Identification and Functional Analysis of foxo Genes in Chinese Tongue Sole (Cynoglossus semilaevis)
Abstract
1. Introduction
2. Results
2.1. Identification and Analysis of Csfoxo Genes
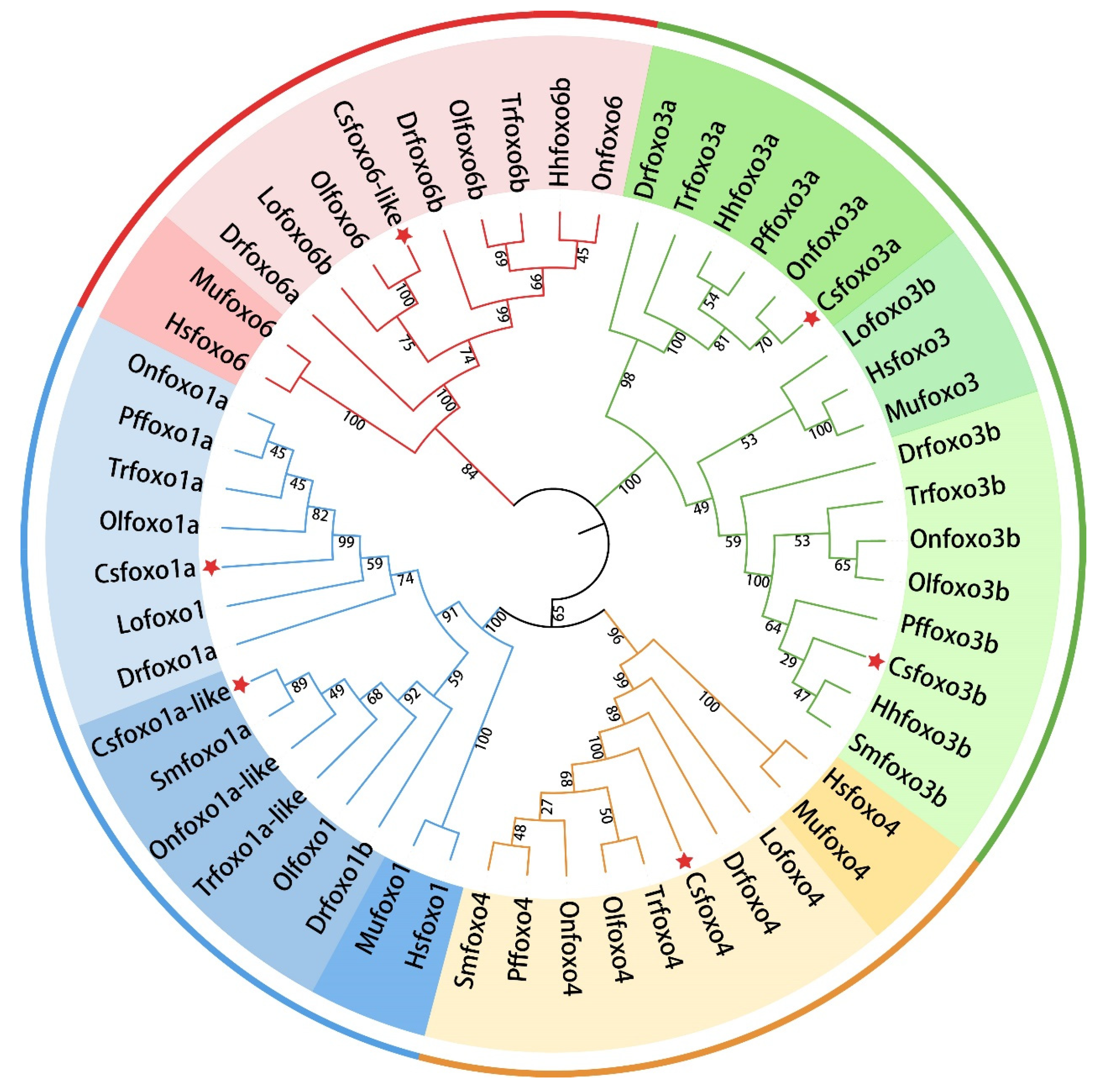
2.2. Phylogenetic Tree Analysis
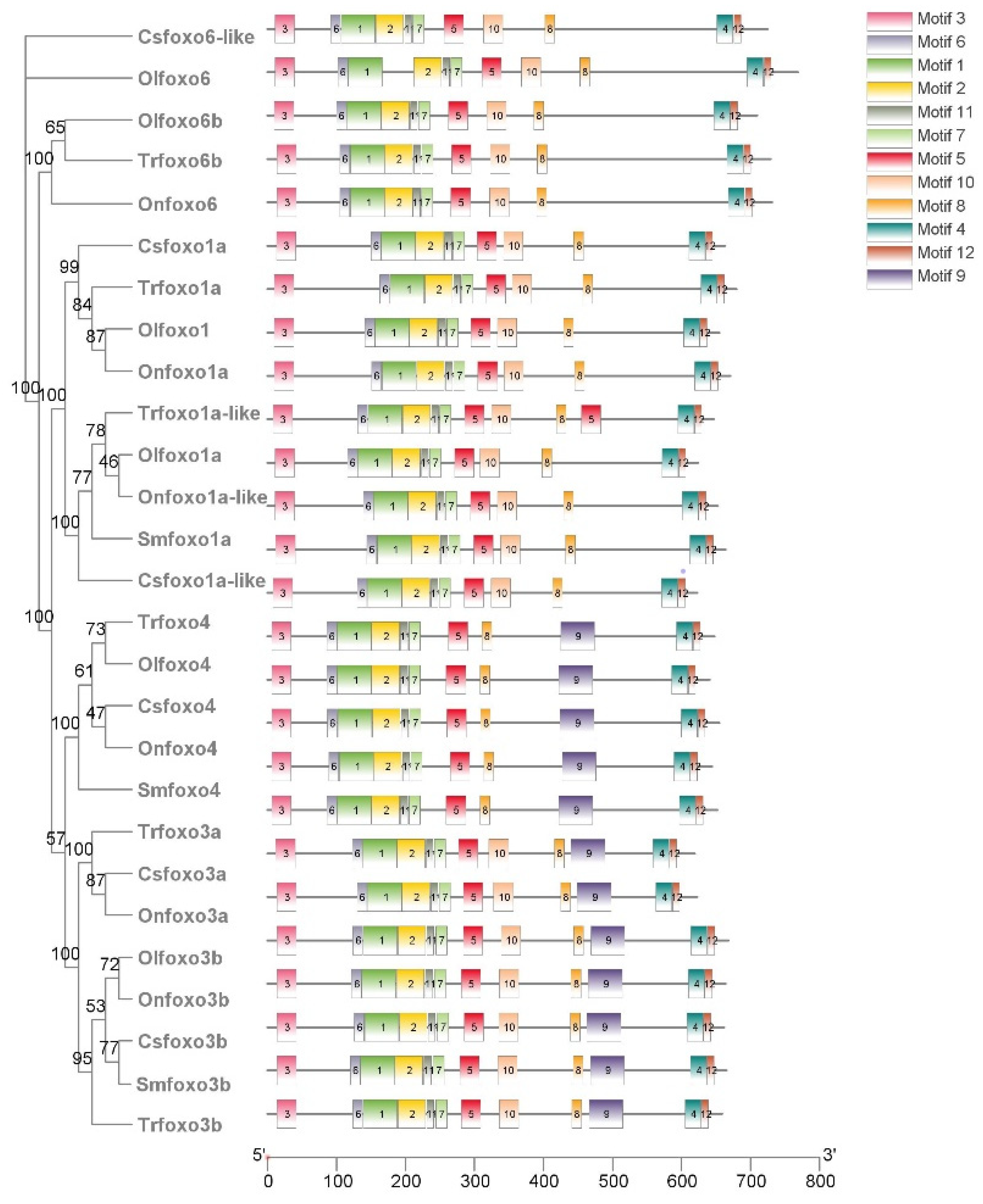
2.3. Structure Analysis of Foxo
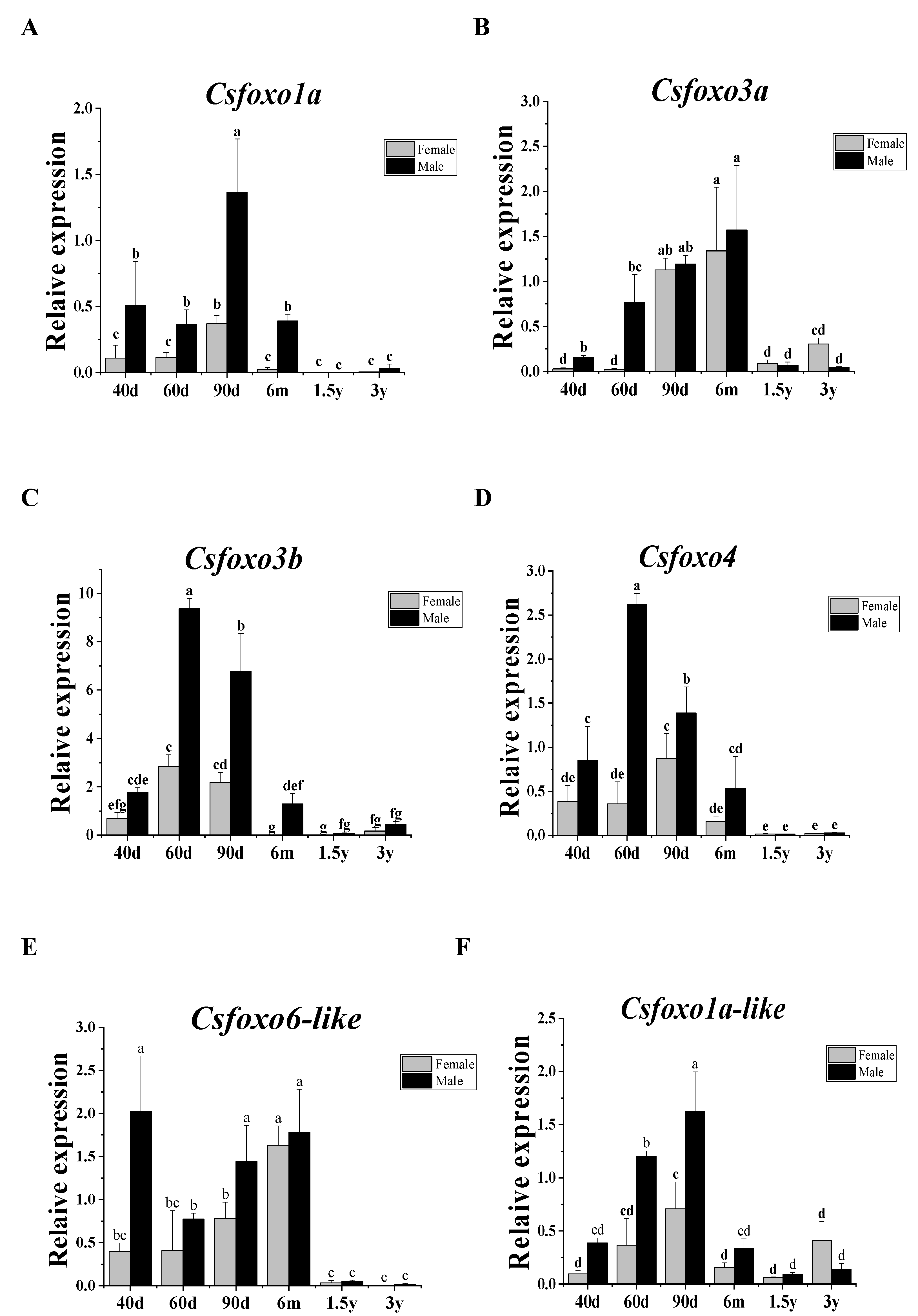
2.4. Analysis of Csfoxo Expression Patterns at Different Developmental Stages
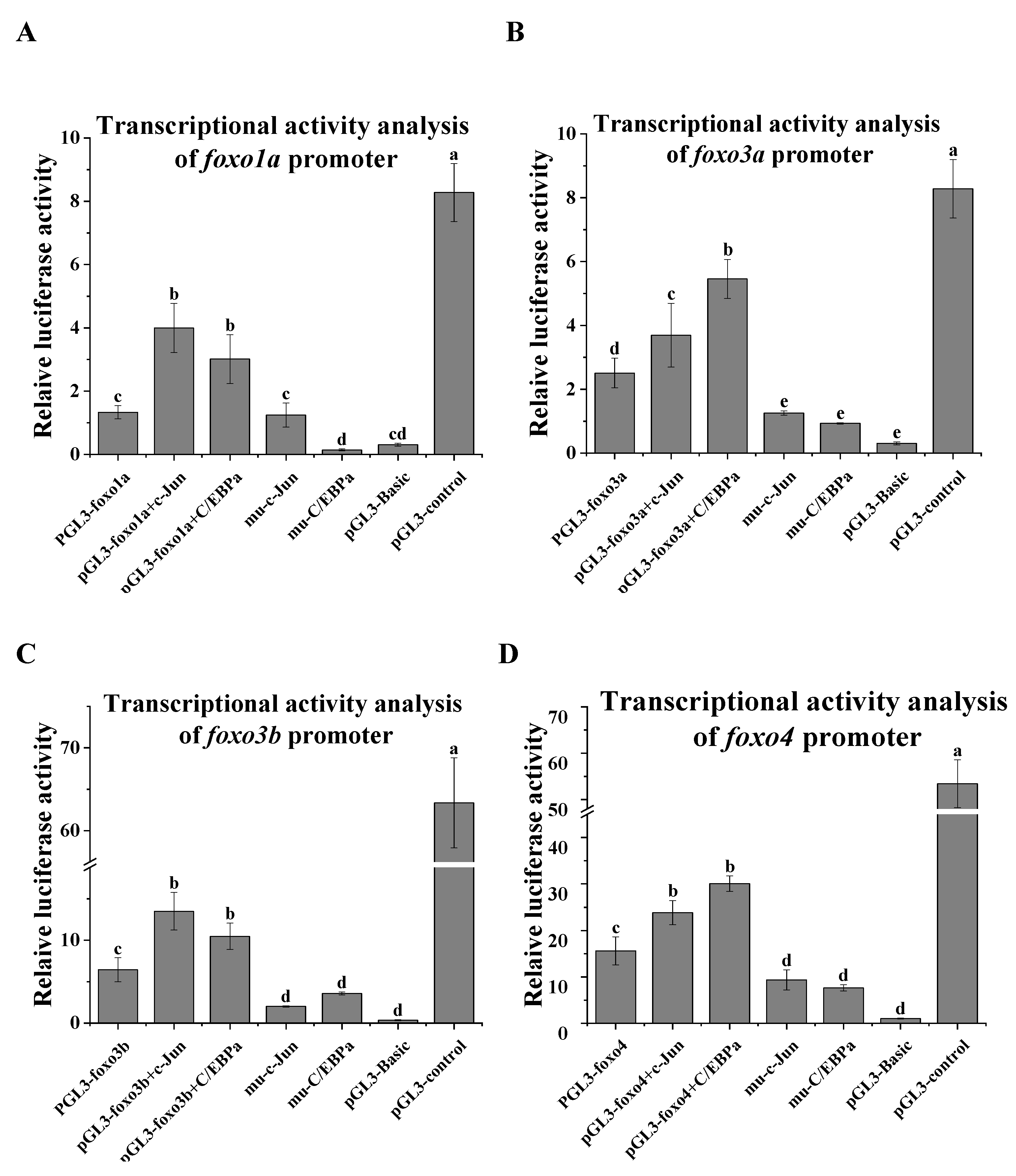
2.5. Promoter Activity Analysis
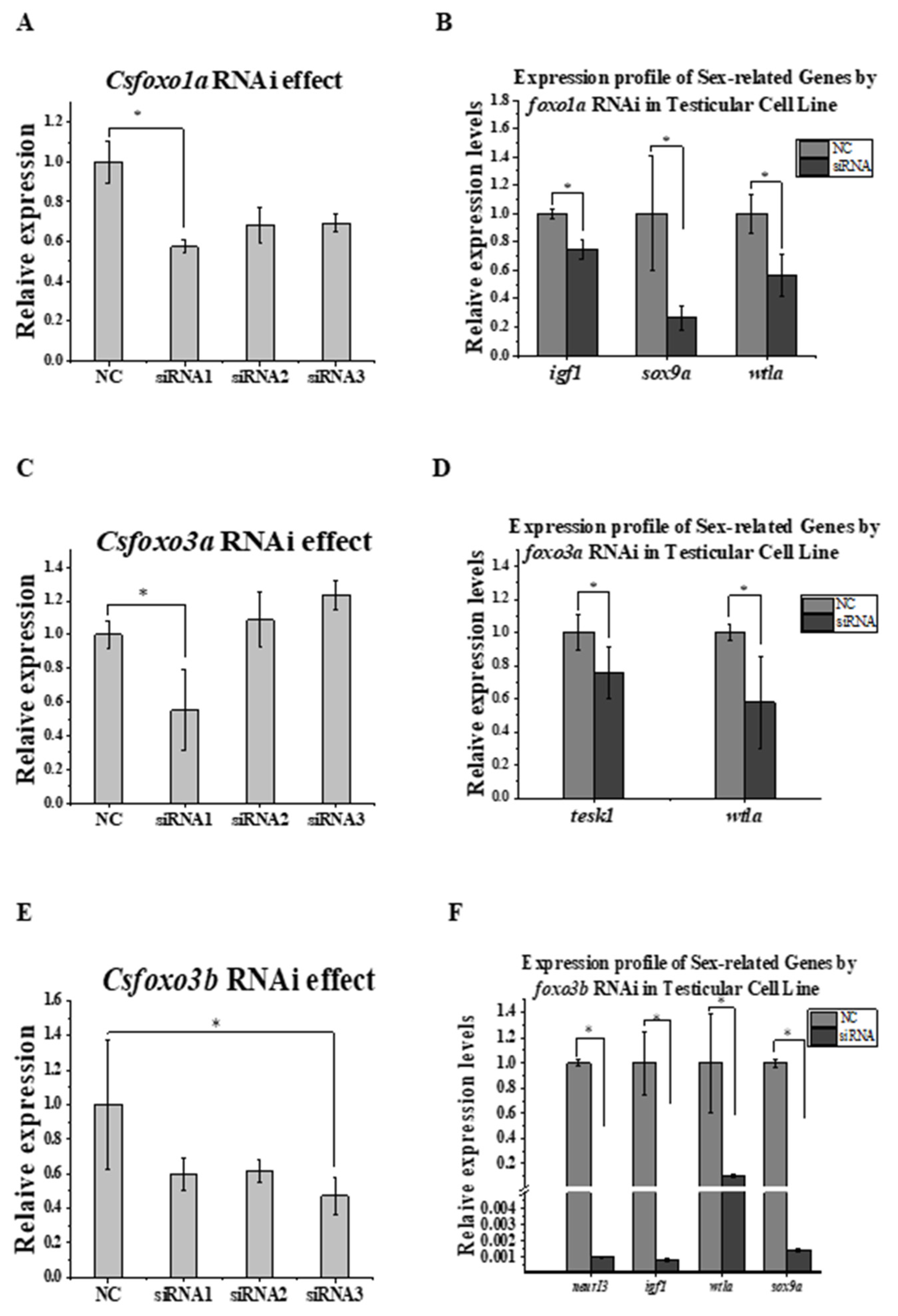
2.6. Csfoxo Knockdown and the Effect on Other Genes
3. Discussion
4. Materials and Methods
4.1. Identification of Csfoxo Members and Sequence Analysis
4.2. Phylogenetic Tree and Structural Characterization
4.3. Experimental Animals and Ethics Approval
4.4. Gene Expression Pattern Analysis
4.5. Promoter Activity Analysis
4.6. siRNA-Mediated Knockdown of Csfoxo in Testicular Cells
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hannenhalli, S.; Kaestner, K.H. The evolution of Fox genes and their role in development and disease. Nat. Rev. Genet. 2009, 10, 233. [Google Scholar] [CrossRef] [PubMed]
- Hosaka, T.; Biggs, W.R.; Tieu, D.; Boyer, A.D.; Varki, N.M.; Cavenee, W.K.; Arden, K.C. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc. Natl. Acad. Sci. USA 2004, 101, 2975. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, H.; Daitoku, H.; Hatta, M.; Aoyama, H.; Yoshimochi, K.; Fukamizu, A. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc. Natl. Acad. Sci. USA 2005, 102, 11278. [Google Scholar] [CrossRef]
