TOR1AIP1-Associated Nuclear Envelopathies
Abstract
1. Introduction
2. Methods and Search Strategy
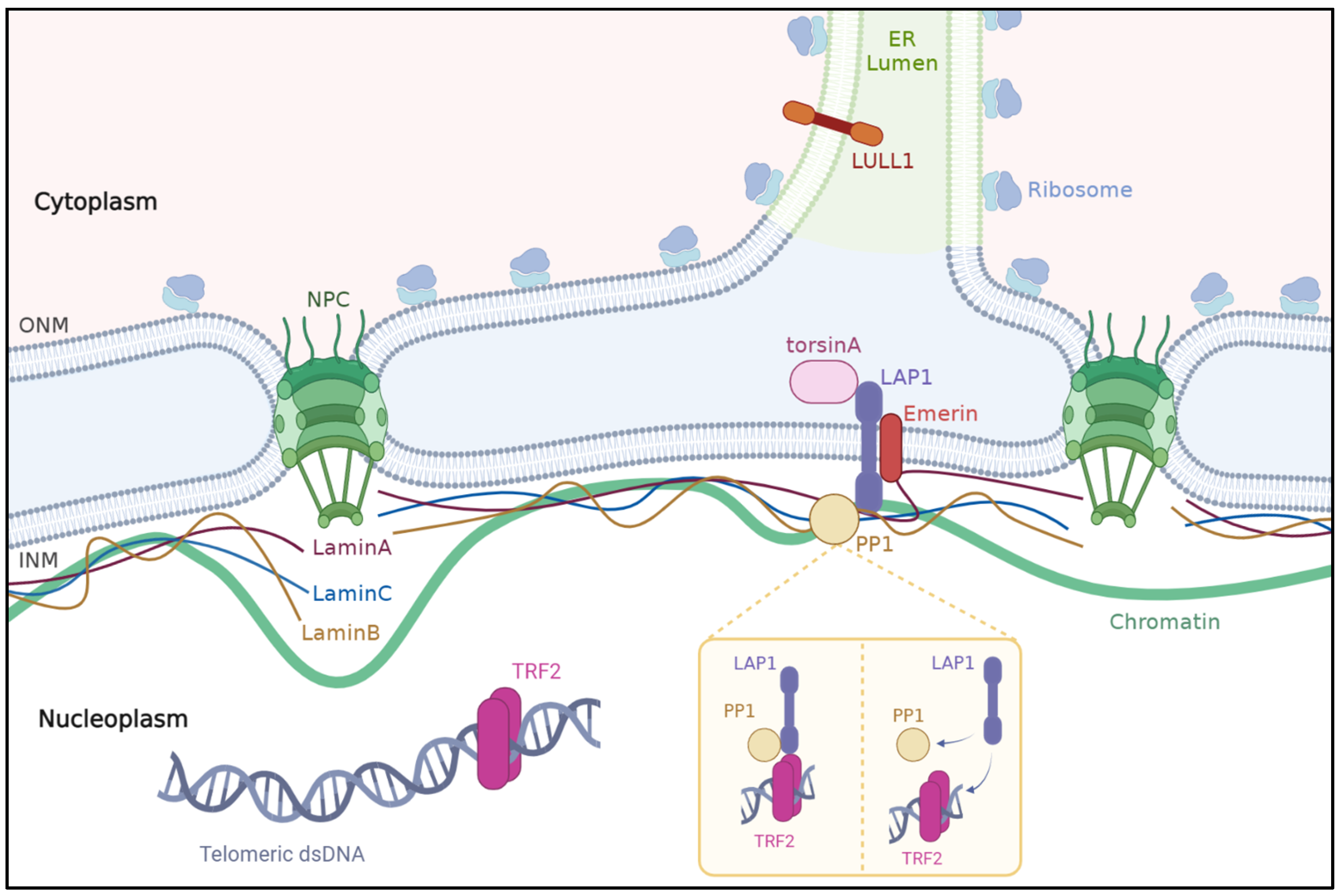
3. LAP1 an Integral Protein of the Inner Nuclear Membrane
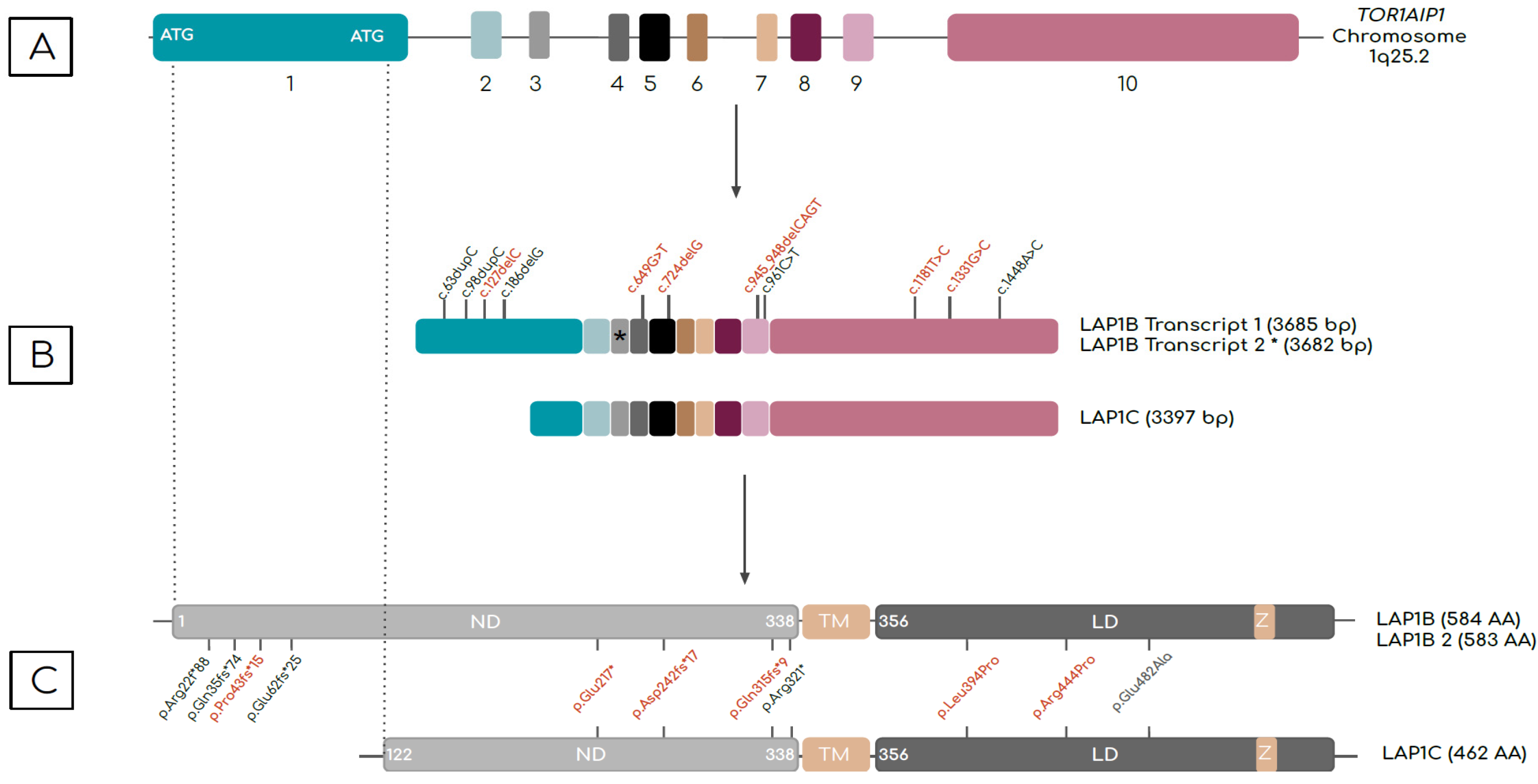
3.1. TOR1AIP1 Encodes Different LAP1 Isoforms
3.2. Interactions of LAP1 with Other Proteins in the Nuclear Envelope
3.2.1. Lamins
3.2.2. Emerin
3.2.3. TorsinA
3.2.4. PP1 and TRF2
4. The Biological Processes Involving LAP1 in Humans
4.1. Nuclear Envelope Structure and Chromatin Positioning
4.2. Mitosis and Cellular Dynamics
4.3. Cell Migration
4.4. DNA Damage Response and Genome Integrity
4.5. Spermatogenesis
4.6. Skeletal Muscle Maintenance and Growth
4.7. Cardiac Muscle Function
4.8. Neuromuscular Transmission
5. Mutations in the TOR1AIP1 and Associated Phenotypes
5.1. Cardiomyopathies and Congenital Heart Defects
5.2. Muscular Dystrophies and Myopathies
5.3. Congenital Myasthenic Syndrome
5.4. Multisystemic Disorders
5.4.1. Multisystemic Disorders with Progeroid Features
5.4.2. Other Multisystemic Presentations
5.5. Pathology and Imaging in TOR1AIP1-Related Disorders
5.6. Evidence for Phenotype–Genotype Correlations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Senior, A.; Gerace, L. Integral membrane proteins specific to the inner nuclear membrane and associated with the nuclear lamina. J. Cell Biol. 1988, 107, 2029–2036. [Google Scholar] [CrossRef] [PubMed]
- Kondo, Y.; Kondoh, J.; Hayashi, D.; Ban, T.; Takagi, M.; Kamei, Y.; Tsuji, L.; Kim, J.; Yoneda, Y. Molecular cloning of one isotype of human lamina-associated polypeptide 1s and a topological analysis using its deletion mutants. Biochem. Biophys. Res. Commun. 2002, 294, 770–778. [Google Scholar] [CrossRef] [PubMed]
- Dorboz, I.; Coutelier, M.; Bertrand, A.T.; Caberg, J.-H.; Elmaleh-Bergès, M.; Lainé, J.; Stevanin, G.; Bonne, G.; Boespflug-Tanguy, O.; Servais, L. Severe dystonia, cerebellar atrophy, and cardiomyopathy likely caused by a missense mutation in TOR1AIP1. Orphanet J. Rare Dis. 2014, 9, 174. [Google Scholar] [CrossRef] [PubMed]
- Malfatti, E.; Catchpool, T.; Nouioua, S.; Sihem, H.; Fournier, E.; Carlier, R.Y.; Cardone, N.; Davis, M.R.; Laing, N.G.; Sternberg, D.; et al. A TOR1AIP1 variant segregating with an early onset limb girdle myasthenia—Support for the role of LAP1 in NMJ function and disease. Neuropathol. Appl. Neurobiol. 2021, 48, e12743. [Google Scholar] [CrossRef] [PubMed]
- Lornage, X.; Mallaret, M.; Silva-Rojas, R.; Biancalana, V.; Giovannini, D.; Dieterich, K.; Saker, S.; Deleuze, J.-F.; Wuyam, B.; Laporte, J.; et al. Selective loss of a LAP1 isoform causes a muscle-specific nuclear envelopathy. Neurogenetics 2021, 22, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Wu, J.; Xian, W.; Liao, B.; Liao, S.; Yao, X.; Zhang, W. Muscular involvement and tendon contracture in limb-girdle muscular dystrophy 2Y: A mild adult phenotype and literature review. BMC Musculoskelet. Disord. 2020, 21, 588. [Google Scholar] [CrossRef]
- Cossins, J.; Webster, R.; Maxwell, S.; Cruz, P.M.R.; Knight, R.; Llewelyn, J.G.; Shin, J.-Y.; Palace, J.; Beeson, D. Congenital myasthenic syndrome due to a TOR1AIP1 mutation: A new disease pathway for impaired synaptic transmission. Brain Commun. 2020, 2, fcaa174. [Google Scholar] [CrossRef]
- Ghaoui, R.; Benavides, T.; Lek, M.; Waddell, L.B.; Kaur, S.; North, K.N.; MacArthur, D.G.; Clarke, N.F.; Cooper, S.T. TOR1AIP1 as a cause of cardiac failure and recessive limb-girdle muscular dystrophy. Neuromuscul. Disord. 2016, 26, 500–503. [Google Scholar] [CrossRef]
