Abstract
Some progress has been made in understanding the pathways related to rice heading, but their applications to breeding japonica rice varieties adapted to grow in low-latitude areas (“indica to japonica”) are limited. We edited eight adaptation-related genes via a lab-established CRISPR/Cas9 system in a japonica variety, Shennong265 (SN265). All T0 plants and their progeny bearing random mutation permutations were planted in southern China and screened for changes in heading date. We found that the double mutant of Days to heading 2 (DTH2) and CONSTANS 3 (OsCO3) (dth2-osco3), two CONSTANS-like (COL) genes, showed significantly delayed heading under both short-day (SD) and long-day (LD) conditions in Guangzhou and manifested great yield increase under SD conditions. We further demonstrated that the heading-related Hd3a-OsMADS14 pathway was down-regulated in the dth2-osco3 mutant lines. The editing of the COL genes DTH2 and OsCO3 greatly improves the agronomic performance of japonica rice in Southern China.
1. Introduction
Rice (Oryza sativa L.) is the staple crop for more than one-fifth of the world’s population in the world, which stresses the importance of enhancing its productivity and yield for food security. Indica and japonica are two subspecies of Asian cultivated rice that originated in Asia and have been grown worldwide. The grain quality of japonica varieties is superior to that of indica varieties, and thus japonica rice dominates the market in China. To meet demand, “indica to japonica” projects have been proposed for the cultivation of japonica rice in Southern China [1]. Traditionally, japonica rice is mainly grown in the temperate zones of the mid-latitudes, whereas indica rice is distributed in the tropics and subtropics [2]. Elite japonica varieties are vulnerable to short photoperiods and high temperature when growing in low-latitude areas, which greatly shortens their vegetative growth period and culminates in yield reduction and quality degradation [3,4].
Heading Date is an important agronomic trait reflecting regional adaptability and yield potential of rice cultivars. At the molecular level, rice heading date is regulated by a complicated regulatory network involving a large number of genes [5]. The relevant signaling pathways have been extensively investigated, and a few flowering time genes have been cloned, such as a CONSTANS-like (COL) gene Heading date 1 (Hd1) and Early heading date 1 (Ehd1) [6,7]. Hd1 is an important regulator of photoperiod regulation of heading in rice. Early studies have shown that Hd1 has a dual function, promoting heading under short-day (SD) conditions and delaying heading under long-day (LD) conditions [8]. Ehd1 is a unique B-type response regulator in rice, which plays a role in promoting heading independently of Hd1 under SD conditions [9].
Heading date 3a (Hd3a) and Rice Flowering Locus T 1 (RFT1) are two florigen genes in rice, acting downstream of most flowering regulators, for example, Hd1, Ehd1, and several COL genes [10,11]. Under SD conditions, the expression of Hd3a is induced by activated Hd1 and Ehd1 and repressed by a COL gene, OsCO3 [12]. Under LD conditions, Hd3a is down-regulated by Hd1 but up-regulated by a COL gene, DTH2. DTH2 and Ehd1 also up-regulate RFT1 [9,13], the major flowering promoter gene under LD conditions.
Despite the advances in molecular dissection of flowering pathways, there are few studies on the application of these molecular findings to the “indica to japonica” projects. We previously identified adaptation-related genes associated with the heading date in SN265, growing in northern and southern China [14,15]. In this study, we selected the eight most heading-relevant genes and aimed to identify the mutation permutations that may improve the agronomic performance of SN265 grown in Guangzhou (southern China). A lab-established CRISPR/Cas9 multiplex genome editing system was applied to generate mutation permutations [16], followed by agronomic trait evaluation. We found that the double mutation of DTH2 and OsCO3 (dth2-osco3), two COL genes [12,13], significantly delayed heading under both SD and LD conditions in Guangzhou and gained remarkable yield increase under SD conditions. Overall, these findings provide new resources for the “indica to japonica” breeding programs in South China and improve our understanding of the molecular mechanisms supporting agronomic performance in rice.
