Molecular Characterization of Resistance to Nicosulfuron in Setaria viridis
Abstract
1. Introduction
2. Results
2.1. Identification of Resistance-Associated Mutations in ALS
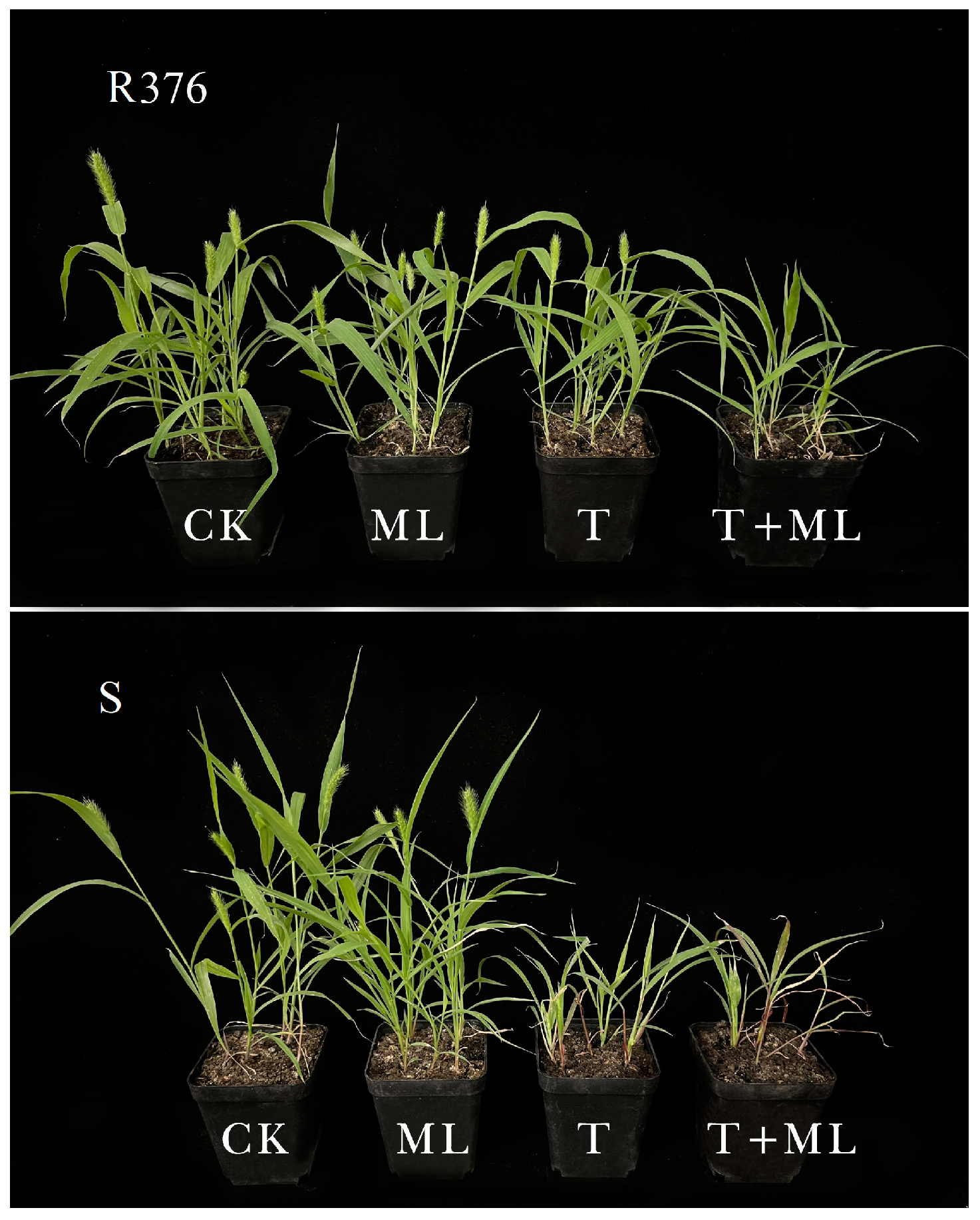
2.2. Effect of Pre-Treatment with Malathion on Nicosulfuron Resistance
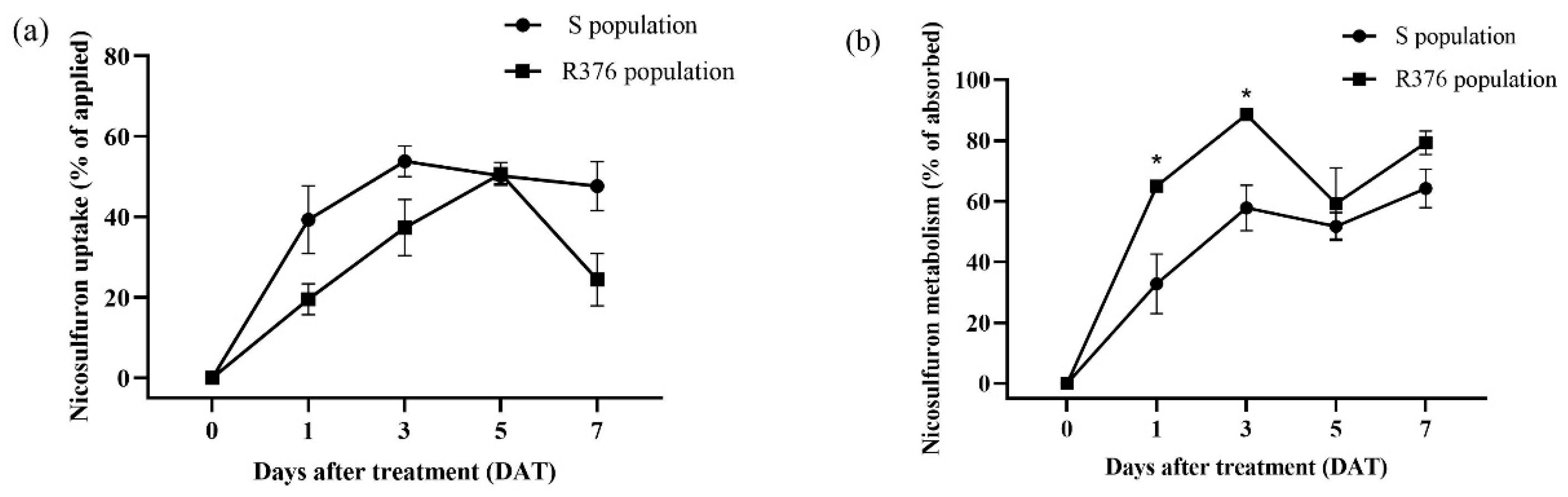
2.3. Nicosulfuron Metabolism in S. viridis Plants
2.4. Transcriptome Sequencing and Assembly
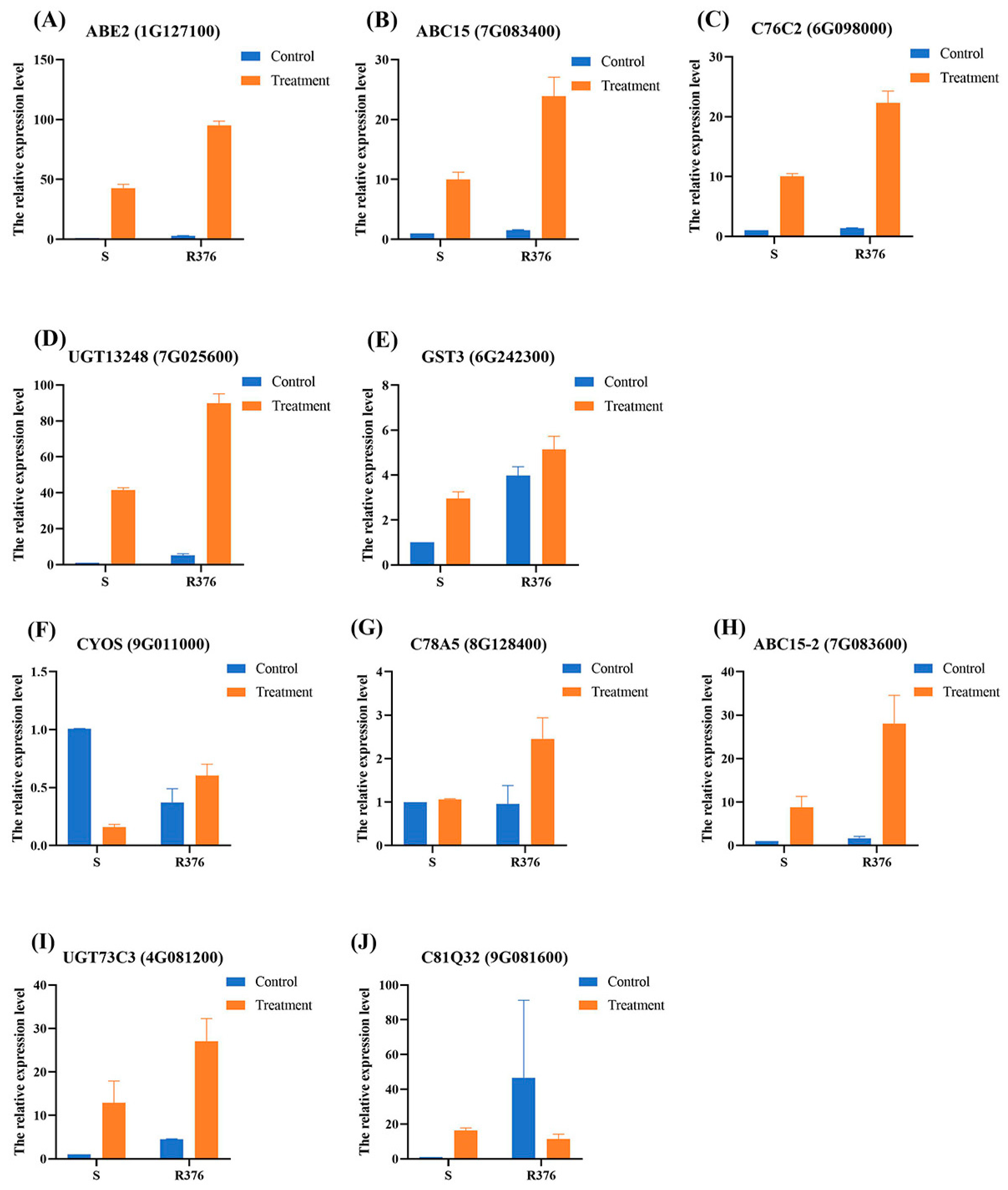
2.5. Selection of Candidate Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Sequencing of the ALS Gene
4.3. Effect of Malathion on Nicosulfuron Resistance
4.4. HPLC-MS/MS Analysis of Nicosulfuron
4.5. RNA Extraction, Library Construction, and Sequencing
4.6. De Novo Transcriptome Assembly and Gene Function Annotation
4.7. Identification and Analysis of Differentially Expressed Genes
4.8. The qPCR Validation
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Fu, Y.; Piao, H.; Mu, R. Scientific notes on some biological characteristics of Setaria viridis. J. Plant Prot. 1986, 3, 186–200. [Google Scholar] [CrossRef]
- Li, X.; Cui, H.; Chen, J.; He, K.; Wang, F.; Zhang, S. The ways to minimize dosages and increase efficacy of herbicides in corn in North China. J. Maize Sci. 2021, 29, 92–99. [Google Scholar] [CrossRef]
- Mazur, B.J.; Chui, C.F.; Smith, J.K. Isolation and characterization of plant genes coding for acetolactate synthase, the target enzyme for two classes of herbicides. Plant Physiol. 1987, 85, 1110–1117. [Google Scholar] [CrossRef]
- Wu, C.; Zhang, H.; Zhang, J.; Song, M.; Kong, F.; Lu, X. Resistance of weeds to nicosulfuron in corn fields in China. Plant Prot. 2016, 43, 198–203. [Google Scholar]
- Huang, Z.; Lu, Z.; Huang, H.; Li, W.; Cao, Y.; Wei, S. Target site mutations and cytochrome P450s-involved metabolism confer resistance to nicosulfuron in green foxtail (Setaria viridis). Pestic. Biochem. Physiol. 2021, 179, 104956. [Google Scholar] [CrossRef] [PubMed]
- Powles, S.B.; Yu, Q. Evolution in action: Plants resistant to herbicides. Annu. Rev. Plant Biol. 2010, 61, 317–347. [Google Scholar] [CrossRef]