- Fu, Z.; Tindall, D.J. FOXOs, cancer and regulation of apoptosis. Oncogene 2008, 27, 2312. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tang, N.; Hadden, T.J.; Rishi, A.K. Akt, FoxO and regulation of apoptosis. Biochim. Biophys. Acta 2011, 1813, 1978. [Google Scholar] [CrossRef]
- Eijkelenboom, A.; Burgering, B.M. FOXOs: Signalling integrators for homeostasis maintenance. Nat. Rev. Mol. Cell. Biol. 2013, 14, 83. [Google Scholar] [CrossRef]
- Gao, L.; Yuan, Z.; Zhou, T.; Yang, Y.; Gao, D.; Dunham, R.; Liu, Z. FOXO genes in channel catfish and their response after bacterial infection. Dev. Comp. Immunol. 2019, 97, 38. [Google Scholar] [CrossRef]
- Ni, Y.G.; Berenji, K.; Wang, N.; Oh, M.; Sachan, N.; Dey, A.; Cheng, J.; Lu, G.; Morris, D.J.; Castrillon, D.H.; et al. Foxo transcription factors blunt cardiac hypertrophy by inhibiting calcineurin signaling. Circulation 2006, 114, 1159. [Google Scholar] [CrossRef]
- Arden, K.C.; Biggs, W.R. Regulation of the FoxO family of transcription factors by phosphatidylinositol-3 kinase-activated signaling. Arch. Biochem. Biophys. 2002, 403, 292. [Google Scholar] [CrossRef]
- Hacker, U.; Grossniklaus, U.; Gehring, W.J.; Jackle, H. Developmentally regulated Drosophila gene family encoding the fork head domain. Proc. Natl. Acad. Sci. USA 1992, 89, 8754. [Google Scholar] [CrossRef]