- Lessel, I.; Chen, M.-J.; Lüttgen, S.; Arndt, F.; Fuchs, S.; Meien, S.; Thiele, H.; Jones, J.R.; Shaw, B.R.; Crossman, D.K.; et al. Two novel cases further expand the phenotype of TOR1AIP1-associated nuclear envelopathies. Hum. Genet. 2020, 139, 483–498. [Google Scholar] [CrossRef]
- Fichtman, B.; Zagairy, F.; Biran, N.; Barsheshet, Y.; Chervinsky, E.; Ben Neriah, Z.; Shaag, A.; Assa, M.; Elpeleg, O.; Harel, A.; et al. Combined loss of LAP1B and LAP1C results in an early onset multisystemic nuclear envelopathy. Nat. Commun. 2019, 10, 605. [Google Scholar] [CrossRef]
- Bhatia, A.; Mobley, B.C.; Cogan, J.; Koziura, M.E.; Brokamp, E.; Phillips, J.; Newman, J.; Moore, S.A.; Hamid, R.; Acosta, M.T.; et al. Magnetic Resonance Imaging characteristics in case of TOR1AIP1 muscular dystrophy. Clin. Imaging 2019, 58, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Kayman-Kurekci, G.; Talim, B.; Korkusuz, P.; Sayar, N.; Sarioglu, T.; Oncel, I.; Sharafi, P.; Gundesli, H.; Balci-Hayta, B.; Purali, N.; et al. Mutation in TOR1AIP1 encoding LAP1B in a form of muscular dystrophy: A novel gene related to nuclear envelopathies. Neuromuscul. Disord. 2014, 24, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.; Domingues, S.C.; Costa, P.; Muller, T.; Galozzi, S.; Marcus, K.; da Cruz e Silva, E.F.; da Cruz e Silva, O.A.; Rebelo, S. Identification of a Novel Human LAP1 Isoform That Is Regulated by Protein Phosphorylation. PLoS ONE 2014, 9, e113732. [Google Scholar] [CrossRef] [PubMed]
- Tadokoro, K.; Yamazaki-Inoue, M.; Tachibana, M.; Fujishiro, M.; Nagao, K.; Toyoda, M.; Ozaki, M.; Ono, M.; Miki, N.; Miyashita, T.; et al. Frequent occurrence of protein isoforms with or without a single amino acid residue by subtle alternative splicing: The case of Gln in DRPLA affects subcellular localization of the products. J. Hum. Genet. 2005, 50, 382–394. [Google Scholar] [CrossRef] [PubMed]
- Foisner, R.; Gerace, L. Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell 1993, 73, 1267–1279. [Google Scholar] [CrossRef]
- Martin, L.; Crimaudo, C.; Gerace, L. cDNA Cloning and Characterization of Lamina-associated Polypeptide 1C (LAP1C), an Integral Protein of the Inner Nuclear Membrane. J. Biol. Chem. 1995, 270, 8822–8828. [Google Scholar] [CrossRef]
- Goodchild, R.E.; Dauer, W. The AAA+ protein torsinA interacts with a conserved domain present in LAP1 and a novel ER protein. J. Cell Biol. 2005, 168, 855–862. [Google Scholar] [CrossRef]
- Santos, M.; Rebelo, S.; Van Kleeff, P.J.M.; Kim, C.E.; Dauer, W.T.; Fardilha, M.; da Cruz e Silva, O.A.; da Cruz e Silva, E.F. The Nuclear Envelope Protein, LAP1B, Is a Novel Protein Phosphatase 1 Substrate. PLoS ONE 2013, 8, e76788. [Google Scholar] [CrossRef]
- Shin, J.-Y.; Méndez-López, I.; Hong, M.; Wang, Y.; Tanji, K.; Wu, W.; Shugol, L.; Krauss, R.S.; Dauer, W.T.; Worman, H.J. Lamina-associated polypeptide 1 is dispensable for embryonic myogenesis but required for postnatal skeletal muscle growth. Hum. Mol. Genet. 2016, 26, 65–78. [Google Scholar] [CrossRef]
- Shin, J.-Y.; Dauer, W.T.; Worman, H.J. Lamina-associated polypeptide 1: Protein interactions and tissue-selective functions. Semin. Cell Dev. Biol. 2014, 29, 164–168. [Google Scholar] [CrossRef]
- Gruenbaum, Y.; Wilson, K.L.; Harel, A.; Goldberg, M.; Cohen, M. Review: Nuclear Lamins—Structural Proteins with Fundamental Functions. J. Struct. Biol. 2000, 129, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Gruenbaum, Y.; Goldman, R.D.; Meyuhas, R.; Mills, E.; Margalit, A.; Fridkin, A.; Dayani, Y.; Prokocimer, M.; Enosh, A. The Nuclear Lamina and Its Functions in the Nucleus. Int. Rev. Cytol. 2003, 226, 1–62. [Google Scholar] [PubMed]