- Ashigh, J.; Tardif, F.J. An amino acid substitution at position 205 of acetohydroxyacid synthase reduces fitness under optimal light in resistant populations of Solanum ptychanthum. Weed Res. 2009, 49, 479–489. [Google Scholar] [CrossRef]
- Palmieri, V.E.; Alvarez, C.E.; Permingeat, H.R.; Perotti, V.E. A122S, A205V, D376E, W574L and S653N substitutions in acetolactate synthase (ALS) from Amaranthus palmeri show different functional impacts on herbicide resistance. Pest Manag. Sci. 2022, 78, 749–757. [Google Scholar] [CrossRef]
- Vital Silva, V.; Mendes, R.; Suzukawa, A.; Adegas, F.; Marcelino-Guimaraes, F.; Oliveira, R. A target-site mutation confers cross-resistance to ALS-inhibiting herbicides in Erigeron sumatrensis from Brazil. Plants 2022, 11, 467. [Google Scholar] [CrossRef]
- Hada, Z.; Menchari, Y.; Rojano-Delgado, A.M.; Torra, J.; Menéndez, J.; Palma-Bautista, C.; de Prado, R.; Souissi, T. Point mutations as main resistance mechanism together with P450-based metabolism confer broad resistance to different ALS-Inhibiting herbicides in Glebionis coronaria from Tunisia. Front. Plant Sci. 2021, 12, 626702. [Google Scholar] [CrossRef]
- Yuan, J.S.; Tranel, P.J.; Stewart, C.N. Non-target-site herbicide resistance: A family business. Trends Plant Sci. 2007, 12, 6–13. [Google Scholar] [CrossRef]
- Dayan, F.E.; Barker, A.; Tranel, P.J. Origins and structure of chloroplastic and mitochondrial plant protoporphyrinogen oxidases: Implications for the evolution of herbicide resistance. Pest Manag. Sci. 2018, 74, 2226–2234. [Google Scholar] [CrossRef] [PubMed]
- Saika, H.; Horita, J.; Taguchi-Shiobara, F.; Nonaka, S.; Nishizawa-Yokoi, A.; Iwakami, S.; Hori, K.; Matsumoto, T.; Tanaka, T.; Itoh, T.; et al. A novel rice cytochrome P450 gene, CYP72A31, confers tolerance to acetolactate synthase-inhibiting herbicides in rice and Arabidopsis. Plant Physiol. 2014, 166, 1232–1240. [Google Scholar] [CrossRef]