- Sun, Y.; Li, M.; Cui, Z.; Zhang, M.; Zhang, T.; Li, L.; Wang, N.; Xu, X.; Wei, M.; Xu, W. Transcriptomic Analysis Revealed Candidate Genes Involved in Pseudomale Sperm Abnormalities in Chinese Tongue Sole (Cynoglossus semilaevis). Biology 2022, 11, 1716. [Google Scholar] [CrossRef] [PubMed]
- Goertz, M.J.; Wu, Z.; Gallardo, T.D.; Hamra, F.K.; Castrillon, D.H. Foxo1 is required in mouse spermatogonial stem cells for their maintenance and the initiation of spermatogenesis. J. Clin. Investig. 2011, 121, 3456. [Google Scholar] [CrossRef] [PubMed]
- Tarnawa, E.D.; Baker, M.D.; Aloisio, G.M.; Carr, B.R.; Castrillon, D.H. Gonadal expression of Foxo1, but not Foxo3, is conserved in diverse Mammalian species. Biol. Reprod. 2013, 88, 103. [Google Scholar] [CrossRef]
- Lansard, M.; Panserat, S.; Plagnes-Juan, E.; Seiliez, I.; Skiba-Cassy, S. Integration of insulin and amino acid signals that regulate hepatic metabolism-related gene expression in rainbow trout: Role of TOR. Amino. Acids 2010, 39, 801. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, M.; Cheng, P.; Gong, Z.; Li, X.; Wang, N.; Wei, M.; Xu, X.; Xu, W. pitpbeta_w Encoding Phosphatidylinositol Transfer Protein Is Involved in Female Differentiation of Chinese Tongue Sole, Cynoglossus semilaevis. Front. Genet. 2022, 13, 861763. