Assessing Kidney Injury Induced by Mercuric Chloride in Guinea Pigs with In Vivo and In Vitro Experiments
Abstract
1. Introduction
2. Results and Discussion
2.1. Dose Optimization of Mercuric Chloride in Guinea Pigs
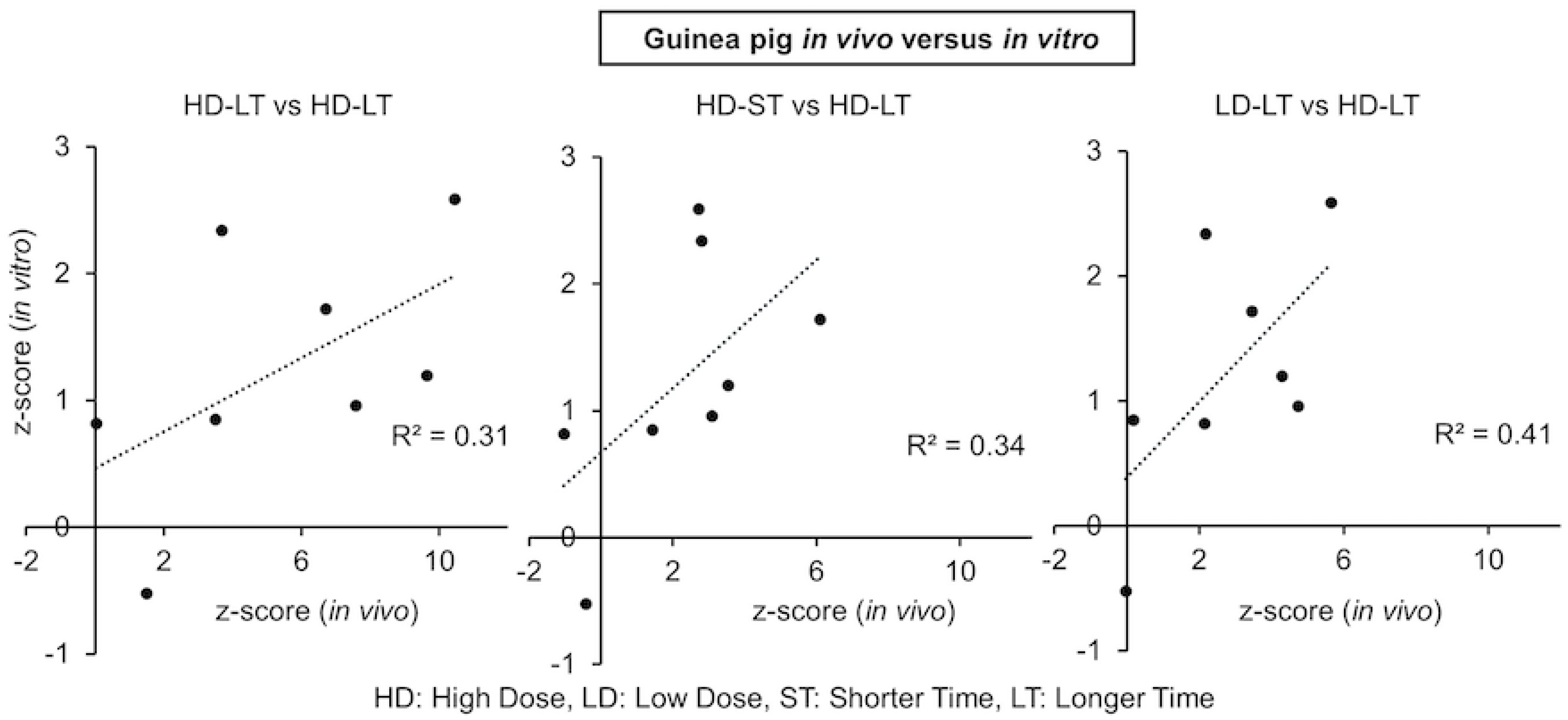
2.2. Gene-Level Analysis
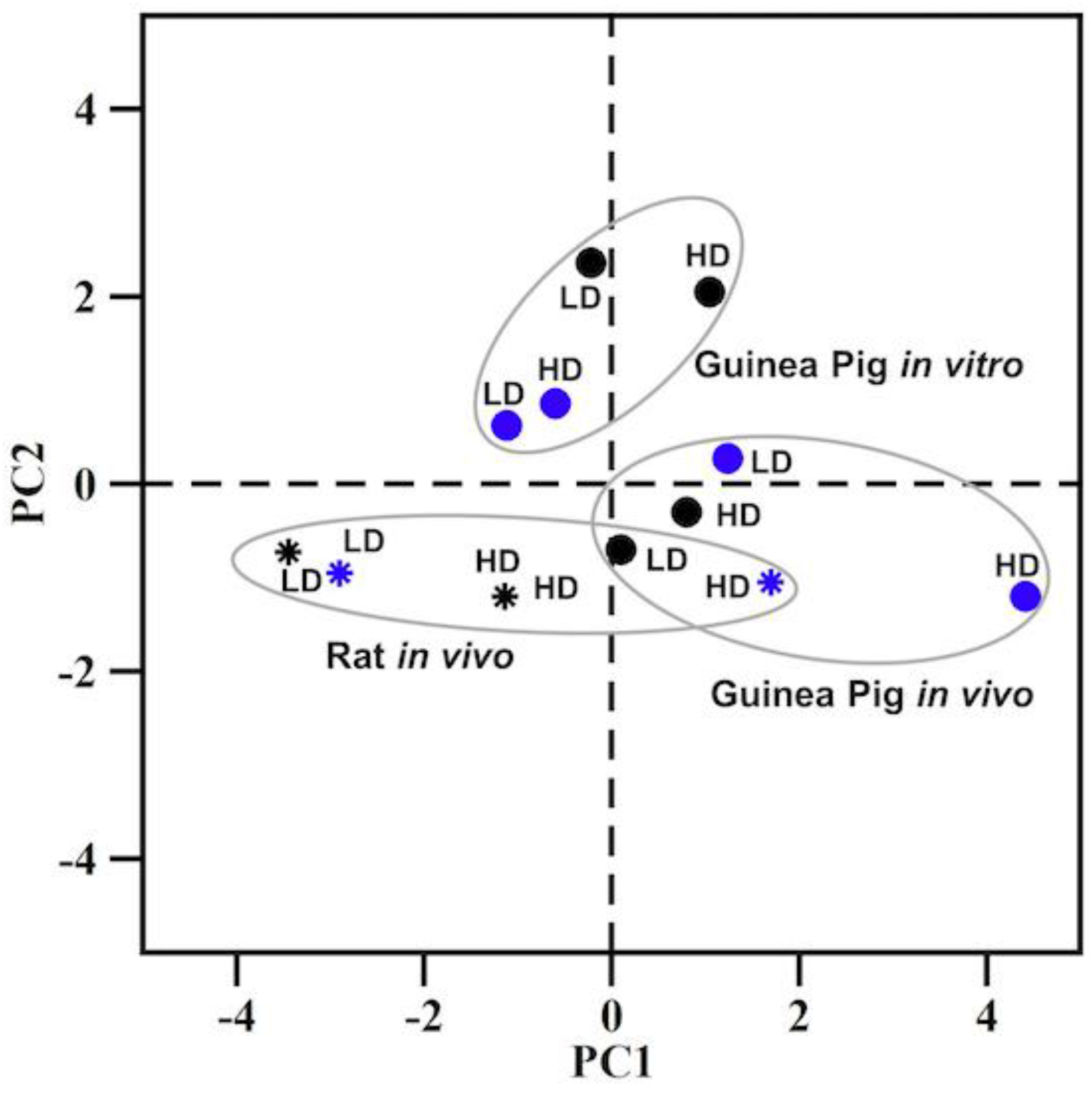
2.3. Kidney Injury Module Activation Analysis
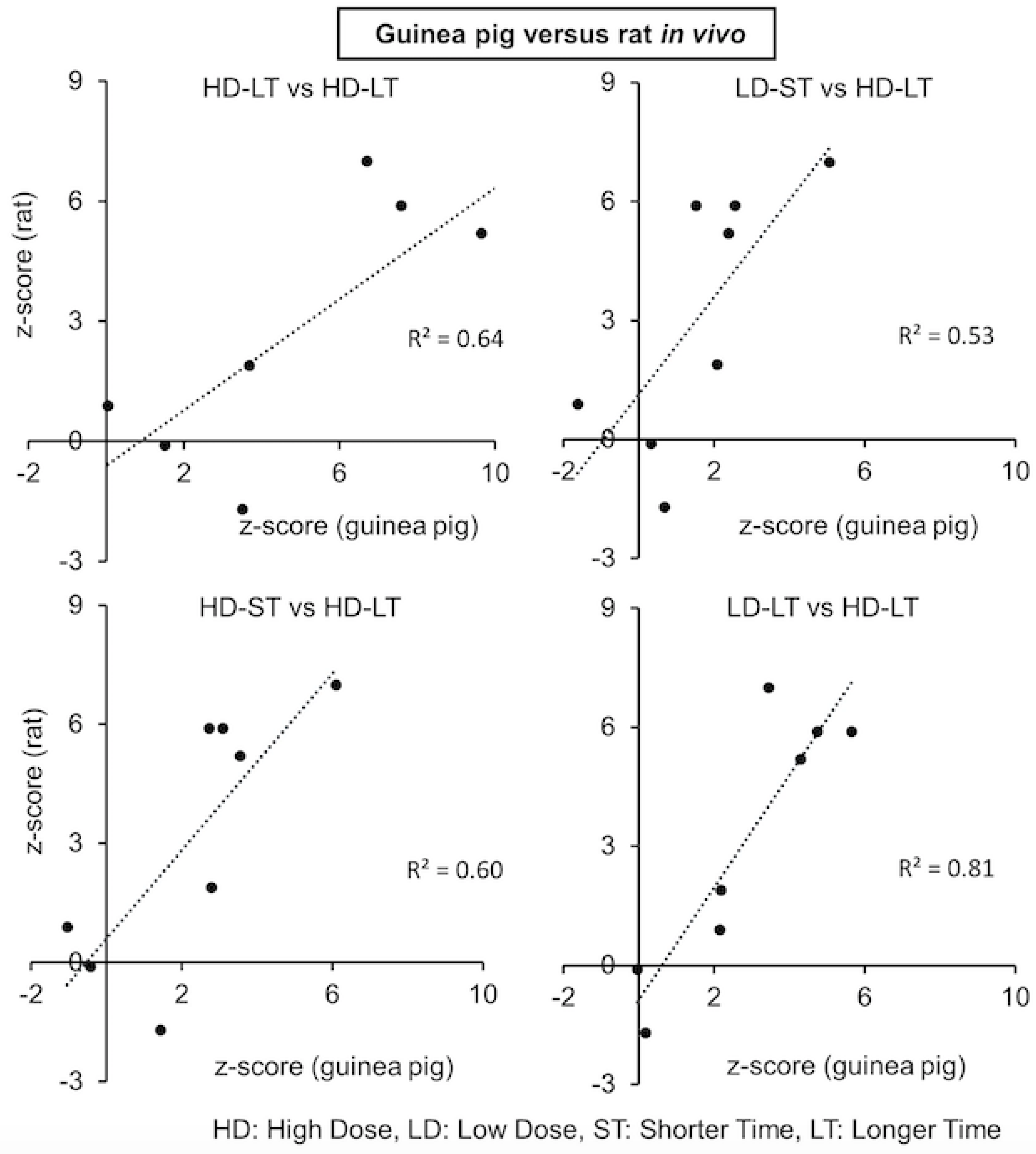
2.4. Correlation between Guinea Pig and Rat Mercuric Chloride Exposures
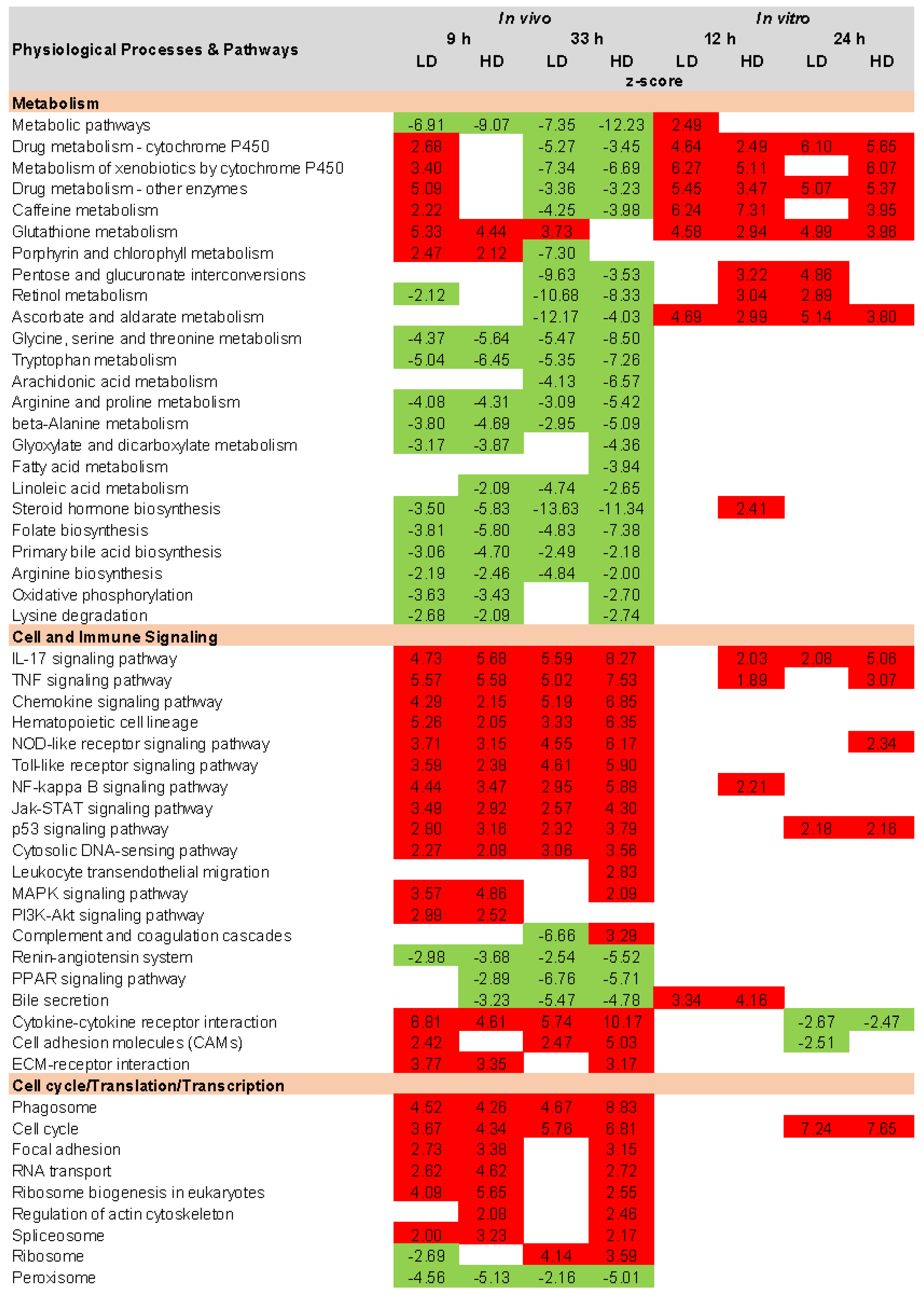
2.5. KEGG Pathway Analysis
3. Materials and Methods
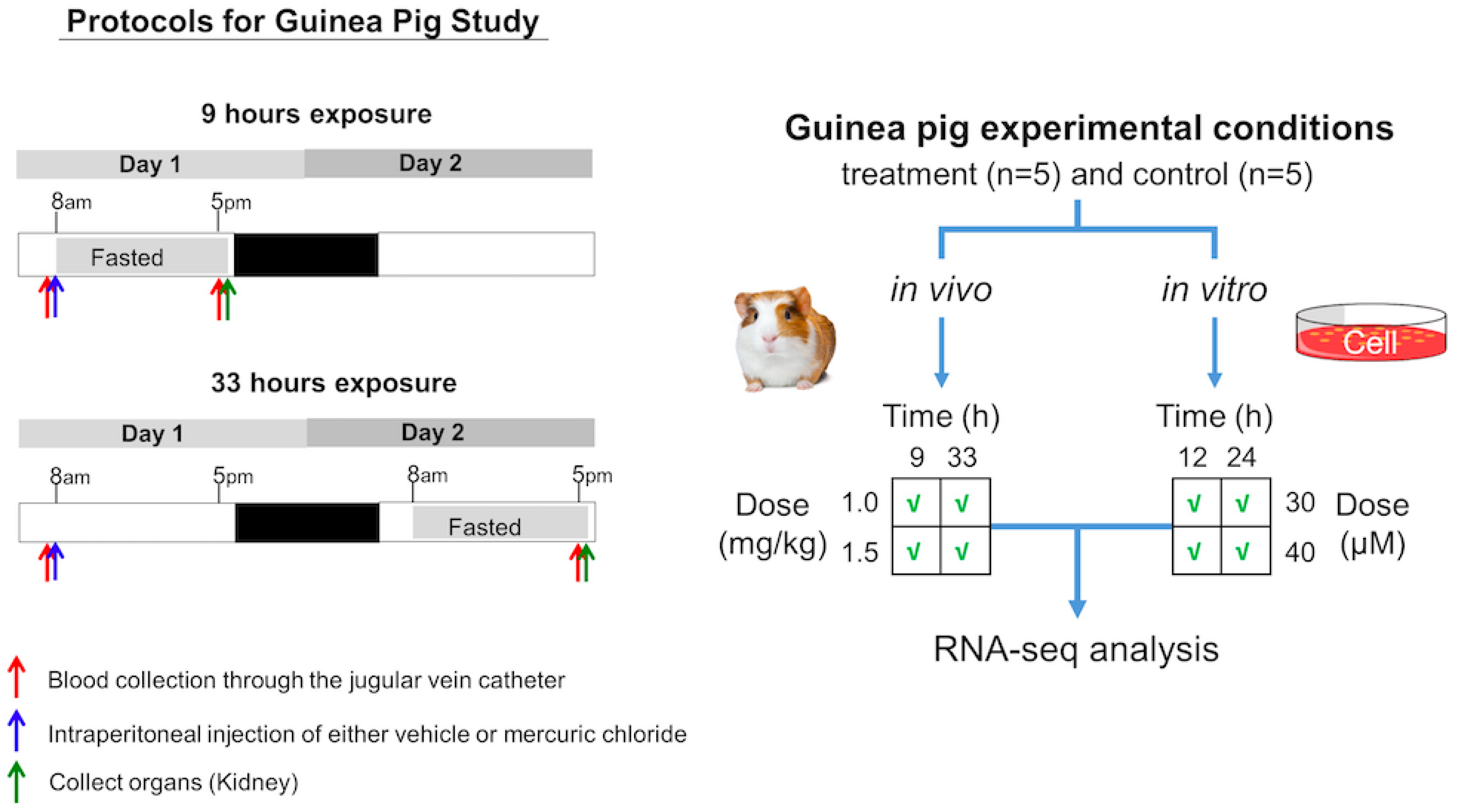
3.1. Experimental Design for In Vivo Studies
3.1.1. Animals
3.1.2. Preliminary Studies for Optimization of Dose and Time after Exposure