2. Results
2.1. dth2-osco3 Delays Heading in Japonica
Selecting appropriate allele combinations facilitates breeding programs targeting broad adaptation. Here, we aim to improve the yield potential of japonica varieties growing in South China by pinpointing beneficial mutation permutations conferring delayed heading in Guangzhou, a subtropical region. The eight most heading-relevant genes, DTH2, OsCO3, LATE-FLOWERING 1 (OsLF1), SET DOMAIN GROUP PROTEIN 725 (SDG725), GLUTAMINE SYNTHETASE 1(OsGS1), Dof zinc factor 12 (OsDof12), Nuclear factor Y C2 subunit (OsNF-YC2), and PURINE PERMEASE 7 (OsPUP7), were selected for editing from the genes differentiated between japonica and indica, which were identified in a previous genome-wide study [15]. The CRISPR/Cas9-based multiplex gene editing vectors containing the gRNAs of these genes were introduced into SN265, a japonica cultivar, via Agrobacterium-mediated transformation. Among all the homozygous T2 lines obtained, two lines, #19 and #21, carrying double mutations of DTH2 and OsCO3, showed a significant decrease in days to heading (DTH) under both SD and LD conditions (Figure 1a). Single-base insertions were observed in the coding regions of both DTH2 and OsCO3 in both #19 and #21, resulting in frameshift mutations (Figure S1a,b). Under SD conditions, the DTHs of the mutant lines were approximately 12 days later than that of SN265 (late season in Guangzhou, 113° E/23° N), whilst under LD conditions, the DTHs of the mutant lines were delayed by approximately 17 days compared to that of SN265 (Figure 1a,b).
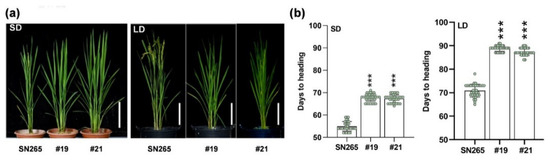
Figure 1.
Double mutation of DTH2 and OSCO3 delayed heading of japonica rice under both SD and LD conditions. (a) Phenotypes of 60-d-old (the left panel) and 89-d-old (the right panel) plants of SN265 (wild type, japonica rice) and the double mutant dth2-osco3 (two homozygous T2 lines #19 and #21) growing under SD conditions (the left panel) and LD conditions (the right panel). Scale bars, 20 cm (b) Days to heading of SN265 and dth2-osco3 lines under SD (the left panel) and LD (the right panel). Error bars represent SD of three biological replicates. Values are means ± SD. ***, p < 0.001.
To further explore how DTH2 and OsCO3 regulate heading in rice, we first examined the expression patterns of DTH2 and OsCO3 by qRT-PCR in SN265 grown in Guangzhou. Under SD conditions, both DTH2 and OsCO3 showed rhythmic expression patterns, whereas under LD conditions, only DTH2 showed rhythmic expression patterns (Figure 2a). We further found that the transcript levels of Hd3a, a key flowering activator in rice [10], and its downstream effector OsMADS14 [9,17] in #19 and #21 were significantly lower than those in SN265 under both SD and LD conditions (Figure 2b,c). Our results indicated that the disruption of both DTH2 and OsCO3 delays heading by down-regulating the Hd3a-OsMADS14 pathway.
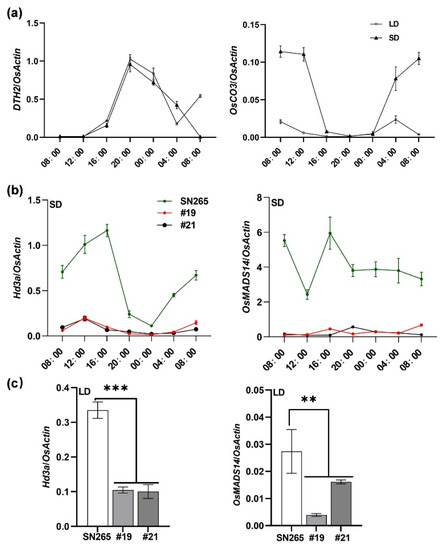
Figure 2.
Expression analysis in SN265 and dth2-osco3 lines via qRT-PCR under SD and LD conditions in Guangzhou. Under SD, leaves of 53-d-old plants were used for the expression analysis. Under LD, leaves of 80-d-old plants were used for the expression analysis. (a) expression of DTH2 and OsCO3 in SN265 under SD and LD, respectively. (b,c) expression of Hd3a and OsMADS14 in SN265 and dth2-osco3 lines under SD and LD, respectively. Error bars represent SD of three biological replicates. Values are means ± SD. **, p < 0.01. ***, p < 0.001.