- Thyssen, G.N.; Naoumkina, M.; McCarty, J.C.; Jenkins, J.N.; Florane, C.; Li, P.; Fang, D.D. The P450 gene CYP749A16 is required for tolerance to the sulfonylurea herbicide trifloxysulfuron sodium in cotton (Gossypium hirsutum L.). BMC Plant Biol. 2018, 18, 186. [Google Scholar] [CrossRef] [PubMed]
- Iwakami, S.; Endo, M.; Saika, H.; Okuno, J.; Nakamura, N.; Yokoyama, M.; Watanabe, H.; Toki, S.; Uchino, A.; Inamura, T. Cytochrome P450 CYP81A12 and CYP81A21 are associated with resistance to two acetolactate synthase inhibitors in Echinochloa phyllopogon. Plant Physiol. 2014, 165, 618–629. [Google Scholar] [CrossRef]
- Chen, J.; Yu, Q.; Patterson, E.; Sayer, C.; Powles, S. Dinitroaniline herbicide resistance and mechanisms in weeds. Front. Plant Sci. 2021, 12, 634018. [Google Scholar] [CrossRef]
- Torra, J.; Osuna, M.D.; Merotto, A.; Vila-Aiub, M. Editorial: Multiple Herbicide-resistant weeds and non-target site resistance mechanisms: A global challenge for food production. Front. Plant Sci. 2021, 12, 763212. [Google Scholar] [CrossRef]
- Barua, R.; Malone, J.; Boutsalis, P.; Gill, G.; Preston, C. Inheritance and mechanism of glyphosate resistance in annual bluegrass (Poa annua L.). Pest Manag. Sci. 2022, 78, 1377–1385. [Google Scholar] [CrossRef]
- Chen, J.; Goggin, D.; Han, H.; Busi, R.; Yu, Q.; Powles, S. Enhanced trifluralin metabolism can confer resistance in Lolium rigidum. J. Agric. Food Chem. 2018, 66, 7589–7596. [Google Scholar] [CrossRef]
- Obenland, O.A.; Ma, R.; O’Brien, S.R.; Lygin, A.V.; Riechers, D.E. Carfentrazone-ethyl resistance in an Amaranthus tuberculatus population is not mediated by amino acid alterations in the PPO2 protein. PLoS ONE 2019, 14, e0215431. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Picard, J.C.; Tian, X.; Darmency, H. A herbicide-resistant ACCase 1781 Setaria mutant shows higher fitness than wild type. Heredity 2010, 105, 394–400. [Google Scholar] [CrossRef]
- Darmency, H.; Picard, J.C.; Wang, T. Fitness costs linked to dinitroaniline resistance mutation in Setaria. Heredity 2011, 107, 80–86. [Google Scholar] [CrossRef]