- Burke, B.; Stewart, C.L. Life at the edge: The nuclear envelope and human disease. Nat. Rev. Mol. Cell Biol. 2002, 3, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, E.; Guan, T.; Gerace, L. Involvement of the Lamin Rod Domain in Heterotypic Lamin Interactions Important for Nuclear Organization. J. Cell Biol. 2001, 153, 479–490. [Google Scholar] [CrossRef]
- Goldberg, M.; Harel, A.; Gruenbaum, Y. The Nuclear Lamina: Molecular Organization and Interaction with Chromatin. Crit. Rev. Eukaryot. Gene Expr. 1999, 9, 285–293. [Google Scholar] [CrossRef]
- Schirmer, E.C.; Florens, L.; Guan, T.; Yates, J.R.; Gerace, L. Nuclear Membrane Proteins with Potential Disease Links Found by Subtractive Proteomics. Science 2003, 301, 1380–1382. [Google Scholar] [CrossRef]
- Serrano, J.B.; Da Cruz e Silva, O.A.; Rebelo, S. Lamina Associated Polypeptide 1 (LAP1) Interactome and Its Functional Features. Membranes 2016, 6, 8. [Google Scholar] [CrossRef]
- Burke, B. On the cell-free association of lamins A and C with metaphase chromosomes. Exp. Cell Res. 1990, 186, 169–176. [Google Scholar] [CrossRef]
- Glass, J.R.; Gerace, L. Lamins A and C bind and assemble at the surface of mitotic chromosomes. J. Cell Biol. 1990, 111, 1047–1057. [Google Scholar] [CrossRef]
- Burke, B.; Stewart, C.L. The nuclear lamins: Flexibility in function. Nat. Rev. Mol. Cell Biol. 2012, 14, 13–24. [Google Scholar] [CrossRef]
- Gerace, L.; Tapia, O. Messages from the voices within: Regulation of signaling by proteins of the nuclear lamina. Curr. Opin. Cell Biol. 2018, 52, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Worman, H.J.; Bonne, G. “Laminopathies”: A wide spectrum of human diseases. Exp. Cell Res. 2007, 313, 2121–2133. [Google Scholar] [CrossRef] [PubMed]
- Burke, B.; Stewart, C.L. Functional Architecture of the Cell’s Nucleus in Development, Aging, and Disease. Curr. Top. Dev. Biol. 2014, 109, 1–52. [Google Scholar] [PubMed]
- Evangelisti, C.; Rusciano, I.; Mongiorgi, S.; Ramazzotti, G.; Lattanzi, G.; Manzoli, L.; Cocco, L.; Ratti, S. The wide and growing range of lamin B-related diseases: From laminopathies to cancer. Cell. Mol. Life Sci. 2022, 79, 126. [Google Scholar] [CrossRef] [PubMed]
- Storey, E.C.; Fuller, H.R. Genotype-Phenotype Correlations in Human Diseases Caused by Mutations of LINC Complex-Associated Genes: A Systematic Review and Meta-Summary. Cells 2022, 11, 4065. [Google Scholar] [CrossRef]
- Maison, C.; Pyrpasopoulou, A.; Theodoropoulos, P.A.; Georgatos, S.D. The inner nuclear membrane protein LAP1 forms a native complex with B-type lamins and partitions with spindle-associated mitotic vesicles. EMBO J. 1997, 16, 4839–4850. [Google Scholar] [CrossRef]
- Gant, T.M.; Wilson, K.L. Nuclear assembly. Annu. Rev. Cell Dev. Biol. 1997, 13, 669–695. [Google Scholar] [CrossRef]
- Shin, J.-Y.; Méndez-López, I.; Wang, Y.; Hays, A.P.; Tanji, K.; Lefkowitch, J.H.; Schulze, P.C.; Worman, H.J.; Dauer, W.T. Lamina-Associated Polypeptide-1 Interacts with the Muscular Dystrophy Protein Emerin and Is Essential for Skeletal Muscle Maintenance. Dev. Cell 2013, 26, 591–603. [Google Scholar] [CrossRef]
- Nagano, A.; Koga, R.; Ogawa, M.; Kurano, Y.; Kawada, J.; Okada, R.; Hayashi, Y.K.; Tsukahara, T. Emerin deficiency at the nuclear membrane in patients with Emery-Dreifuss muscular dystrophy. Nat. Genet. 1996, 12, 254–259. [Google Scholar] [CrossRef]