3.1.3. Studies for Measuring Changes in Gene Expression
3.2. Experimental Design for In Vitro Studies
3.2.1. Animals
3.2.2. Guinea Pig Renal Proximal Tubular Epithelial Cell Isolation and Culture
3.2.3. Preliminary Studies to Ascertain the Optimal Exposure of Toxicant
3.2.4. Exposure of Cells to Toxicant for RNA Isolation
3.3. RNA Sequencing
3.4. Analysis of RNA-seq Data
3.5. Injury Module Activation and Pathway Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Disclaimer
References
- Barbier, O.; Jacquillet, G.; Tauc, M.; Cougnon, M.; Poujeol, P. Effect of heavy metals on, and handling by, the kidney. Nephron Physiol. 2005, 99, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Tchounwou, P.B.; Yedjou, C.G.; Patlolla, A.K.; Sutton, D.J. Heavy metal toxicity and the environment. Mol. Clin. Environ. Toxicol. 2012, 101, 133–164. [Google Scholar]
- Jaishankar, M.; Tseten, T.; Anbalagan, N.; Mathew, B.B.; Beeregowda, K.N. Toxicity, mechanism and health effects of some heavy metals. Interdiscip. Toxicol. 2014, 7, 60. [Google Scholar] [CrossRef] [PubMed]
- Balali-Mood, M.; Naseri, K.; Tahergorabi, Z.; Khazdair, M.R.; Sadeghi, M. Toxic mechanisms of five heavy metals: Mercury, lead, chromium, cadmium, and arsenic. Front. Pharmacol. 2021, 12, 643972. [Google Scholar] [CrossRef]
- Agency for Toxic Substances and Disease Registry. National Priorities List from the Agency for Toxic Substances and Disease Registry (ATSDR). Available online: https://www.atsdr.cdc.gov/spl/resources/2015_atsdr_substance_priority_list.html (accessed on 26 January 2023).