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wen, H.; Li, Y.; Zhang, Z.; Wang, L.; Mao, X.; Li, J.; Qi, X. FOXO1A promotes neuropeptide FF transcription subsequently regulating the expression of feeding-related genes in spotted sea bass (Lateolabrax maculatus). Mol. Cell. Endocrinol. 2020, 517, 110871. [Google Scholar] [CrossRef]
- Hedrick, S.M. The cunning little vixen: Foxo and the cycle of life and death. Nat. Immunol. 2009, 10, 1057. [Google Scholar] [CrossRef]
- Liu, Q.; Zhang, Y.; Shi, B.; Lu, H.; Zhang, L.; Zhang, W. Foxo3b but not Foxo3a activates cyp19a1a in Epinephelus coioides. J. Mol. Endocrinol. 2016, 56, 337. [Google Scholar] [CrossRef]
- Lin, S.J.; Chiang, M.C.; Shih, H.Y.; Chiang, K.C.; Cheng, Y.C. Spatiotemporal expression of foxo4, foxo6a, and foxo6b in the developing brain and retina are transcriptionally regulated by PI3K signaling in zebrafish. Dev. Genes Evol. 2017, 227, 219–230. [Google Scholar] [CrossRef]
- Bertho, S.; Pasquier, J.; Pan, Q.; Le Trionnaire, G.; Bobe, J.; Postlethwait, J.H.; Pailhoux, E.; Schartl, M.; Herpin, A.; Guiguen, Y. Foxl2 and Its Relatives Are Evolutionary Conserved Players in Gonadal Sex Differentiation. Sex. Dev. 2016, 10, 111–129. [Google Scholar] [CrossRef]
- Barthel, A.; Schmoll, D.; Unterman, T.G. FoxO proteins in insulin action and metabolism. Trends Endocrinol. Metab. 2005, 16, 183. [Google Scholar] [CrossRef] [PubMed]
- Accili, D.; Arden, K.C. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell 2004, 117, 421. [Google Scholar] [CrossRef] [PubMed]