- Manilal, S.; Man, N.T.; Sewry, C.A.; Morris, G.E. The Emery-Dreifuss muscular dystrophy protein, emerin, is a nuclear membrane protein. Hum. Mol. Genet. 1996, 5, 801–808. [Google Scholar] [CrossRef]
- Bione, S.; Maestrini, E.; Rivella, S.; Mancini, M.; Regis, S.; Romeo, G.; Toniolo, D. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat. Genet. 1994, 8, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Melcon, G.; Kozlov, S.; Cutler, D.A.; Sullivan, T.; Hernandez, L.; Zhao, P.; Mitchell, S.; Nader, G.; Bakay, M.; Rottman, J.N.; et al. Loss of emerin at the nuclear envelope disrupts the Rb1/E2F and MyoD pathways during muscle regeneration. Hum. Mol. Genet. 2006, 15, 637–651. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, R.; Hayashi, Y.K.; Ogawa, M.; Kurokawa, R.; Matsumoto, H.; Noguchi, S.; Nonaka, I.; Nishino, I. Emerin-Lacking Mice Show Minimal Motor and Cardiac Dysfunctions with Nuclear-Associated Vacuoles. Am. J. Pathol. 2006, 168, 907–917. [Google Scholar] [CrossRef] [PubMed]
- Burdette, A.J.; Churchill, P.F.; Caldwell, G.A.; Caldwell, K.A. The early-onset torsion dystonia-associated protein, torsinA, displays molecular chaperone activity in vitro. Cell Stress Chaperones 2010, 15, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Konakova, M.; Huynh, D.P.; Yong, W.; Pulst, S.M. Cellular distribution of torsin A and torsin B in normal human brain. Arch. Neurol. 2001, 58, 921–927. [Google Scholar] [CrossRef] [PubMed]
- Naismith, T.V.; Dalal, S.; Hanson, P.I. Interaction of torsinA with its major binding partners is impaired by the dystonia-associated DeltaGAG deletion. J. Biol. Chem. 2009, 284, 27866–27874. [Google Scholar] [CrossRef]
- Jungwirth, M.; Dear, M.L.; Brown, P.; Holbrook, K.; Goodchild, R. Relative tissue expression of homologous torsinB correlates with the neuronal specific importance of DYT1 dystonia-associated torsinA. Hum. Mol. Genet. 2009, 19, 888–900. [Google Scholar] [CrossRef]
- Kim, C.E.; Perez, A.; Perkins, G.; Ellisman, M.H.; Dauer, W.T. A molecular mechanism underlying the neural-specific defect in torsinA mutant mice. Proc. Natl. Acad. Sci. USA 2010, 107, 9861–9866. [Google Scholar] [CrossRef]
- Zhao, C.; Brown, R.S.H.; Chase, A.R.; Eisele, M.R.; Schlieker, C. Regulation of Torsin ATPases by LAP1 and LULL1. Proc. Natl. Acad. Sci. USA 2013, 110, E1545–E1554. [Google Scholar] [CrossRef]
- Brown, R.S.H.; Zhao, C.; Chase, A.R.; Wang, J.; Schlieker, C. The mechanism of Torsin ATPase activation. Proc. Natl. Acad. Sci. USA 2014, 111, E4822–E4831. [Google Scholar] [CrossRef]
- A Sosa, B.; Demircioglu, F.E.; Chen, J.Z.; Ingram, J.; Ploegh, H.L.; Schwartz, T.U. How lamina-associated polypeptide 1 (LAP1) activates Torsin. eLife 2014, 3, e03239. [Google Scholar] [CrossRef]
- Luithle, N.; de Bos, J.U.; Hovius, R.; Maslennikova, D.; Lewis, R.T.M.; Ungricht, R.; Fierz, B.; Kutay, U. Torsin ATPases influence chromatin interaction of the Torsin regulator LAP1. eLife 2020, 9, 15. [Google Scholar] [CrossRef] [PubMed]
- Ozelius, L.J.; Hewett, J.W.; Page, C.E.; Bressman, S.B.; Kramer, P.L.; Shalish, C.; De Leon, D.; Brin, M.F.; Raymond, D.; Corey, D.P.; et al. The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat. Genet. 1997, 17, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Ozelius, L.J.; Page, C.E.; Klein, C.; Hewett, J.W.; Mineta, M.; Leung, J.; Shalish, C.; Bressman, S.B.; de Leon, D.; Brin, M.F.; et al. The TOR1A (DYT1) Gene Family and Its Role in Early Onset Torsion Dystonia. Genomics 1999, 62, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Millen, L.; Mendoza, J.L.; Thomas, P.J. A Unique Redox-sensing Sensor II Motif in TorsinA Plays a Critical Role in Nucleotide and Partner Binding*. J. Biol. Chem. 2010, 285, 37271–37280. [Google Scholar] [CrossRef]