- Martin, A.P.; Palmer, W.M.; Brown, C.; Abel, C.; Lunn, J.E.; Furbank, R.T.; Grof, C.P.L. A developing Setaria viridis internode: An experimental system for the study of biomass generation in a C4 model species. Biotechnol. Biofuels 2016, 9, 45. [Google Scholar] [CrossRef] [PubMed]
- Finley, T.; Chappell, H.; Veena, V. Agrobacterium-mediated transformation of Setaria viridis, a model system for cereals and bioenergy crops. Curr. Protoc. 2021, 1, e127. [Google Scholar] [CrossRef] [PubMed]
- Sada, Y.; Ikeda, H.; Yamato, S.; Kizawa, S. Characterization of sulfonylurea-resistant Schoenoplectus juncoides having a target-site Asp376Glu mutation in the acetolactate synthase. Pestic. Biochem. Physiol. 2013, 107, 106–111. [Google Scholar] [CrossRef]
- Menegat, A.; Bailly, G.C.; Aponte, R.; Heinrich, G.M.T.; Sievernich, B.; Gerhards, R. Acetohydroxyacid synthase (AHAS) amino acid substitution Asp376Glu in Lolium perenne: Effect on herbicide efficacy and plant growth. J. Plant Dis. Prot. 2016, 123, 145–153. [Google Scholar] [CrossRef]
- Zakaria, N.; Ruzmi, R.; Moosa, S.; Asib, N.; Zulperi, D.; Ismail, S.I.; Ahmad-Hamdani, M.S. Asp-376-Glu substitution endows target-site resistance to AHAS inhibitors in Limnocharis flava, an invasive weed in tropical rice fields. Physiol. Mol. Biol. Plants 2021, 27, 969–983. [Google Scholar] [CrossRef]
- Yu, Q.; Powles, S. Metabolism-based herbicide resistance and cross-resistance in crop weeds: A threat to herbicide sustainability and global crop production. Plant Physiol. 2014, 166, 1106–1118. [Google Scholar] [CrossRef]
- Dimaano, N.G.; Iwakami, S. Cytochrome P450-mediated herbicide metabolism in plants: Current understanding and prospects. Pest Manag. Sci. 2021, 77, 22–32. [Google Scholar] [CrossRef]
- Zhao, N.; Yan, Y.; Liu, W.; Wang, J. Cytochrome P450 CYP709C56 metabolizing mesosulfuron-methyl confers herbicide resistance in Alopecurus aequalis. Cell. Mol. Life Sci. 2022, 79, 205. [Google Scholar] [CrossRef]
- Nakka, S.; Godar, A.S.; Thompson, C.R.; Peterson, D.E.; Jugulam, M. Rapid detoxification via glutathione S-transferase (GST) conjugation confers a high level of atrazine resistance in Palmer amaranth (Amaranthus palmeri). Pest Manag. Sci. 2017, 73, 2236–2243. [Google Scholar] [CrossRef]