- Benedict, R.T.; Alman, B.; Klotzbach, J.M.; Citra, M.; Diamond, G.L.; Herber, D.; Ingerman, L.; Nieman, S.; Tariq, S.; Zaccaria, K. Toxicological Profile for Mercury: Draft for Public Comment: April 2022. Available online: https://stacks.cdc.gov/view/cdc/117563 (accessed on 26 January 2023).
- Kromer, E.; Friedrich, G.; Wallner, P. Mercury and mercury compounds in surface air, soil gas, soils and rocks. J. Geochem. Explor. 1981, 15, 51–62. [Google Scholar] [CrossRef]
- Boffetta, P.; Merler, E.; Vainio, H. Carcinogenicity of mercury and mercury compounds. Scand. J. Work. Environ. Health 1993, 19, 1–7. [Google Scholar] [CrossRef]
- Vaidya, V.; Mehendale, H. Mercuric Chloride (HgCl2); Academic Press: Oxford, UK, 2014; pp. 203–206. [Google Scholar]
- Hassett-Sipple, B.; Swartout, J.; Schoeny, R. Health Effects of Mercury and Mercury Compounds. In Mercury Study Report to Congress; Environmental Protection Agency: Research Triangle Park, NC, USA, 1997; Volume 5. [Google Scholar]
- Magos, L.; Clarkson, T.W. Overview of the clinical toxicity of mercury. Ann. Clin. Biochem. 2006, 43, 257–268. [Google Scholar] [CrossRef]
- Hussain, S.; Rodgers, D.A.; Duhart, H.M.; Ali, S.F. Mercuric chloride-induced reactive oxygen species and its effect on antioxidant enzymes in different regions of rat brain. J. Environ. Sci. Health Part B 1997, 32, 395–409. [Google Scholar] [CrossRef]
- Pollard, K.; Hultman, P. Effects of mercury on the immune system. Met. Ions Biol. Syst. 1997, 34, 421–440. [Google Scholar]
- Stacchiotti, A.; Borsani, E.; Rodella, L.; Rezzani, R.; Bianchi, R.; Lavazza, A. Dose-dependent mercuric chloride tubular injury in rat kidney. Ultrastruct. Pathol. 2003, 27, 253–259. [Google Scholar] [CrossRef]
- Stacchiotti, A.; Lavazza, A.; Rezzani, R.; Borsani, E.; Rodella, L.; Bianchi, R. Mercuric chloride-induced alterations in stress protein distribution in rat kidney. Histol. Histopathol. 2004, 19, 1209–1218. [Google Scholar]
- Bernhoft, R.A. Mercury toxicity and treatment: A review of the literature. J. Environ. Public Health 2012, 2012, 460508. [Google Scholar] [CrossRef]
- Thukral, S.K.; Nordone, P.J.; Hu, R.; Sullivan, L.; Galambos, E.; Fitzpatrick, V.D.; Healy, L.; Bass, M.B.; Cosenza, M.E.; Afshari, C.A. Prediction of nephrotoxicant action and identification of candidate toxicity-related biomarkers. Toxicol. Pathol. 2005, 33, 343–355. [Google Scholar] [CrossRef]
- Trebucobich, M.S.; Hazelhoff, M.H.; Chevalier, A.A.; Passamonti, S.; Brandoni, A.; Torres, A.M. Protein expression of kidney and liver bilitranslocase in rats exposed to mercuric chloride—A potential tissular biomarker of toxicity. Toxicol. Lett. 2014, 225, 305–310. [Google Scholar] [CrossRef]
- Tokumoto, M.; Lee, J.Y.; Shimada, A.; Tohyama, C.; Satoh, M. Glutathione has a more important role than metallothionein-I/II against inorganic mercury-induced acute renal toxicity. J. Toxicol. Sci. 2018, 43, 275–280. [Google Scholar] [CrossRef]
- Caglayan, C.; Kandemir, F.M.; Yildirim, S.; Kucukler, S.; Eser, G. Rutin protects mercuric chloride-induced nephrotoxicity via targeting of aquaporin 1 level, oxidative stress, apoptosis and inflammation in rats. J. Trace Elem. Med. Biol. 2019, 54, 69–78. [Google Scholar] [CrossRef]
- Rojas-Franco, P.; Franco-Colin, M.; Torres-Manzo, A.P.; Blas-Valdivia, V.; Thompson-Bonilla, M.D.R.; Kandir, S.; Cano-Europa, E. Endoplasmic reticulum stress participates in the pathophysiology of mercury-caused acute kidney injury. Ren. Fail. 2019, 41, 1001–1010. [Google Scholar] [CrossRef]
- Ippolito, D.L.; AbdulHameed, M.D.M.; Tawa, G.J.; Baer, C.E.; Permenter, M.G.; McDyre, B.C.; Dennis, W.E.; Boyle, M.H.; Hobbs, C.A.; Streicker, M.A.; et al. Gene expression patterns associated with histopathology in toxic liver fibrosis. Toxicol. Sci. 2015, 149, 67–88. [Google Scholar] [CrossRef]
- AbdulHameed, M.D.; Ippolito, D.L.; Stallings, J.D.; Wallqvist, A. Mining kidney toxicogenomic data by using gene co-expression modules. BMC Genom. 2016, 17, 790. [Google Scholar] [CrossRef]
- McDyre, B.C.; AbdulHameed, M.D.M.; Permenter, M.G.; Dennis, W.E.; Baer, C.E.; Koontz, J.M.; Boyle, M.H.; Wallqvist, A.; Lewis, J.A.; Ippolito, D.L. Comparative proteomic analysis of liver steatosis and fibrosis after oral hepatotoxicant administration in Sprague-Dawley rats. Toxicol. Pathol. 2018, 46, 202–223. [Google Scholar] [CrossRef]
- Te, J.A.; AbdulHameed, M.D.M.; Wallqvist, A. Systems toxicology of chemically induced liver and kidney injuries: Histopathology-associated gene co-expression modules. J. Appl. Toxicol. 2016, 36, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Tawa, G.J.; AbdulHameed, M.D.M.; Yu, X.; Kumar, K.; Ippolito, D.L.; Lewis, J.A.; Stallings, J.D.; Wallqvist, A. Characterization of chemically induced liver injuries using gene co-expression modules. PloS ONE 2014, 9, e107230. [Google Scholar] [CrossRef] [PubMed]