- Greer, E.L.; Brunet, A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene 2005, 24, 7410. [Google Scholar] [CrossRef] [PubMed]
- Coletta, A.; Pinney, J.W.; Solis, D.Y.; Marsh, J.; Pettifer, S.R.; Attwood, T.K. Low-complexity regions within protein sequences have position-dependent roles. BMC Syst. Biol. 2010, 4, 43. [Google Scholar] [CrossRef]
- Ren, W.; Guo, J.; Jiang, F.; Lu, J.; Ding, Y.; Li, A.; Liang, X.; Jia, W. CCAAT/enhancer-binding protein alpha is a crucial regulator of human fat mass and obesity associated gene transcription and expression. Biomed. Res. Int. 2014, 2014, 406909. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, R.; Feng, B.; Li, S.; Mahboob, S.; Shao, C. Cloning and functional analysis of c/ebpalpha as negative regulator of dmrt1 in Chinese tongue sole (Cynoglossus semilaevis). Gene 2021, 768, 145321. [Google Scholar] [CrossRef]
- Hilberg, F.; Aguzzi, A.; Howells, N.; Wagner, E.F. c-jun is essential for normal mouse development and hepatogenesis. Nature 1993, 365, 179. [Google Scholar] [CrossRef]
- Berishvili, G.; D’Cotta, H.; Baroiller, J.F.; Segner, H.; Reinecke, M. Differential expression of IGF-I mRNA and peptide in the male and female gonad during early development of a bony fish, the tilapia Oreochromis niloticus. Gen. Comp. Endocrinol. 2006, 146, 204. [Google Scholar] [CrossRef]