- Saunders, C.A.; Harris, N.J.; Willey, P.T.; Woolums, B.M.; Wang, Y.; McQuown, A.J.; Schoenhofen, A.; Worman, H.J.; Dauer, W.T.; Gundersen, G.G.; et al. TorsinA controls TAN line assembly and the retrograde flow of dorsal perinuclear actin cables during rearward nuclear movement. J. Cell Biol. 2017, 216, 657–674. [Google Scholar] [CrossRef]
- Shin, J.Y.; Hernandez-Ono, A.; Fedotova, T.; Östlund, C.; Lee, M.J.; Gibeley, S.B.; Liang, C.-C.; Dauer, W.T.; Ginsberg, H.N.; Worman, H.J. Nuclear envelope-localized torsinA-LAP1 complex regulates hepatic VLDL secretion and steatosis. J. Clin. Investig. 2019, 129, 4885–4900. [Google Scholar] [CrossRef]
- Östlund, C.; Hernandez-Ono, A.; Shin, J.-Y. The Nuclear Envelope in Lipid Metabolism and Pathogenesis of NAFLD. Biology 2020, 9, 338. [Google Scholar] [CrossRef]
- Rebelo, S.; Santos, M.; Martins, F.; Da Cruz e Silva, E.F.; Da Cruz e Silva, E.F. Protein phosphatase 1 is a key player in nuclear events. Cell. Signal. 2015, 27, 2589–2598. [Google Scholar] [CrossRef]
- Pereira, C.D.; Martins, F.; Santos, M.; Müeller, T.; Da Cruz e Silva, O.A.; Rebelo, S. Nuclear Accumulation of LAP1:TRF2 Complex During DNA Damage Response Uncovers a Novel Role for LAP1. Cells 2020, 9, 1804. [Google Scholar] [CrossRef]
- Zhang, D.; Yan, P.; Han, T.; Cheng, X.; Li, J. Identification of key genes and biological processes contributing to colitis associated dysplasia in ulcerative colitis. PeerJ 2021, 9, e11321. [Google Scholar] [CrossRef] [PubMed]
- Cali-Daylan, A.E.; Dincer, P. Gene co-expression network analysis of dysferlinopathy: Altered cellular processes and functional prediction of TOR1AIP1, a novel muscular dystrophy gene. Neuromuscul. Disord. 2016, 27, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.-T.; Liu, X.-Y.; Sun, H.-D.; Xu, J.-Y.; Sun, J.-M.; Liu, W.; Chen, T.; Liu, J.-W.; Tan, Y.; Sun, W.; et al. Urinary Proteomics Analysis of Active Vitiligo Patients: Biomarkers for Steroid Treatment Efficacy Prediction and Monitoring. Front. Mol. Biosci. 2022, 9, 761562. [Google Scholar] [CrossRef]
- Roux, K.J.; Kim, D.I.; Raida, M.; Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012, 196, 801. [Google Scholar] [CrossRef] [PubMed]
- Zhong, N.; Radu, G.; Ju, W.; Brown, W.T. Novel progerin-interactive partner proteins hnRNP E1, EGF, Mel 18, and UBC9 interact with lamin A/C. Biochem. Biophys. Res. Commun. 2005, 338, 855–861. [Google Scholar] [CrossRef]
- Gadde, S.; Heald, R. Mechanisms and Molecules of the Mitotic Spindle. Curr. Biol. 2004, 14, R797–R805. [Google Scholar] [CrossRef]
- Karsenti, E.; Vernos, I. The Mitotic Spindle: A Self-Made Machine. Science 2001, 294, 543–547. [Google Scholar] [CrossRef]
- Serrano, J.B.; Martins, F.; Marafona, A.M.; da Cruz e Silva, O.A.B.; Rebelo, S. Encyclopedia of Signaling Molecules; Springer: Cham, Switzerland, 2018; Volume 176, pp. 5547–5556. [Google Scholar]
- Neumann, B.; Walter, T.; Hériché, J.-K.; Bulkescher, J.; Erfle, H.; Conrad, C.; Rogers, P.; Poser, I.; Held, M.; Liebel, U.; et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature 2010, 464, 721–727. [Google Scholar] [CrossRef]
- Santos, M.; Costa, P.; Martins, F.; Da Cruz e Silva, E.F.; Da Cruz e Silva, O.A.B.; Rebelo, S. LAP1 is a crucial protein for the maintenance of the nuclear envelope structure and cell cycle progression. Mol. Cell. Biochem. 2014, 399, 143–153. [Google Scholar] [CrossRef]