- Schyman, P.; Printz, R.L.; Estes, S.K.; Boyd, K.L.; Shiota, M.; Wallqvist, A. Identification of the toxicity pathways associated with thioacetamide-induced injuries in rat liver and kidney. Front. Pharmacol. 2018, 9, 1272. [Google Scholar] [CrossRef] [PubMed]
- Schyman, P.; Printz, R.L.; Estes, S.K.; O’Brien, T.P.; Shiota, M.; Wallqvist, A. Assessing chemical-induced liver injury in vivo from in vitro gene expression data in the rat: The case of thioacetamide toxicity. Front. Genet. 2019, 10, 1233. [Google Scholar] [CrossRef] [PubMed]
- Schyman, P.; Printz, R.L.; Estes, S.K.; O’Brien, T.P.; Shiota, M.; Wallqvist, A. Concordance between thioacetamide-induced liver injury in rat and human in vitro gene expression data. Int. J. Mol. Sci. 2020, 21, 4017. [Google Scholar] [CrossRef]
- Schyman, P.; Printz, R.L.; Pannala, V.R.; AbdulHameed, M.D.M.; Estes, S.K.; Shiota, C.; Boyd, K.L.; Shiota, M.; Wallqvist, A. Genomics and metabolomics of early-stage thioacetamide-induced liver injury: An interspecies study between guinea pig and rat. Toxicol. Appl. Pharmacol. 2021, 430, 115713. [Google Scholar] [CrossRef]
- Schyman, P.; Printz, R.L.; AbdulHameed, M.D.M.; Estes, S.K.; Shiota, C.; Shiota, M.; Wallqvist, A. A toxicogenomic approach to assess kidney injury induced by mercuric chloride in rats. Toxicology 2020, 442, 152530. [Google Scholar] [CrossRef]
- Pannala, V.R.; Estes, S.K.; Rahim, M.; Trenary, I.; O’Brien, T.P.; Shiota, C.; Printz, R.L.; Reifman, J.; Shiota, M.; Young, J.D. Toxicant-induced metabolic alterations in lipid and amino acid pathways are predictive of acute liver toxicity in rats. Int. J. Mol. Sci. 2020, 21, 8250. [Google Scholar] [CrossRef]
- Schyman, P.; Xu, Z.; Desai, V.; Wallqvist, A. TOXPANEL: A gene-set analysis tool to assess liver and kidney injuries. Front. Pharmacol. 2021, 12, 601511. [Google Scholar] [CrossRef]
- Igarashi, Y.; Nakatsu, N.; Yamashita, T.; Ono, A.; Ohno, Y.; Urushidani, T.; Yamada, H. Open TG-GATEs: A large-scale toxicogenomics database. Nucleic Acids Res. 2015, 43, D921–D927. [Google Scholar] [CrossRef]
- Ganter, B.; Tugendreich, S.; Pearson, C.I.; Ayanoglu, E.; Baumhueter, S.; Bostian, K.A.; Brady, L.; Browne, L.J.; Calvin, J.T.; Day, G.-J.; et al. Development of a large-scale chemogenomics database to improve drug candidate selection and to understand mechanisms of chemical toxicity and action. J. Biotechnol. 2005, 119, 219–244. [Google Scholar] [CrossRef]
- Nielsen, J.B.; Andersen, H.R.; Andersen, O.; Starklint, H. Mercuric chloride-induced kidney damage in mice: Time course and effect of dose. J. Toxicol. Environ. Health 1991, 34, 469–483. [Google Scholar] [CrossRef]
- Al-Madani, W.; Siddiqi, N.; Alhomida, A. Renal toxicity of mercuric chloride at different time intervals in rats. Biochem. Insights 2009, 2, BCI-S2928. [Google Scholar] [CrossRef]
- Oliveira, V.A.; Favero, G.; Stacchiotti, A.; Giugno, L.; Buffoli, B.; de Oliveira, C.S.; Lavazza, A.; Albanese, M.; Rodella, L.F.; Pereira, M.E.; et al. Acute mercury exposition of virgin, pregnant, and lactating rats: Histopathological kidney and liver evaluations. Environ. Toxicol. 2017, 32, 1500–1512. [Google Scholar] [CrossRef]
- Vaidya, V.S.; Ferguson, M.A.; Bonventre, J.V. Biomarkers of acute kidney injury. Annu. Rev. Pharmacol. Toxicol. 2008, 48, 463. [Google Scholar] [CrossRef]
- Ronco, C.; Bellomo, R.; Kellum, J.A. Acute kidney injury. Lancet 2019, 394, 1949–1964. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Collins, J.R.; Alvord, W.G.; Roayaei, J.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. The DAVID Gene Functional Classification Tool: A novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007, 8, 1–16. [Google Scholar] [CrossRef]
- Price, P.M.; Safirstein, R.L.; Megyesi, J. The cell cycle and acute kidney injury. Kidney Int. 2009, 76, 604–613. [Google Scholar] [CrossRef]
- Thomasova, D.; Anders, H.-J. Cell cycle control in the kidney. Nephrol. Dial. Transplant. 2015, 30, 1622–1630. [Google Scholar] [CrossRef]
- Agarwal, A.; Dong, Z.; Harris, R.; Murray, P.; Parikh, S.M.; Rosner, M.H.; Kellum, J.A.; Ronco, C. Cellular and molecular mechanisms of AKI. J. Am. Soc. Nephrol. 2016, 27, 1288–1299. [Google Scholar] [CrossRef]
- Sharfuddin, A.A.; Molitoris, B.A. Pathophysiology of ischemic acute kidney injury. Nat. Rev. Nephrol. 2011, 7, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Correa-Costa, M.; Azevedo, H.; Amano, M.T.; Gonçalves, G.M.; Hyane, M.I.; Cenedeze, M.A.; Renesto, P.G.; Pacheco-Silva, A.; Moreira-Filho, C.A.; Câmara, N.O.S. Transcriptome analysis of renal ischemia/reperfusion injury and its modulation by ischemic pre-conditioning or hemin treatment. PloS ONE 2012, 7, e49569. [Google Scholar] [CrossRef] [PubMed]