- Reinecke, M. Insulin-like growth factors and fish reproduction. Biol. Reprod. 2010, 82, 656. [Google Scholar] [CrossRef]
- Reinecke, M.; Collet, C. The phylogeny of the insulin-like growth factors. Int. Rev. Cytol. 1998, 183, 1–94. [Google Scholar]
- Reinecke, M.; Bjornsson, B.T.; Dickhoff, W.W.; McCormick, S.D.; Navarro, I.; Power, D.M.; Gutierrez, J. Growth hormone and insulin-like growth factors in fish: Where we are and where to go. Gen. Comp. Endocrinol. 2005, 142, 20. [Google Scholar] [CrossRef] [PubMed]
- Jakob, S.; Lovell-Badge, R. Sex determination and the control of Sox9 expression in mammals. Febs. J. 2011, 278, 1002. [Google Scholar] [CrossRef] [PubMed]
- Takamatsu, N.; Kanda, H.; Ito, M.; Yamashita, A.; Yamashita, S.; Shiba, T. Rainbow trout SOX9: cDNA cloning, gene structure and expression. Gene 1997, 202, 167. [Google Scholar] [CrossRef] [PubMed]
- Chiang, E.F.; Pai, C.I.; Wyatt, M.; Yan, Y.L.; Postlethwait, J.; Chung, B. Two sox9 genes on duplicated zebrafish chromosomes: Expression of similar transcription activators in distinct sites. Dev. Biol. 2001, 231, 149. [Google Scholar] [CrossRef]
- Kluver, N.; Herpin, A.; Braasch, I.; Driessle, J.; Schartl, M. Regulatory back-up circuit of medaka Wt1 co-orthologs ensures PGC maintenance. Dev. Biol. 2009, 325, 179. [Google Scholar] [CrossRef]
