaryotic mRNA subcellular localization by using model fusion strategy. Mol. Ther. 2021, 29, 2617–2623. [Google Scholar] [CrossRef] [PubMed]
- Zu, Y. Research and Implementation of Clustering Method Based on Feature Extraction. Master’s Thesis, Jiangnan University, Wuxi, China, 2018. [Google Scholar]
- Orozco-Arias, S.; Candamil-Cortes, M.S.; Jaimes, P.A.; Pina, J.S.; Tabares-Soto, R.; Guyot, R.; Isaza, G. K-mer-based machine learning method to classify LTR-retrotransposons in plant genomes. PeerJ 2021, 9, e11456. [Google Scholar] [CrossRef] [PubMed]
- Fletez-Brant, C.; Lee, D.; McCallion, A.S.; Beer, M.A. kmer-SVM: A web server for identifying predictive regulatory sequence features in genomic data sets. Nucleic Acids Res. 2013, 41, W544–W556. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Feng, P.M.; Lin, H.; Chou, K.C. iRSpot-PseDNC: Identify recombination spots with pseudo dinucleotide composition. Nucleic Acids Res. 2013, 41, e68. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, X.; Chen, F.; Fang, L.; Li, J. Accurate prediction of DNA N4-methylcytosine sites via boost-learning various types of sequence features. BMC Genom. 2020, 21, 627. [Google Scholar] [CrossRef]
- Chen, X.; Xiong, Y.; Liu, Y.; Chen, Y.; Bi, S.; Zhu, X. m5CPred-SVM: A novel method for predicting m5C sites of RNA. BMC Bioinform. 2020, 21, 489. [Google Scholar] [CrossRef]
- Liu, W.Y.; Chen, L.Y.; Huang, Y.Y.; Fu, L.; Song, L.Y.; Wang, Y.Y.; Bai, Z.; Meng, F.F.; Bi, Y.F. Antioxidation and active constituents analysis of flower residue of Rosa damascena. Chin. Herb. Med. 2020, 12, 336–341. [Google Scholar] [CrossRef]
- Kumar, P.S.; K, A.K.; Mohapatra, S.; Naik, B.; Nayak, J.; Mishra, M. CatBoost Ensemble Approach for Diabetes Risk Prediction at Early Stages. In Proceedings of the 2021 1st Odisha International Conference on Electrical Power Engineering, Communication and Computing Technology (ODICON), Bhubaneswar, India, 8–9 January 2021; pp. 1–6. [Google Scholar]
- Prokhorenkova, L.; Gusev, G.; Vorobev, A.; Dorogush, A.V.; Gulin, A. CatBoost: Unbiased boosting with categorical features. In Proceedings of the 32nd Conference on Neural Information Processing Systems, Montreal, QC, Canada, 2–8 December 2018. [Google Scholar]
- Dorogush, A.V.; Ershov, V.; Gulin, A. CatBoost: Gradient boosting with categorical features support. arXiv 2018, arXiv:1810.11363. [Google Scholar]