- Pozzi, A.; Zent, R. Integrins in kidney disease. J. Am. Soc. Nephrol. 2013, 24, 1034–1039. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Dong, Z. Regulation and pathological role of p53 in cisplatin nephrotoxicity. J. Pharmacol. Exp. Ther. 2008, 327, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Liu, Y.; Wei, Q.; Huo, Y.; Liu, K.; Liu, F.; Dong, Z. Tubular p53 regulates multiple genes to mediate AKI. J. Am. Soc. Nephrol. 2014, 25, 2278–2289. [Google Scholar] [CrossRef]
- Scaduto, R., Jr.; Gattone, V., 2nd; Grotyohann, L.; Wertz, J.; Martin, L. Effect of an altered glutathione content on renal ischemic injury. Am. J. Physiol. Ren. Physiol. 1988, 255, F911–F921. [Google Scholar] [CrossRef]
- Ratliff, B.B.; Abdulmahdi, W.; Pawar, R.; Wolin, M.S. Oxidant mechanisms in renal injury and disease. Antioxid. Redox Signal. 2016, 25, 119–146. [Google Scholar] [CrossRef]
- Lin, Y.-H.; Yang-Yen, H.-F. The osteopontin-CD44 survival signal involves activation of the phosphatidylinositol 3-kinase/Akt signaling pathway. J. Biol. Chem. 2001, 276, 46024–46030. [Google Scholar] [CrossRef]
- Al-Lamki, R.S.; Mayadas, T.N. TNF receptors: Signaling pathways and contribution to renal dysfunction. Kidney Int. 2015, 87, 281–296. [Google Scholar] [CrossRef]
- Li, N.; Chen, J.; Wang, P.; Fan, H.; Hou, S.; Gong, Y. Major signaling pathways and key mediators of macrophages in acute kidney injury. Mol. Med. Rep. 2021, 23, 455. [Google Scholar] [CrossRef]
- Ortega-Loubon, C.; Martínez-Paz, P.; García-Morán, E.; Tamayo-Velasco, Á.; López-Hernández, F.J.; Jorge-Monjas, P.; Tamayo, E. Genetic susceptibility to acute kidney injury. J. Clin. Med. 2021, 10, 3039. [Google Scholar] [CrossRef]
- Shi, Y.; Chen, G.; Teng, J. Network-based expression analyses and experimental verifications reveal the involvement of STUB1 in acute kidney injury. Front. Mol. Biosci. 2021, 8, 655361. [Google Scholar] [CrossRef]
- Zhang, L.; Cai, J.; Xiao, J.; Ye, Z. Identification of core genes and pathways between geriatric multimorbidity and renal insufficiency: Potential therapeutic agents discovered using bioinformatics analysis. BMC Med. Genom. 2022, 15, 212. [Google Scholar] [CrossRef]
- Agrawal, S.; Tripathi, G.; Khan, F.; Sharma, R.; Pandirikkal Baburaj, V. Relationship between GSTs gene polymorphism and susceptibility to end stage renal disease among North Indians. Ren. Fail. 2007, 29, 947–953. [Google Scholar] [CrossRef]
- Imig, J.D. Epoxide hydrolase and epoxygenase metabolites as therapeutic targets for renal diseases. Am. J. Physiol. Ren. Physiol. 2005, 289, F496–F503. [Google Scholar] [CrossRef]
- Rouillard, A.D.; Gundersen, G.W.; Fernandez, N.F.; Wang, Z.; Monteiro, C.D.; McDermott, M.G.; Ma’ayan, A. The harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016, 2016, baw100. [Google Scholar] [CrossRef]
- Shin, Y.J.; Kim, K.A.; Kim, E.S.; Kim, J.H.; Kim, H.S.; Ha, M.; Bae, O.N. Identification of aldo-keto reductase (AKR7A1) and glutathione S-transferase pi (GSTP1) as novel renal damage biomarkers following exposure to mercury. Hum. Exp. Toxicol. 2018, 37, 1025–1036. [Google Scholar] [CrossRef]
- Kojima, I.; Tanaka, T.; Inagi, R.; Nishi, H.; Aburatani, H.; Kato, H.; Miyata, T.; Fujita, T.; Nangaku, M. Metallothionein is upregulated by hypoxia and stabilizes hypoxia-inducible factor in the kidney. Kidney Int. 2009, 75, 268–277. [Google Scholar] [CrossRef]
- Özcelik, D.; Nazıroglu, M.; Tunçdemir, M.; Çelik, Ö.; Öztürk, M.; Flores-Arce, M. Zinc supplementation attenuates metallothionein and oxidative stress changes in kidney of streptozotocin-induced diabetic rats. Biol. Trace Elem. Res. 2012, 150, 342–349. [Google Scholar] [CrossRef]
- Leierer, J.; Rudnicki, M.; Braniff, S.-J.; Perco, P.; Koppelstaetter, C.; Mühlberger, I.; Eder, S.; Kerschbaum, J.; Schwarzer, C.; Schroll, A. Metallothioneins and renal ageing. Nephrol. Dial. Transplant. 2016, 31, 1444–1452. [Google Scholar] [CrossRef]
- Liu, J.; Kumar, S.; Dolzhenko, E.; Alvarado, G.F.; Guo, J.; Lu, C.; Chen, Y.; Li, M.; Dessing, M.C.; Parvez, R.K. Molecular characterization of the transition from acute to chronic kidney injury following ischemia/reperfusion. JCI Insight 2017, 2, e94716. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Bai, Y.; Jayne, L.A.; Abdulkader, F.; Gandhi, M.; Perreau, T.; Parikh, S.V.; Gardner, D.S.; Davidson, A.J.; Sander, V. SOX9 promotes stress-responsive transcription of VGF nerve growth factor inducible gene in renal tubular epithelial cells. J. Biol. Chem. 2020, 295, 16328–16341. [Google Scholar] [CrossRef] [PubMed]
- Viñas, J.L.; Porter, C.J.; Douvris, A.; Spence, M.; Gutsol, A.; Zimpelmann, J.A.; Tailor, K.; Campbell, P.A.; Burns, K.D. Sex diversity in proximal tubule and endothelial gene expression in mice with ischemic acute kidney injury. Clin. Sci. 2020, 134, 1887–1909. [Google Scholar] [CrossRef]