- Lu, Y.F.; Liu, Q.; Liu, K.Q.; Wang, H.Y.; Li, C.H.; Wang, Q.; Shao, C.W. Identification of global alternative splicing and sex-specific splicing via comparative transcriptome analysis of gonads of Chinese tongue sole (Cynoglossus semilaevis). Zool. Res. 2022, 43, 319. [Google Scholar] [CrossRef]
- Toshima, J.; Koji, T.; Mizuno, K. Stage-specific expression of testis-specific protein kinase 1 (TESK1) in rat spermatogenic cells. Biochem. Biophys. Res. Commun. 1998, 249, 107. [Google Scholar] [CrossRef]
- Meng, L.; Zhu, Y.; Zhang, N.; Liu, W.; Liu, Y.; Shao, C.; Wang, N.; Chen, S. Cloning and characterization of tesk1, a novel spermatogenesis-related gene, in the tongue sole (Cynoglossus semilaevis). PLoS ONE 2014, 9, e107922. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic. Acids Res. 2015, 43, W39. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402. [Google Scholar] [CrossRef]
- Wang, N.; Gong, Z.; Wang, J.; Xu, W.; Yang, Q.; Chen, S. Characterization of Chinese tongue sole (Cynoglossus semilaevis) 24-dehydrocholesterol reductase: Expression profile, epigenetic modification, and its knock-down effect. Gen. Comp. Endocrinol. 2021, 312, 113870. [Google Scholar] [CrossRef] [PubMed]

| Name | Gene ID | Chr | Genomic Location | ORF | AA | MW (kDa) | pI |
|---|---|---|---|---|---|---|---|
| Csfoxo1a | 103378152 | 4 | 12505667-12537512 | 1992 | 663 | 71.49 | 6.29 |
| Csfoxo3a | 103379355 | 1 | 11149558-11159268 | 1872 | 623 | 66.52 | 5.02 |
| Csfoxo3b | 103380652 | 7 | 1249234-1287747 | 1989 | 662 | 70.12 | 4.79 |