- Hutchins, J.R.A.; Toyoda, Y.; Hegemann, B.; Poser, I.; Hériché, J.-K.; Sykora, M.M.; Augsburg, M.; Hudecz, O.; Buschhorn, B.A.; Bulkescher, J.; et al. Systematic Analysis of Human Protein Complexes Identifies Chromosome Segregation Proteins. Science 2010, 328, 593–599. [Google Scholar] [CrossRef]
- Nicholson, J.; Scherl, A.; Way, L.; Blackburn, E.A.; Walkinshaw, M.D.; Ball, K.L.; Hupp, T.R. A systems wide mass spectrometric based linear motif screen to identify dominant in-vivo interacting proteins for the ubiquitin ligase MDM2. Cell. Signal. 2014, 26, 1243–1257. [Google Scholar] [CrossRef] [PubMed]
- Gomes, E.R.; Jani, S.; Gundersen, G.G. Nuclear Movement Regulated by Cdc42, MRCK, Myosin, and Actin Flow Establishes MTOC Polarization in Migrating Cells. Cell 2005, 121, 451–463. [Google Scholar] [CrossRef] [PubMed]
- Gundersen, G.G.; Worman, H.J. Nuclear Positioning. Cell 2013, 152, 1376–1389. [Google Scholar] [CrossRef]
- Luxton, G.G.; Gundersen, G.G. Orientation and function of the nuclear–centrosomal axis during cell migration. Curr. Opin. Cell Biol. 2011, 23, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, S.; Ballif, B.A.; Smogorzewska, A.; McDonald, E.R., III; Hurov, K.E.; Luo, J.; Bakalarski, C.E.; Zhao, Z.; Solimini, N.; Lerenthal, Y.; et al. ATM and ATR Substrate Analysis Reveals Extensive Protein Networks Responsive to DNA Damage. Science 2007, 316, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Shiloh, Y. The ATM-mediated DNA-damage response: Taking shape. Trends Biochem. Sci. 2006, 31, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Kastan, M.B.; Bartek, J. Cell-cycle checkpoints and cancer. Nature 2004, 432, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Taylor, P.; Moran, M.F. Proteomic Analysis of the Epidermal Growth Factor Receptor (EGFR) Interactome and Post-translational Modifications Associated with Receptor Endocytosis in Response to EGF and Stress. Mol. Cell. Proteom. 2014, 13, 1644–1658. [Google Scholar] [CrossRef]
- Emdal, K.B.; Hernandez-Ono, A.; Lee, M.J.; Gibeley, S.B.; Liang, C.-C.; Dauer, W.T.; Ginsberg, H.N.; Worman, H.J. Temporal proteomics of NGF-TrkA signaling identifies an inhibitory role for the E3 ligase Cbl-b in neuroblastoma cell differentiation. Sci. Signal. 2015, 8, ra40. [Google Scholar] [CrossRef]
- Martin, I.; Kim, J.W.; Lee, B.D.; Kang, H.C.; Xu, J.-C.; Jia, H.; Stankowski, J.; Kim, M.-S.; Zhong, J.; Kumar, M.; et al. Ribosomal Protein s15 Phosphorylation Mediates LRRK2 Neurodegeneration in Parkinson’s Disease. Cell 2014, 157, 472–485. [Google Scholar] [CrossRef]
- Lee, O.-H.; Kim, H.; He, Q.; Baek, H.J.; Yang, D.; Chen, L.-Y.; Liang, J.; Chae, H.K.; Safari, A.; Liu, D.; et al. Genome-wide YFP Fluorescence Complementation Screen Identifies New Regulators for Telomere Signaling in Human Cells. Mol. Cell. Proteom. 2011, 10, S1–S11. [Google Scholar] [CrossRef] [PubMed]
- Havugimana, P.C.; Hart, G.T.; Nepusz, T.; Yang, H.; Turinsky, A.L.; Li, Z.; Wang, P.I.; Boutz, D.R.; Fong, V.; Phanse, S.; et al. A Census of Human Soluble Protein Complexes. Cell 2012, 150, 1068–1081. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Liu, D.; Songyang, Z. The telosome/shelterin complex and its functions. Genome Biol. 2008, 9, 232. [Google Scholar] [CrossRef] [PubMed]
- de Lange, T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 2005, 19, 2100–2110. [Google Scholar] [CrossRef]