- Marrs, K.A. The functions and regulation of glutathione s-transferases in plants. Annu. Rev. Plant Biol. 1996, 47, 127–158. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, J.; Li, X.; Li, D.; Li, Z.; Cui, H. Pro-197-Ser mutation in ALS and high-level GST activities: Multiple resistance to ALS and ACCase inhibitors in Beckmannia syzigachne. Front. Plant Sci. 2020, 11, 572610. [Google Scholar] [CrossRef]
- Shaner, D.L. Role of translocation as a mechanism of resistance to Glyphosate. Weed Sci. 2009, 57, 118–123. [Google Scholar] [CrossRef]
- Martinoia, E.; Klein, M.; Geisler, M.; Bovet, L.; Forestier, C.; Kolukisaoglu, Ü.; Müller-Röber, B.; Schulz, B. Multifunctionality of plant ABC transporters—More than just detoxifiers. Planta 2002, 214, 345–355. [Google Scholar] [CrossRef]
- Frelet-Barrand, A.; Kolukisaoglu, H.Ü.; Plaza, S.; Rüffer, M.; Azevedo, L.; Hörtensteiner, S.; Marinova, K.; Weder, B.; Schulz, B.; Klein, M. Comparative mutant analysis of arabidopsis ABCC-type ABC transporters: AtMRP2 contributes to detoxification, vacuolar organic anion transport and chlorophyll degradation. Plant Cell Physiol. 2008, 49, 557–569. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Baldauf, S.; Lim, E.K.; Bowles, D.J. Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J. Biol. Chem. 2001, 276, 4338–4343. [Google Scholar] [CrossRef]
- Nguyen, D.Q.; Eamens, A.L.; Grof, C.P.L. Reference gene identification for reliable normalisation of quantitative RT-PCR data in Setaria viridis. Plant Methods 2018, 14, 24. [Google Scholar] [CrossRef]

| Treatments | S | R376 | RI d | ||||
|---|---|---|---|---|---|---|---|
| GR50 a (g a.i. ha−1) ± (SE) b | 95% CI c | p | GR50 (g a.i. ha−1) ± (SE) | 95% CI | p | ||
| Nicosulfuron | 7.5 ± 0.56 | 6.4–8.5 | 0.2075 | 268.6 ± 12.3 | 176.6–361.3 | 0.8014 | 35.8 |
| Malathion + nicosulfuron | 7.1 ± 0.81 | 5.7–9.0 | 0.0632 | 159.8 ± 5.9 | 93.4–226.2 | 0.4970 | 22.5 |
| Samples | Clean Reads a | Clean Bases b | GC Content c | ≥Q30% d |
|---|---|---|---|---|
| Rck1 | 21,357,755 | 6,394,783,796 | 55.20% | 92.56% |