2.2. dth2-osco3 Increased Panicle Size and Grain Yield under SD Conditions
To test the yield potential of the late-heading dth2-osco3 mutants, some yield-related agronomic traits, including plant height (PH), panicle length (PL), the number of primary branches (NPB), the number of secondary branches (NSB), the number of tillers (NT), and yield per plant (YPP), were evaluated under both SD and LD conditions. Compared to SN265, the dth2-osco3 lines (#19, #21) demonstrated a significant increase in PH, PL, NPB, NSB, and YPP but no difference in NT under SD conditions (Figure 3a–g). However, under LD conditions (early season in Guangzhou), the double mutant lines showed no significant differences in NSB, NT, and YPP along with reduced PH and PL, although there was still a significant increase in NPB (Figure 3h–n). These observations suggest that impaired DTH2 and OsCO3 improve the agronomic performance of SN265 in the indica rice-growing areas under SD conditions.
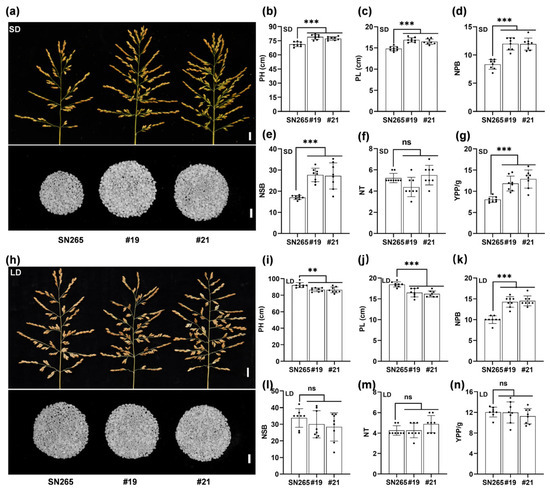
Figure 3.
Double mutation of DTH2 and OsCO3 improves the agronomic performance of japonica rice under SD and LD conditions. (a) and (h) The panicle shape and number of polished grains of SN265 and dth2-osco3 lines growing under SD and LD conditions. Scale bar for the panicles, 1 cm; scale bar for the polished grains, 10 mm. (b) and (i) the plant height, PH. (c) and (j) the panicle length, PL. (d) and (k) the number of primary branches, NPB. (e) and (l) the number of secondary branches, NSB. (f) and (m) the number of tillers, NT. (g) and (n) the yield per plant, YPP. YPP(g) = (the number of spikelets per plant × seed setting rate × per 1000-grain weight)/1000. Values are means ± SD. **, p < 0.01. ***, p < 0.001. ns, no significance.
Taken together, our data suggest that DTH2 and OsCO3 negatively regulate the adaptation of agronomic traits, especially for DTH and YPP, in japonica rice growing at low-latitudes.
2.3. Haplotype Analysis of DTH2 and OsCO3
To investigate the distribution of the haplotypes of DTH2 and OsCO3 in japonica and indica rice and validate their contribution to regional adaptation, we searched rice genomic databases for the functional nucleotide polymorphisms at the DTH2 and OsCO3 loci. We used RFGB v2.0 (https://www.rmbreeding.cn/, accessed on 11 November 2022) and RiceVarMap v2.0 (http://ricevarmap.ncpgr.cn/, accessed on 11 November 2022) [18,19,20] to identify the haplotypes of DTH2 and OsCO3 in 3000 rice germplasms. The data revealed that both DTH2 and OsCO3 had three major haplotypes, D-HapI~D-HapIII, and C-HapI~C-HapIII, respectively. D-HapI and C-HapI were mainly distributed in indica varieties, whereas most of the others were found in japonica varieties (Figure 4a,b). These three haplotypes of OsCO3 and DTH2 showed significant differences in yield-related traits such as heading date, plant height, and panicle number and may be associated with the phenotypic differentiation between indica and japonica. For example, C-HapIII-carrying varieties exhibited prolonged heading date, higher plant height, and reduced panicle number (Figure 4c–e), whereas the varieties with D-HapIII showed the opposite (Figure 4f–h). These data suggest that the adaptation of the aforementioned agronomic traits may be regulated by OsCO3 and DTH2 in japonica rice. We therefore postulate that the functional nucleotide polymorphisms at DTH2 and OsCO3 may serve as the target sites of genome editing in the “indica to japonica” projects.