- Pham, T.D.; Yokoya, N.; Xia, J.; Ha, N.T.; Le, N.N.; Nguyen, T.T.T.; Dao, T.H.; Vu, T.T.P.; Pham, T.D.; Takeuchi, W. Comparison of Machine Learning Methods for Estimating Mangrove Above-Ground Biomass Using Multiple Source Remote Sensing Data in the Red River Delta Biosphere Reserve, Vietnam. Remote Sens. 2020, 12, 1334. [Google Scholar] [CrossRef]
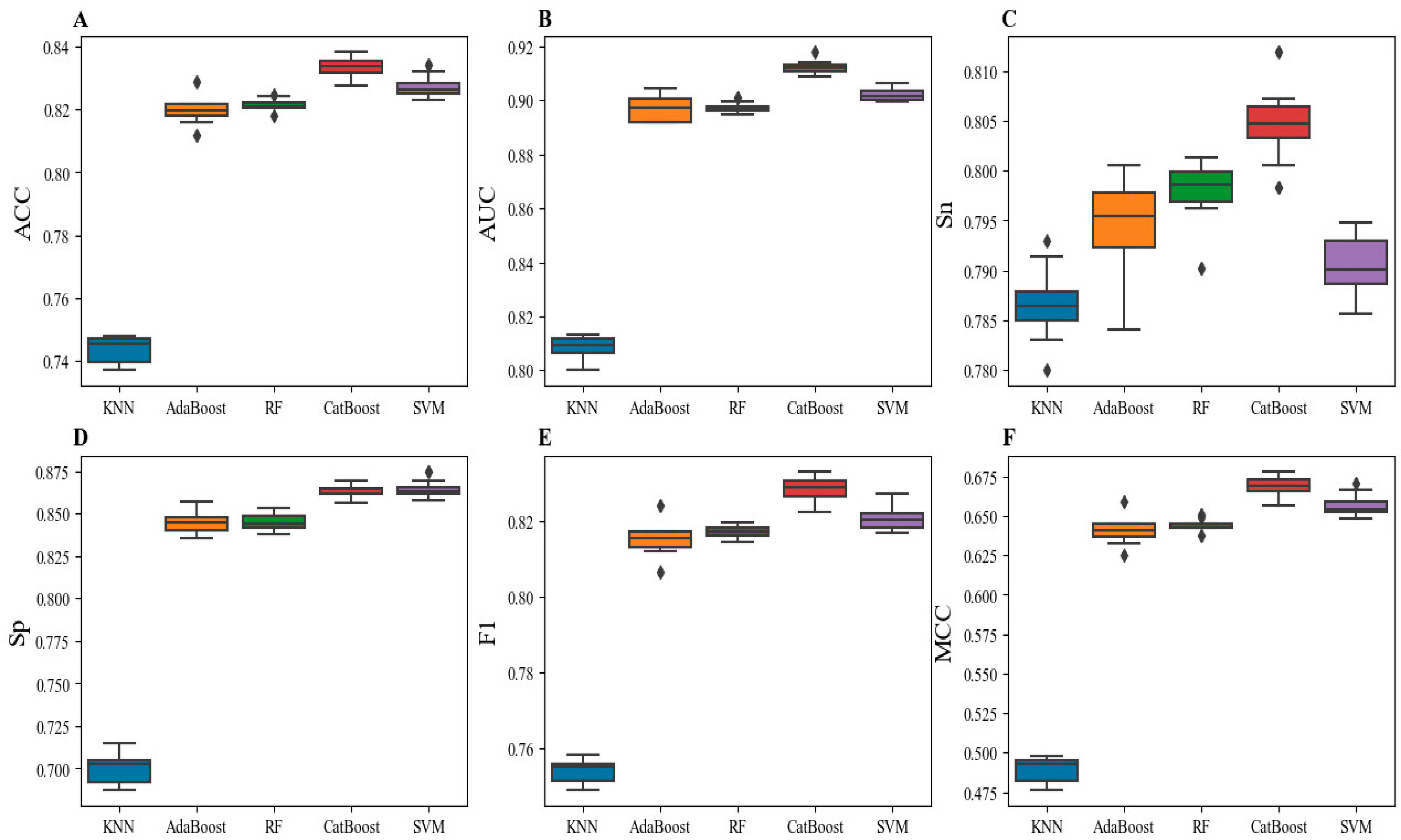

| Classifier | ACC | AUC | Sn | Sp | F1 | MCC |
|---|---|---|---|---|---|---|
| AdaBoost | 0.820 ± 0.004 | 0.897 ± 0.004 | 0.794 ± 0.005 | 0.845 ± 0.006 | 0.815 ± 0.004 | 0.641 ± 0.009 |
| RF | 0.821 ± 0.002 | 0.897 ± 0.002 | 0.798 ± 0.003 | 0.845 ± 0.005 | 0.817 ± 0.002 | 0.644 ± 0.004 |
| KNN | 0.743 ± 0.004 | 0.809 ± 0.004 | 0.787 ± 0.004 | 0.700 ± 0.009 | 0.754 ± 0.003 | 0.489 ± 0.008 |
| SVM | 0.827 ± 0.003 | 0.902 ± 0.002 | 0.791 ± 0.003 | 0.864 ± 0.005 | 0.821 ± 0.003 | 0.657 ± 0.007 |
| CatBoost | 0.834 ± 0.003 | 0.912 ± 0.002 | 0.805 ± 0.004 | 0.863 ± 0.004 | 0.829 ± 0.003 | 0.669 ± 0.006 |
| Feature | ACC | AUC | Sn | Sp | F1 | MCC |
|---|---|---|---|---|---|---|
| PseEIIP | 0.831 ± 0.003 | 0.910 ± 0.002 | 0.804 ± 0.002 | 0.859 ± 0.006 | 0.827 ± 0.003 | 0.664 ± 0.006 |
| DBE | 0.811 ± 0.003 | 0.892 ± 0.003 | 0.800 ± 0.002 | 0.822 ± 0.004 | 0.809 ± 0.003 | 0.623 ± 0.006 |
| Hash | 0.813 ± 0.003 | 0.891 ± 0.003 | 0.788 ± 0.004 | 0.838 ± 0.004 | 0.808 ± 0.003 | 0.627 ± 0.007 |
| NCP | 0.808 ± 0.003 | 0.890 ± 0.003 | 0.794 ± 0.002 | 0.822 ± 0.005 | 0.805 ± 0.003 | 0.617 ± 0.006 |
| PseKNC | 0.804 ± 0.003 | 0.885 ± 0.003 | 0.788 ± 0.004 | 0.820 ± 0.006 | 0.801 ± 0.003 | 0.609 ± 0.006 |
| Kmer | 0.811 ± 0.002 | 0.881 ± 0.002 | 0.776 ± 0.003 | 0.845 ± 0.005 | 0.804 ± 0.002 | 0.623 ± 0.005 |
| SCPseTNC | 0.808 ± 0.004 | 0.878 ± 0.003 | 0.779 ± 0.002 | 0.838 ± 0.006 | 0.802 ± 0.003 | 0.618 ± 0.007 |
| DNM | 0.805 ± 0.004 | 0.875 ± 0.003 | 0.773 ± 0.005 | 0.838 ± 0.006 | 0.799 ± 0.004 | 0.612 ± 0.007 |
| Ksnpf | 0.806 ± 0.003 | 0.874 ± 0.002 | 0.771 ± 0.004 | 0.842 ± 0.006 | 0.799 ± 0.003 | 0.614 ± 0.006 |
| PseEIIP + DBE | 0.832 ± 0.003 | 0.910 ± 0.002 | 0.805 ± 0.002 | 0.858 ± 0.005 | 0.827 ± 0.003 | 0.665 ± 0.006 |
| PseEIIP + DBE + Hash | 0.832 ± 0.002 | 0.910 ± 0.003 | 0.803 ± 0.002 | 0.862 ± 0.004 | 0.827 ± 0.002 | 0.666 ± 0.005 |