- Feng, D.; Ngov, C.; Henley, N.; Boufaied, N.; Gerarduzzi, C. Characterization of matricellular protein expression signatures in mechanistically diverse mouse models of kidney injury. Sci. Rep. 2019, 9, 16736. [Google Scholar] [CrossRef]
- Rowland, J.; Akbarov, A.; Eales, J.; Xu, X.; Dormer, J.P.; Guo, H.; Denniff, M.; Jiang, X.; Ranjzad, P.; Nazgiewicz, A. Uncovering genetic mechanisms of kidney aging through transcriptomics, genomics, and epigenomics. Kidney Int. 2019, 95, 624–635. [Google Scholar] [CrossRef] [PubMed]
- Elshemy, M.; Zahran, F.; Omran, M.; Nabil, A. DPPD ameliorates renal fibrosis induced by HgCl 2 in rats. Biosci. Res. 2018, 15, 2416–2425. [Google Scholar]
- Ackermann, M.; Strimmer, K. A general modular framework for gene set enrichment analysis. BMC Bioinform. 2009, 10, 47. [Google Scholar] [CrossRef]
- Yu, C.; Woo, H.J.; Yu, X.; Oyama, T.; Wallqvist, A.; Reifman, J. A strategy for evaluating pathway analysis methods. BMC Bioinform. 2017, 18, 453. [Google Scholar] [CrossRef]
- Zhao, M.; Ma, J.; Li, M.; Zhang, Y.; Jiang, B.; Zhao, X.; Huai, C.; Shen, L.; Zhang, N.; He, L. Cytochrome P450 enzymes and drug metabolism in humans. Int. J. Mol. Sci. 2021, 22, 12808. [Google Scholar] [CrossRef]
- Nolin, T.; Naud, J.; Leblond, F.; Pichette, V. Emerging evidence of the impact of kidney disease on drug metabolism and transport. Clin. Pharmacol. Ther. 2008, 83, 898–903. [Google Scholar] [CrossRef]
- Dreisbach, A.W. The influence of chronic renal failure on drug metabolism and transport. Clin. Pharmacol. Ther. 2009, 86, 553–556. [Google Scholar] [CrossRef]
- Shang, Y.; Siow, Y.L.; Isaak, C.K. Downregulation of glutathione biosynthesis contributes to oxidative stress and liver dysfunction in acute kidney injury. Oxidative Med. Cell. Longev. 2016, 2016, 9707292. [Google Scholar] [CrossRef]
- Lash, L.H. Role of glutathione transport processes in kidney function. Toxicol. Appl. Pharmacol. 2005, 204, 329–342. [Google Scholar] [CrossRef]
- Muntané-Relat, J.; Ourlin, J.-C.; Domergue, J.; Maurel, P. Differential effects of cytokines on the inducible expression of CYP1A1, CYP1A2, and CYP3A4 in human hepatocytes in primary culture. Hepatology 1995, 22, 1143–1153. [Google Scholar] [CrossRef]
- Kirwan, C.; MacPhee, I.; Lee, T.; Holt, D.; Philips, B. Acute kidney injury reduces the hepatic metabolism of midazolam in critically ill patients. Intensive Care Med. 2012, 38, 76–84. [Google Scholar] [CrossRef]
- Dixon, J.; Lane, K.; MacPhee, I.; Philips, B. Xenobiotic metabolism: The effect of acute kidney injury on non-renal drug clearance and hepatic drug metabolism. Int. J. Mol. Sci. 2014, 15, 2538–2553. [Google Scholar] [CrossRef]
- Hoke, T.S.; Douglas, I.S.; Klein, C.L.; He, Z.; Fang, W.; Thurman, J.M.; Tao, Y.; Dursun, B.; Voelkel, N.F.; Edelstein, C.L. Acute renal failure after bilateral nephrectomy is associated with cytokine-mediated pulmonary injury. J. Am. Soc. Nephrol. 2007, 18, 155–164. [Google Scholar] [CrossRef]
- Vaillant, A.A.J.; Qurie, A. Interleukin. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Zalups, R.K. Molecular interactions with mercury in the kidney. Pharmacol. Rev. 2000, 52, 113–144. [Google Scholar]
- Basile, D.P.; Anderson, M.D.; Sutton, T.A. Pathophysiology of acute kidney injury. Compr. Physiol. 2012, 2, 1303–1353. [Google Scholar]
- Miller, S.; Pallan, S.; Gangji, A.S.; Lukic, D.; Clase, C.M. Mercury-associated nephrotic syndrome: A case report and systematic review of the literature. Am. J. Kidney Dis. 2013, 62, 135–138. [Google Scholar] [CrossRef]
- Bachir, A.I.; Horwitz, A.R.; Nelson, W.J.; Bianchini, J.M. Actin-based adhesion modules mediate cell interactions with the extracellular matrix and neighboring cells. Cold Spring Harb. Perspect. Biol. 2017, 9, a023234. [Google Scholar] [CrossRef] [PubMed]
- Mehrotra, P.; Collett, J.A.; McKinney, S.D.; Stevens, J.; Ivancic, C.M.; Basile, D.P. IL-17 mediates neutrophil infiltration and renal fibrosis following recovery from ischemia reperfusion: Compensatory role of natural killer cells in athymic rats. Am. J. Physiol. Ren. Physiol. 2017, 312, F385–F397. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Pan, Y.; Chen, H.; Jiang, L.; Liao, Y. Key genes of renal tubular necrosis: A bioinformatics analysis. Transl. Androl. Urol. 2020, 9, 654. [Google Scholar] [CrossRef] [PubMed]
- Kellum, J.A.; Romagnani, P.; Ashuntantang, G.; Ronco, C.; Zarbock, A.; Anders, H.-J. Acute kidney injury. Nat. Rev. Dis. Prim. 2021, 7, 52. [Google Scholar] [CrossRef]
- Shiota, M. Measurement of glucose homeostasis in vivo: Combination of tracers and clamp techniques. In Animal Models in Diabetes Research. Methods in Molecular Biology; Joost, H.G., Al-Hasani, H., Schürmann, A., Eds.; Humana Press: Totowa, NJ, USA, 2012; pp. 229–253. [Google Scholar]