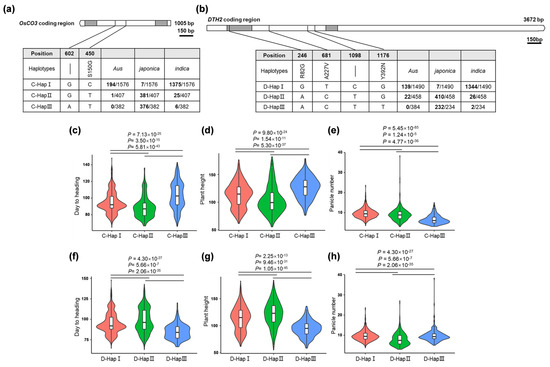
Figure 4.
Haplotypes of OsCO3 and DTH2 in rice and their association with agronomic traits. (a) and (b) Haplotypes of OsCO3 and DTH2 and their distributions in indica, japonica, and Aus varieties. (c–e) The association between the haplotypes of OsCO3 and days to heading, plant height, and panicle number. (f–h) The association between the haplotypes of DTH2 and days to heading, plant height, and panicle number. The haplotype and agronomic trait data were retrieved on RFGB v2.0 (https://www.rmbreeding.cn/, accessed on 11 November 2022) and RiceVarMap v2.0 (http://ricevarmap.ncpgr.cn/, accessed on 11 November 2022).
3. Discussion
During a long-term domestication process, japonica and indica rice have evolved to be adapted to different environments [15]. This study demonstrated that the disruption of two heading-related genes, DTH2 and OsCO3, greatly improved the agronomic performance of japonica cultivars growing in the indica rice cultivation areas, for example, in terms of heading date and yield per plant. The late-heading double mutant dth2-osco3 was identified from a homozygous T2 population consisting of the genomic mutations of eight heading-related genes differentiated between japonica and indica rice. Indeed, a previous study has shown that DTH2 has two functional nucleotide polymorphisms (FNPs) in indica rice IR24 and japonica rice Asominori, which plays an important role in the northward expansion of japonica [13]. We thus postulate that targeted editing of the genes differentiated between japonica and indica rice may help introduce beneficial mutations toward regional adaptation. In this regard, our work informs two critical loci regulating the adaptive traits of japonica varieties. When more and more such genomic loci are collected, especially for those contributing to the adaptation to temperature fluctuations and photoperiodic changes, we may eventually reach our final goal of the “indica to japonica” projects.
4. Materials and Methods
4.1. Plant Materials and Plant Growth Conditions
Rice (Oryza sativa L. ssp. japonica) Shennong265 (SN265) was obtained from Dr. Liang Tang’s lab at Shenyang Agricultural University. All the experiments were performed on an experimental farm at South China Agricultural University in Guangzhou (GZ, 113° E/23° N), China, during the normal rice-growing early season (12–13.5 h day length from mid-March to July) and the late season (11.5–13.5 h day length from mid-July to November) (Figure S2).
4.2. Genetic Transformation
We designed eight sgRNAs specifically targeting the coding regions of DTH2, OsCO3, OsLF1, SDG725, OsGS1, OsDof12, OsNF-YC2, and OsPUP7 using CRISPR/Cas9 systems [16]. The sgRNAs were cloned into the binary vector pYLCRISPR/Cas9Pubi-H2, carrying the Cas9 gene and an sgRNA expression module for editing. The primer design and vector construction were following the CRISPR-GE (http://skl.scau.edu.cn/, accessed on 20 July 2019, Guangzhou, China) (Table S1) [16]. The vectors were introduced into SN265 via the Agrobacterium-mediated transformation approach. The targeted genomic regions of the eight genes were examined by PCR using the primers listed in Table S2. The sequencing results were decoded by the Degenerate Sequence Decoding (DSD) method [21].
4.3. Trait Evaluation and Data Analysis
Eight plants in the middle of each row for SN265 and the dth2-osco3 lines were analyzed individually to score PL, NPB, NSB, NT, and YPP after ripening. Thirty-five plants were analyzed individually to score PH and DTH. DTH was scored as time (in d) from seed-soaking to the emergence of the first panicles. PH was measured as the distance from the ground surface to the tip of the tallest panicle, and PL is the distance from the panicle base to the uppermost spikelet. NPB and NSB were measured as described previously [22]. NT refers to the effective tiller number. YPP (g) = (the number of spikelets per plant × seed setting rate × per 1000-grain weight)/1000.