| Rck2 | 20,859,879 | 6,247,437,338 | 54.66% | 92.13% |
| Rck3 | 24,958,030 | 7,473,507,024 | 55.12% | 92.19% |
| Rt1 | 20,964,984 | 6,275,572,248 | 53.72% | 95.19% |
| Rt2 | 21,160,996 | 6,339,218,842 | 50.11% | 93.73% |
| Rt3 | 21,802,295 | 6,530,031,944 | 53.52% | 95.35% |
| Sck1 | 19,886,346 | 5,956,973,016 | 54.92% | 95.40% |
| Sck2 | 25,714,276 | 7,700,027,284 | 54.97% | 95.69% |
| Sck3 | 21,408,426 | 6,411,927,072 | 54.96% | 95.59% |
| St1 | 22,129,252 | 6,625,605,236 | 53.42% | 95.13% |
| St2 | 19,674,122 | 5,885,620,152 | 53.87% | 95.27% |
| St3 | 21,613,683 | 6,467,906,952 | 54.01% | 95.57% |
| Rck1 | 21,357,755 | 6,394,783,796 | 55.20% | 92.56% |
| Annotation Database | Annotated Number | Annotated Percent (%) |
|---|---|---|
| COG | 9863 | 24.5 |
| GO | 24,099 | 59.8 |
| KEGG | 20,174 | 50 |
| KOG | 14,738 | 36.5 |
| Pfam | 25,125 | 62.3 |
| Swiss-Prot | 20,508 | 50.9 |
| eggNOG | 25,648 | 63.6 |
| NR | 38,961 | 96.6 |
| All | 38,986 | 96.7 |
| Groups/Samples | DEG Number | Upregulated | Downregulated |
|---|---|---|---|
| RT vs. RC | 5339 | 2172 | 3167 |
| RC vs. SC | 1582 | 1056 | 526 |
| RT vs. ST | 1691 | 974 | 717 |
| ST vs. SC | 8257 | 3568 | 4689 |
| GeneBank Accession Number | Log2 (RT/ST) | Log2 (RC/SC) | Gene Description | ||
|---|---|---|---|---|---|
| RNA-Seq | qPCR | RNA-Seq | qPCR | ||
| 1G127100 | 2.21 | 1.16 ± 0.04 *** | 1.63 | 1.54 ± 0.1 ** | ABC transporter E family member 2 |
| 7G083400 | 3.39 | 1.25 ± 0.02 * | 3.92 | 0.59 ± 0.08 * | ABC transporter C family member 15 |
| 6G098000 | 6.55 | 1.14 ± 0.05 * | 3.66 | 0.46 ± 0.07 ** | Cytochrome P450 76C2 |
| 7G025600 | - | 1.06 ± 1.22 *** | 3.00 | 2.33 ± 0.18 * | UDP-glucosyltransferase UGT13248 |
| 6G242300 | 8.37 | 0.79 ± 0.11 * | 7.13 | 1.97 ± 0.13 *** | Glutathione S-transferase T3 |
| 9G011000 | 3.31 | 1.92 ± 0.11 * | - | −1.68 ± 0.42 * | Premnaspirodiene oxygenase OS |
| 8G128400 | 2.83 | 1.14 ± 0.20 * | - | −0.66 ± 0.70 | Cytochrome P450 78A5 |
| 7G083600 | 1.16 | 1.79 ± 0.37 * | 2.71 | 0.62 ± 0.27 | ABC transporter C family member 15 |