| Csfoxo4 | 103390850 | 15 | 8667641-8676081 | 1968 | 655 | 69.55 | 5.71 |
| Csfoxo6-like | 103387887 | 13 | 1015113-1050104 | 2178 | 725 | 78.07 | 6.78 |
| Csfoxo1a-like | 103395707 | 19 | 17163967-17178152 | 1872 | 623 | 67.9 | 6.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, T.; Zhang, M.; Sun, Y.; Li, L.; Cheng, P.; Li, X.; Wang, N.; Chen, S.; Xu, W. Identification and Functional Analysis of foxo Genes in Chinese Tongue Sole (Cynoglossus semilaevis). Int. J. Mol. Sci. 2023, 24, 7625. https://doi.org/10.3390/ijms24087625
Zhang T, Zhang M, Sun Y, Li L, Cheng P, Li X, Wang N, Chen S, Xu W. Identification and Functional Analysis of foxo Genes in Chinese Tongue Sole (Cynoglossus semilaevis). International Journal of Molecular Sciences. 2023; 24(8):7625. https://doi.org/10.3390/ijms24087625
Chicago/Turabian StyleZhang, Tingting, Mengqian Zhang, Yuxuan Sun, Lu Li, Peng Cheng, Xihong Li, Na Wang, Songlin Chen, and Wenteng Xu. 2023. "Identification and Functional Analysis of foxo Genes in Chinese Tongue Sole (Cynoglossus semilaevis)" International Journal of Molecular Sciences 24, no. 8: 7625. https://doi.org/10.3390/ijms24087625
APA StyleZhang, T., Zhang, M., Sun, Y., Li, L., Cheng, P., Li, X., Wang, N., Chen, S., & Xu, W. (2023). Identification and Functional Analysis of foxo Genes in Chinese Tongue Sole (Cynoglossus semilaevis). International Journal of Molecular Sciences, 24(8), 7625. https://doi.org/10.3390/ijms24087625