- Firsov, D.; Bonny, O. Circadian rhythms and the kidney. Nat. Rev. Nephrol. 2018, 14, 626–635. [Google Scholar] [CrossRef]
- Taub, M. Primary kidney proximal tubule cells. Basic Cell Cult. Protoc. 2005, 290, 231–247. [Google Scholar]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Achuthan, P.; Akanni, W.; Amode, M.R.; Barrell, D.; Bhai, J.; Billis, K.; Cummins, C.; Gall, A.; Girón, C.G.; et al. Ensembl 2018. Nucleic Acids Res. 2018, 46, D754–D761. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, B.; Lin, L.-L.; Zhao, S. Evaluation and comparison of computational tools for RNA-seq isoform quantification. BMC Genom. 2017, 18, 583. [Google Scholar] [CrossRef]
- Pimentel, H.; Bray, N.L.; Puente, S.; Melsted, P.; Pachter, L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 2017, 14, 687. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]


| Common Differential Expression Genes (DEGs) | ||||||
|---|---|---|---|---|---|---|
| In vivo | In vivo | |||||
| Overlap | 9 h | 33 h | ||||
| LD | HD | LD | HD | |||
| 9 h | LD | 4272 | 3814 | 452 | 3296 | |
| HD | 5665 | 542 | 4211 | |||
| 33 h | LD | 836 | 784 | |||
| HD | 8264 | |||||
| In vitro | In vitro | |||||
| Overlap | 12 h | 24 h | ||||
| LD | HD | LD | HD | |||
| 12 h | LD | 4377 | 3698 | 2240 | 2346 | |
| HD | 7540 | 2588 | 3246 | |||
| 24 h | LD | 3174 | 2397 | |||
| HD | 4286 | |||||
| In vivo | In vitro | |||||
| Overlap | 12 h | 24 h | ||||
| LD | HD | LD | HD | |||
| 9 h | LD | 1808 | 2650 | 1275 | 1534 | |
| HD | 2250 | 3441 | 1676 | 2041 | ||
| 33 h | LD | 332 | 466 | 248 | 289 | |
| HD | 2877 | 4679 | 2171 | 2825 | ||
| KEGG Pathway | Benjamini p-Value |
|---|---|
| Cell cycle | 3.8 × 10−11 |
| ECM-receptor interaction | 1.2 × 10−8 |
| DNA replication | 7.6 × 10−8 |
| Metabolic pathways | 1.8 × 10−6 |
| Focal adhesion | 2.2 × 10−6 |
| PI3K-Akt signaling pathway | 2.4 × 10−6 |
| Fluid shear stress and atherosclerosis | 2.4 × 10−6 |
| p53 signaling pathway | 5.7 × 10−5 |
| Glutathione metabolism | 8.9 × 10−5 |
| Regulation of actin cytoskeleton | 3.9 × 10−3 |
| Drug metabolism—other enzymes | 8.2 × 10−3 |
| TNF signaling pathway | 8.2 × 10−3 |
| Glycine, serine, and threonine metabolism | 1.6 × 10−2 |
| Peroxisome | 1.7 × 10−2 |
| Chemical carcinogenesis—reactive oxygen species | 3.2 × 10−2 |
| Gene (Down) | log2 (FC) | Gene (Up) | log2 (FC) |
|---|---|---|---|
| Slc7a13 | −1.98 | Gsta2 | 7.07 |
| Cbr1 | −1.73 | Gsta5 | 7.07 |
| LOC100360601 | −1.73 | Mt2A | 6.47 |
| LOC102556347 | −1.73 | Mt1m | 6.47 |
| Ppp1r3c | −1.42 | Gsto1 | 3.04 |
| Cltrn | −1.41 | Mt1 | 2.69 |
| Smoc1 | −1.17 | Uchl1 | 2.51 |
| Cd200 | −1.15 | Gstp1 | 1.65 |
| Col11a1 | −1.04 | Rrm2 | 1.61 |
| Bend5 | −1.00 | Aldh1a7 | 1.57 |
| Kidney Injury Modules | In Vivo | In Vitro | ||||||
|---|---|---|---|---|---|---|---|---|
| 9 h | 33 h | 12 h | 24 h | |||||
| LD | HD | LD | HD | LD | HD | LD | HD | |
| Dilatation | 1.51 | 2.72 | 5.65 | 10.45 | 0.51 | 2.94 | 0.63 | 2.59 |
| Hyaline cast | 2.38 | 3.54 | 4.29 | 9.64 | 0.32 | 1.73 | 0.41 | 1.20 |
| Degeneration | 2.55 | 3.09 | 4.74 | 7.58 | 2.74 | 1.10 | 0.31 | 0.96 |
| Necrosis | 5.05 | 6.10 | 3.45 | 6.70 | 1.57 | 2.74 | 1.77 | 1.72 |
| Fibrogenesis | 2.07 | 2.79 | 2.18 | 3.67 | 3.03 | 5.11 | 1.03 | 2.34 |
| Intracytoplasmic inclusion body | 0.68 | 1.43 | 0.17 | 3.50 | 1.51 | 4.88 | 0.39 | 0.85 |
| Hypertrophy | 0.32 | −0.43 | −0.04 | 1.50 | −0.21 | 0.80 | 0.65 | −0.52 |
| Cellular infiltration | −1.63 | −1.04 | 2.15 | 0.04 | 4.06 | 2.05 | 1.35 | 0.82 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goel, H.; Printz, R.L.; Shiota, C.; Estes, S.K.; Pannala, V.; AbdulHameed, M.D.M.; Shiota, M.; Wallqvist, A. Assessing Kidney Injury Induced by Mercuric Chloride in Guinea Pigs with In Vivo and In Vitro Experiments. Int. J. Mol. Sci. 2023, 24, 7434. https://doi.org/10.3390/ijms24087434
Goel H, Printz RL, Shiota C, Estes SK, Pannala V, AbdulHameed MDM, Shiota M, Wallqvist A. Assessing Kidney Injury Induced by Mercuric Chloride in Guinea Pigs with In Vivo and In Vitro Experiments. International Journal of Molecular Sciences. 2023; 24(8):7434. https://doi.org/10.3390/ijms24087434
Chicago/Turabian StyleGoel, Himanshu, Richard L. Printz, Chiyo Shiota, Shanea K. Estes, Venkat Pannala, Mohamed Diwan M. AbdulHameed, Masakazu Shiota, and Anders Wallqvist. 2023. "Assessing Kidney Injury Induced by Mercuric Chloride in Guinea Pigs with In Vivo and In Vitro Experiments" International Journal of Molecular Sciences 24, no. 8: 7434. https://doi.org/10.3390/ijms24087434
APA StyleGoel, H., Printz, R. L., Shiota, C., Estes, S. K., Pannala, V., AbdulHameed, M. D. M., Shiota, M., & Wallqvist, A. (2023). Assessing Kidney Injury Induced by Mercuric Chloride in Guinea Pigs with In Vivo and In Vitro Experiments. International Journal of Molecular Sciences, 24(8), 7434. https://doi.org/10.3390/ijms24087434