4.4. RNA Extraction and Gene Expression Analysis
Sixty-day-old plants grown in the early season and the late season were used for the expression analysis of the flowering-time genes. Leaves were harvested every 4 h within 1 day (7-time-points). Total RNAs were extracted using an Eastep®Super Total RNA Extraction Kit (Promega, Shanghai, China) and reverse-transcribed into cDNA using a Transcript One-Step gDNA Removal and cDNA Synthesis SuperMix (Transgen, Beijing, China) kit. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) assays were performed using a PerfecStart Green qPCR SuperMix (Transgen, Beijing, China) kit on a C1000 CFX96 Real-time PCR Detection System (Bio-rad, Hercules, CA, USA) with the following setting: 95 °C for 2 min, 42 cycles of two-step amplification (95 °C for 30 s, 60 °C for 30 s). PCR was repeated three times for technical repetition. The primers used for gene expression analysis are listed in Table S3.
5. Conclusions
In this study, we found that the double mutations of DTH2 and OsCO3 (dth2-osco3) significantly delayed heading under both SD and LD conditions in Guangzhou and manifested great yield increase under SD conditions. We further demonstrated that the heading-related Hd3a-OsMADS14 pathway was down-regulated in the dth2-osco3 mutant lines. Our findings provide new resources for the “indica to japonica” projects in South China.
Supplementary Materials
The supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24087346/s1.
Author Contributions
H.W. and Y.Z. conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and tables, and wrote the original draft. L.W. participated in agronomic trait investigation, data curation, and validation. C.X. and J.Y. participated in the agronomic trait investigation. Q.Z. and Y.-G.L. simultaneously conceived and designed the experiments, supervised the study, and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the Key Research Program of Guangzhou Science Technology and Innovation Commission (201904020030) and Laboratory of Lingnan Modern Agriculture Project (NT2021002).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Acknowledgments
We thank Liang Tang for providing rice materials and Jiehu Chen for providing the technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhang, H.C.; Zhang, J.; Gong, J.L.; Chang, Y.; Li, M.; Gao, H.; Dai, Q.G.; Huo, Z.Y.; Xu, K.; Wei, H.Y. The productive advantages and formation mechanisms of “indica rice to japonica rice”. Sci. Agric. Sin. 2013, 46, 686–704. [Google Scholar]
- Khush, G.S. Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 1997, 35, 25–34. [Google Scholar] [CrossRef]
- Saito, H.; Yuan, Q.B.; Okumoto, Y.; Doi, K.; Yoshimura, A.; Inoue, H.; Teraishi, M.; Tsukiyama, T.; Tanisaka, T. Multiple alleles at early flowering 1 locus making variation in the basic vegetative growth period in rice (Oryza sativa L.). Theor. Appl. Genet. 2009, 119, 315–323. [Google Scholar] [CrossRef]
- Wei, X.; Jiang, L.; Xu, J.; Liu, X.; Liu, S.; Zhai, H.; Wan, J. The distribution of japonica rice cultivars in the lower region of the Yangtze River valley is determined by its photoperiod-sensitivity and heading date genotypes. J. Integr. Plant Biol. 2009, 51, 922–932. [Google Scholar] [CrossRef] [PubMed]
- Li, X.F.; Liu, H.Z.; Wang, M.Q.; Liu, H.L.; Tian, X.J.; Zhou, W.J.; Lu, T.X.; Wang, Z.Y.; Chu, C.C.; Fang, J.; et al. Combinations of Hd2 and Hd4 genes determine rice adaptability to Heilongjiang Province, northern limit of China. J. Integr. Plant Biol. 2015, 57, 698–707. [Google Scholar] [CrossRef] [PubMed]