- Serrano, J.; Martins, F.; Sousa, J.; Pereira, C.; Van Pelt, A.; Rebelo, S.; da Cruz e Silva, O.A.B. Descriptive Analysis of LAP1 Distribution and That of Associated Proteins throughout Spermatogenesis. Membranes 2017, 7, 22. [Google Scholar] [CrossRef]
- Pereira, C.D.; Serrano, J.B.; Martins, F.; da Cruz e Silva, O.A.B.; Rebelo, S. Nuclear envelope dynamics during mammalian spermatogenesis: New insights on male fertility. Biol. Rev. 2019, 94, 1195–1219. [Google Scholar] [CrossRef]
- Shin, J.-Y.; Le Dour, C.; Sera, F.; Iwata, S.; Homma, S.; Joseph, L.C.; Morrow, J.; Dauer, W.T.; Worman, H.J. Depletion of lamina-associated polypeptide 1 from cardiomyocytes causes cardiac dysfunction in mice. Nucleus 2014, 5, 260–268. [Google Scholar] [CrossRef]
- Nouioua, S.; Malfatti, E.; Gianina, R.; Hellal, S.; Meriem, T.; Urtizberea, J.A. A case of congenital limb girdle myasthenia solved through a tripartite collaboration. Medecine/Sciences 2021, 37, 50–52. [Google Scholar] [CrossRef]
- Cohen, E.; Bonne, G.; Rivier, F.; Hamroun, D. The 2022 version of the gene table of neuromuscular disorders (nuclear genome). Neuromuscul. Disord. 2021, 31, 1313–1357. [Google Scholar] [CrossRef]
- Finlayson, S.; Beeson, D.; Palace, J. Congenital myasthenic syndromes: An update. Pract. Neurol. 2013, 13, 80–91. [Google Scholar] [CrossRef]
- Carrero, D.; Soria-Valles, C.; López-Otín, C. Hallmarks of progeroid syndromes: Lessons from mice and reprogrammed cells. Dis. Model. Mech. 2016, 9, 719–735. [Google Scholar] [CrossRef] [PubMed]
- Kayman-Kurekci, G.; Korkusuz, P.; Dinçer, P. Response (to Sewry and Goebel). Neuromuscul. Disord. 2014, 24, 1122. [Google Scholar] [CrossRef]
- Aoki, Y.; Wood, M.J. Emerging Oligonucleotide Therapeutics for Rare Neuromuscular Diseases. J. Neuromuscul. Dis. 2021, 8, 869–884. [Google Scholar] [CrossRef] [PubMed]
- Markati, T.; Oskoui, M.; Farrar, M.A.; Duong, T.; Goemans, N.; Servais, L. Emerging therapies for Duchenne muscular dystrophy. Lancet Neurol. 2022, 21, 814–829. [Google Scholar] [CrossRef] [PubMed]
- Fisher, G.; Mackels, L.; Markati, T.; Sarkozy, A.; Ochala, J.; Jungbluth, H.; Ramdas, S.; Ser-vais, L. Early clinical and pre-clinical therapy development in Nemaline Myopathy. Expert Opin. Ther. Targets 2022, 26, 853–867. [Google Scholar] [CrossRef] [PubMed]
| Mutation | Phenotype | Mutation Type | Isoform Affected | Genetic Testing | Segregation Study | Population Database | DNA and RNA Analyses | Protein Analysis 1 | Immunofluorescence | Material | Other Evidence | Ref |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| c.63dupC p.(Arg22Glnfs*88) | CMS | Frameshift (premature stop codon and loss of function) | LAP1B | Panel | Yes | NR | NR | NR | Signal from LAP1 antibody (both isoforms), nearly absent signal in myocytes | Muscle biopsy | Skeletal muscle OM, EM; muscle MRI; ENMG | [4] |
| c.63dupC p.(Arg22Glnfs*88) | MD | Frameshift (premature stop codon and loss of function) | LAP1B | WGS | No | Yes | Reduction of 16% in mRNAs encoding LAP1B + LAP1C; TOR1AIP1 cDNA levels at 6% of control’s | Strong reduction of LAP1B levels; normal LAP1C levels | NR | Fibroblasts | Skeletal muscle OM, EM; muscle MRI | [5] |
| c.98dupC p.(Gln35fs*74) | MD | Frameshift (premature stop codon) | LAP1B | WGS | Yes | Yes | NR | NR | NR | NA | Skeletal muscle OM, EM; muscle MRI | [6] |
| c.127delC p. (Pro43fs*15) | CMS | Frameshift (premature stop codon) | LAP1B | WES | Yes | NR | NR | No LAB1B isoform; normal expression of LAP1C | No expression of either LAP1 isoform in myonuclei | Muscle biopsy; cultured muscle cells | Animal model; skeletal muscle OM, EM | [7] |