| ALL | 0.834 ± 0.003 | 0.912 ± 0.002 | 0.805 ± 0.004 | 0.863 ± 0.004 | 0.829 ± 0.003 | 0.669 ± 0.006 |
| Hyper-Parameters | Optimal Values |
|---|---|
| Iterations | 2000 |
| learning_rate | 0.03 |
| Depth | 9 |
| leaf_estimation_method | “Newton” |
| loss_function | “MultiClass” |
| bootstrap_type | “Bayesian” |
| Model | ACC | AUC | Sn | Sp | F1 | MCC |
|---|---|---|---|---|---|---|
| m6Aminer | 0.834 ± 0.003 | 0.913 ± 0.002 | 0.806 ± 0.003 | 0.861 ± 0.005 | 0.829 ± 0.003 | 0.668 ± 0.006 |
| m6AmPred | 0.826 ± 0.003 | 0.905 ± 0.002 | 0.803 ± 0.002 | 0.849 ± 0.006 | 0.822 ± 0.003 | 0.653 ± 0.007 |
| DLm6Am | 0.827 ± 0.002 | 0.897 ± 0.002 | 0.796 ± 0.005 | 0.858 ± 0.005 | 0.821 ± 0.002 | 0.655 ± 0.004 |
| Model | ACC | AUC | Sn | Sp | F1 | MCC |
|---|---|---|---|---|---|---|
| m6Aminer | 0.647 ± 0.009 | 0.754 ± 0.005 | 0.874 ± 0.006 | 0.420 ± 0.024 | 0.713 ± 0.004 | 0.331 ± 0.015 |
| m6AmPred | 0.623 ± 0.013 | 0.735 ± 0.013 | 0.887 ± 0.009 | 0.358 ± 0.024 | 0.702 ± 0.009 | 0.289 ± 0.028 |
| DLm6Am | 0.642 ± 0.014 | 0.730 ± 0.009 | 0.875 ± 0.009 | 0.409 ± 0.029 | 0.710 ± 0.008 | 0.322 ± 0.026 |
| Nucleotides | EIIP Value |
|---|---|
| A | 0.1260 |
| U | 0.1335 |
| G | 0.0806 |
| C | 0.1340 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Lan, P.; Liu, T.; Liu, X.; Liu, T. m6Aminer: Predicting the m6Am Sites on mRNA by Fusing Multiple Sequence-Derived Features into a CatBoost-Based Classifier. Int. J. Mol. Sci. 2023, 24, 7878. https://doi.org/10.3390/ijms24097878
Liu Z, Lan P, Liu T, Liu X, Liu T. m6Aminer: Predicting the m6Am Sites on mRNA by Fusing Multiple Sequence-Derived Features into a CatBoost-Based Classifier. International Journal of Molecular Sciences. 2023; 24(9):7878. https://doi.org/10.3390/ijms24097878
Chicago/Turabian StyleLiu, Ze, Pengfei Lan, Ting Liu, Xudong Liu, and Tao Liu. 2023. "m6Aminer: Predicting the m6Am Sites on mRNA by Fusing Multiple Sequence-Derived Features into a CatBoost-Based Classifier" International Journal of Molecular Sciences 24, no. 9: 7878. https://doi.org/10.3390/ijms24097878
APA StyleLiu, Z., Lan, P., Liu, T., Liu, X., & Liu, T. (2023). m6Aminer: Predicting the m6Am Sites on mRNA by Fusing Multiple Sequence-Derived Features into a CatBoost-Based Classifier. International Journal of Molecular Sciences, 24(9), 7878. https://doi.org/10.3390/ijms24097878