| 4G081200 | - | 1.38 ± 0.50 | 3.99 | 2.14 ± 0.04 ** | UDP-glycosyltransferase 73C3 |
| 9G081600 | - | −0.62 ± 0.22 | 1.98 | 1.08 ± 0.12 * | Cytochrome P450 81Q32 |
| 9G351200 | 1.66 | 1.90 ± 0.16 | - | −0.27 ± 0.08 | Glutathione S-transferase GSTU6 |
| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| PP2A | ATGTGACACGGAGAACACCA | TGTTTCTGACCAGCAACCAC |
| 1G127100 | AGAACTCATGGACAGGCAAT | GGTAAATATCAGCAGGCTTTCCG |
| 7G083400 | CCCCAATGTCTTTCTTTGACTCC | GTCCCCAGGATTTGTATGACTGA |
| 6G098000 | AGCCATTCATTGAAGAGTCCGA | TGTAGCCGTTGACCTCGAT |
| 7G025600 | TTGGCACACAAGGCAACAGG | TGCCACCAACGGTACACCA |
| 6G242300 | AGGCCAAACCATAAGAGATCCAA | TTTGCCTACGACCGGATCCA |
| 9G011000 | TCAACATCCCGGACCTGT | CCGTCCTGTGGATGTCCT |
| 8G128400 | CAAGCCCGTAAAGCTTGCAG | TAAGCAGCTTCTCCACGACG |
| 7G083600 | CAAAGCGAAACGGGACTGTCA | ATTTCCGAATAGAATGTTGTCCCT |
| 4G081200 | CACCTTCGAGGAGATGGAGC | GCTGGTGGTAGAGTGACACC |
| 9G081600 | AGGCCAACGAGTTCAAGC | CGCCGAACACGTCGAACCA |
| 9G351200 | CTGAAGCTGCTCGGGATGTG | GCTCTTGTTGCGTAGGTTCTCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, Y.; Lan, Y.; Huang, H.; Wei, S.; Li, X.; Sun, Y.; Wang, R.; Huang, Z. Molecular Characterization of Resistance to Nicosulfuron in Setaria viridis. Int. J. Mol. Sci. 2023, 24, 7105. https://doi.org/10.3390/ijms24087105
Cao Y, Lan Y, Huang H, Wei S, Li X, Sun Y, Wang R, Huang Z. Molecular Characterization of Resistance to Nicosulfuron in Setaria viridis. International Journal of Molecular Sciences. 2023; 24(8):7105. https://doi.org/10.3390/ijms24087105
Chicago/Turabian StyleCao, Yi, Yuning Lan, Hongjuan Huang, Shouhui Wei, Xiangju Li, Ying Sun, Ruolin Wang, and Zhaofeng Huang. 2023. "Molecular Characterization of Resistance to Nicosulfuron in Setaria viridis" International Journal of Molecular Sciences 24, no. 8: 7105. https://doi.org/10.3390/ijms24087105
APA StyleCao, Y., Lan, Y., Huang, H., Wei, S., Li, X., Sun, Y., Wang, R., & Huang, Z. (2023). Molecular Characterization of Resistance to Nicosulfuron in Setaria viridis. International Journal of Molecular Sciences, 24(8), 7105. https://doi.org/10.3390/ijms24087105