| c.127delC p.(Pro43fs*15) | MD and/or dilated cardiomyopathy | Frameshift (premature stop codon) | LAP1B | WES | Yes | Yes | NR | No LAP1B isoform; normal levels of Lamin A/C and emerin. | Normal to slightly elevated Lamin A/C, LAP2B, emerin | Muscle biopsy; cardiac biopsy | Skeletal muscle OM, EM; cardiac muscle OM, EM; muscle MRI | [8] |
| c.1181 T > C p.(Leu394Pro) | Missense (change in sequence in exon 10) | LAP1B + LAP1C | ||||||||||
| c.186delG p.(Glu62fs*25) | MD | Frameshift (premature stop codon) | LAP1B | WGS | Yes | Yes | Fivefold decrease in TOR1AIP1 mRNA levels compared to control’s | No LAP1B; Increased expression of 50-kDA isoform (LAP1C?); increased expression of LULL1. | No LAP1B staining in Myonuclei and faint staining in endomysial cell nuclei; normal staining for LULL1, torsinA, and lamin B | Muscle biopsy | Skeletal muscle OM, EM | [12] |
| c.649G > T p.(Glu217*) | MS (progeroid) | Nonsense | LAP1B + LAP1C | WES | Yes | Yes | NR | NR | NR | NA | Skeletal muscle OM; brain MRI | [9] |
| c.724delG p.(Asp242fs*17) | Frameshift (premature stop codon) | LAP1B + LAP1C | ||||||||||
| c.945_ 948delCAGT p.(Gln315fs*9) | MS (progeroid) | Frameshift (premature stop codon) | LAP1B + LAP1C | WES | Yes | Yes | No difference in TOR1AIP1 mRNA levels between patients and controls; reductions in LMNA and LMNB1 mRNA levels | Absence of LAP1B and LAP1C; reduction in lamin B1 (senescence marker) and lamin A/C levels | Loss of lamin A/C staining in 18% of analyzed nuclei; lamin A/C aggregates at the nuclear periphery and nucleoplasm; NE invagination (~90% of abnormalities highlighted); complex nuclear lobulation; nuclear blebs; nuclear holes (4%) resembling cytoplasmic channels | SF | Brain CT; ENMG | [9] |
| c.1331G >C p.(Arg444Pro) | Missense (affects conserved arginine residue) | LAP1B + LAP1C | ||||||||||
| c.961C > T p.(Arg321*) | MS (progeroid) | Nonsense (premature stop codon) | LAP1B + LAP1C | WES | Yes | Yes | Decreased levels of mRNA encoding LAP1 and slightly elevated emerin mRNA | Absence of LAPB1B and LAP1C | Reduced intensity of lamin A/C nuclear rim staining; distorted nuclei; cytoplasmic channels in fibroblasts nuclei (6.3%) | SF | Brain MRI | [10] |
| c.1448A > C p.(Glu482Ala) | MS | Missense (affects highly conserved glutamic acid) | LAP1B + LAP1C | WGS | Yes | Yes | NR | Reduced levels of LAP1 isoforms | Reduction in LAP1 staining in the NE; mislocation and aggregation of LAP1 in the ER | SF | Bioinformatic prediction | [3] |
| Unspecified | MD | NR | NR | WGS | NR | NR | NR | NR | NR | NR | Skeletal muscle OM, EM; muscle MRI | [11] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mackels, L.; Liu, X.; Bonne, G.; Servais, L. TOR1AIP1-Associated Nuclear Envelopathies. Int. J. Mol. Sci. 2023, 24, 6911. https://doi.org/10.3390/ijms24086911
Mackels L, Liu X, Bonne G, Servais L. TOR1AIP1-Associated Nuclear Envelopathies. International Journal of Molecular Sciences. 2023; 24(8):6911. https://doi.org/10.3390/ijms24086911
Chicago/Turabian StyleMackels, Laurane, Xincheng Liu, Gisèle Bonne, and Laurent Servais. 2023. "TOR1AIP1-Associated Nuclear Envelopathies" International Journal of Molecular Sciences 24, no. 8: 6911. https://doi.org/10.3390/ijms24086911
APA StyleMackels, L., Liu, X., Bonne, G., & Servais, L. (2023). TOR1AIP1-Associated Nuclear Envelopathies. International Journal of Molecular Sciences, 24(8), 6911. https://doi.org/10.3390/ijms24086911