- Leng, Y.J.; Gao, Y.H.; Chen, L.; Yang, Y.L.; Huang, L.C.; Dai, L.P.; Ren, D.Y.; Xu, Q.K.; Zhang, Y.; Ponce, K.; et al. Using heading date 1 preponderant alleles from indica cultivars to breed high-yield, high-quality japonica rice varieties for cultivation in south China. Plant Biotechnol. J. 2020, 18, 119–128. [Google Scholar] [CrossRef]
- Wu, M.J.; Liu, H.Q.; Lin, Y.; Chen, J.M.; Fu, Y.P.; Luo, J.M.; Zhang, Z.J.; Liang, K.J.; Chen, S.B.; Wang, F. In-frame and frame-shift editing of the Ehd1 gene to develop japonica rice with prolonged basic vegetative growth periods. Front. Plant Sci. 2020, 11, 307. [Google Scholar] [CrossRef]
- Yano, M.; Katayose, Y.; Ashikari, M.; Yamanouchi, U.; Monna, L.; Fuse, T.; Baba, T.; Yamamoto, K.; Umehara, Y.; Nagamura, Y.; et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene constans. Plant Cell 2000, 12, 2473–2483. [Google Scholar] [CrossRef] [PubMed]
- Doi, K.; Izawa, T.; Fuse, T.; Yamanouchi, U.; Kubo, T.; Shimatani, Z.; Yano, M.; Yoshimura, A. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 2004, 18, 926–936. [Google Scholar] [CrossRef]
- Tamaki, S.; Matsuo, S.; Wong, H.L.; Yokoi, S.; Shimamoto, K. Hd3a protein is a mobile flowering signal in rice. Science 2007, 316, 1033–1036. [Google Scholar] [CrossRef] [PubMed]
- Komiya, R.; Yokoi, S.; Shimamoto, K. A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 2009, 136, 3443–3450. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Yun, C.H.; Lee, J.H.; Jang, Y.H.; Park, H.Y.; Kim, J.K. OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice. Planta 2008, 228, 355–365. [Google Scholar] [CrossRef]
- Wu, W.X.; Zheng, X.M.; Lu, G.W.; Zhong, Z.Z.; Gao, H.; Chen, L.P.; Wu, C.Y.; Wang, H.J.; Wang, Q.; Zhou, K.N.; et al. Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc. Natl. Acad. Sci. USA 2013, 110, 2775–2780. [Google Scholar] [CrossRef] [PubMed]
- Li, X.K.; Wu, L.; Wang, J.H.; Sun, J.; Xia, X.H.; Geng, X.; Wang, X.H.; Xu, Z.J.; Xu, Q. Genome sequencing of rice subspecies and genetic analysis of recombinant lines reveals regional yield- and quality-associated loci. BMC Biol. 2018, 16, 102. [Google Scholar] [CrossRef]
- Wang, M.; Chen, J.; Zhou, F.; Yuan, J.; Chen, L.; Wu, R.; Liu, Y.; Zhang, Q. The ties of brotherhood between japonica and indica rice for regional adaptation. Sci. China Life Sci. 2022, 65, 1369–1379. [Google Scholar] [CrossRef]
- Ma, X.L.; Zhang, Q.Y.; Zhu, Q.L.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.F.; Li, H.Y.; Lin, Y.R.; et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Simpson, G.G.; Dean, C. Arabidopsis, the Rosetta stone of flowering time? Science 2002, 296, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.C.; Yu, H.; Huang, J.; Wang, W.S.; Faruquee, M.; Zhang, F.; Zhao, X.Q.; Fu, B.Y.; Chen, K.; Zhang, H.L.; et al. Towards a deeper haplotype mining of complex traits in rice with RFGB v2.0. Plant Biotechnol. J. 2020, 18, 14–16. [Google Scholar] [CrossRef]
- Zhao, H.; Yao, W.; Ouyang, Y.; Yang, W.; Wang, G.; Lian, X.; Xing, Y.; Chen, L.; Xie, W. RiceVarMap: A comprehensive database of rice genomic variations. Nucleic Acids Res. 2015, 43, D1018–D1022. [Google Scholar] [CrossRef]
- Zhao, H.; Li, J.; Yang, L.; Qin, G.; Xia, C.; Xu, X.; Su, Y.; Liu, Y.; Ming, L.; Chen, L.L.; et al. An inferred functional impact map of genetic variants in rice. Mol. Plant 2021, 14, 1584–1599. [Google Scholar] [CrossRef]
- Liu, W.; Xie, X.; Ma, X.; Li, J.; Chen, J.; Liu, Y.G. DSDecode: A web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Mol. Plant 2015, 8, 1431–1433. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Huang, M.; Zong, W.; Xiao, D.; Lei, C.; Luo, Y.; Song, Y.; Li, S.; Hao, Y.; Luo, W.; et al. Hd1, Ghd7, and DTH8 synergistically determine the rice heading date and yield-related agronomic traits. J. Genet Genom. 2022, 49, 437–447. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).