Clinicopathological Significance of Cell Adhesion Molecule 4 Expression in Gallbladder Cancer and Its Prognostic Role
Abstract
1. Introduction
2. Results
2.1. Clinicopathological Characteristics of Patients
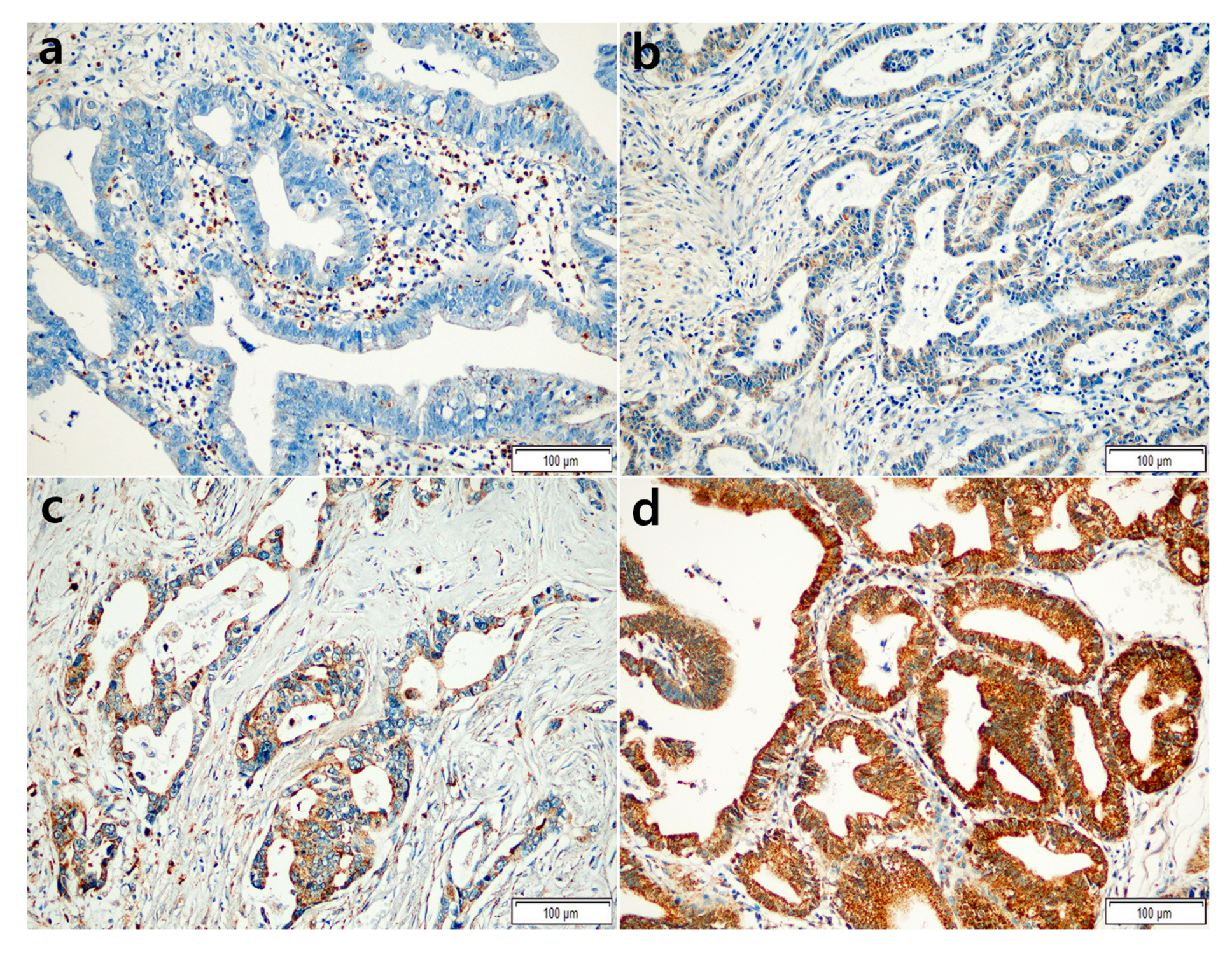
2.2. Correlations between CADM4 Expression and Clinicopathological Characteristics
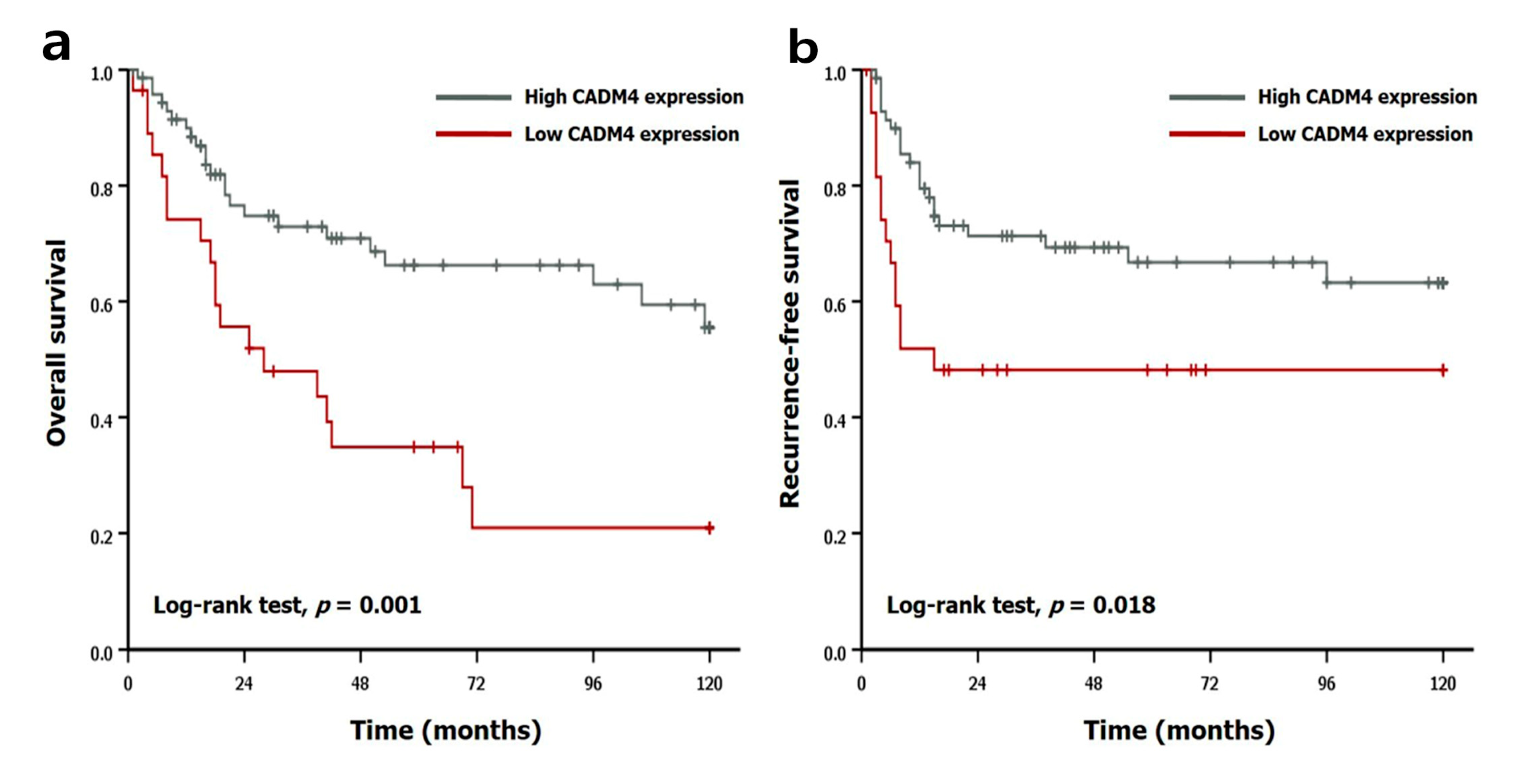
2.3. Prognostic Implication of CADM4 Expression
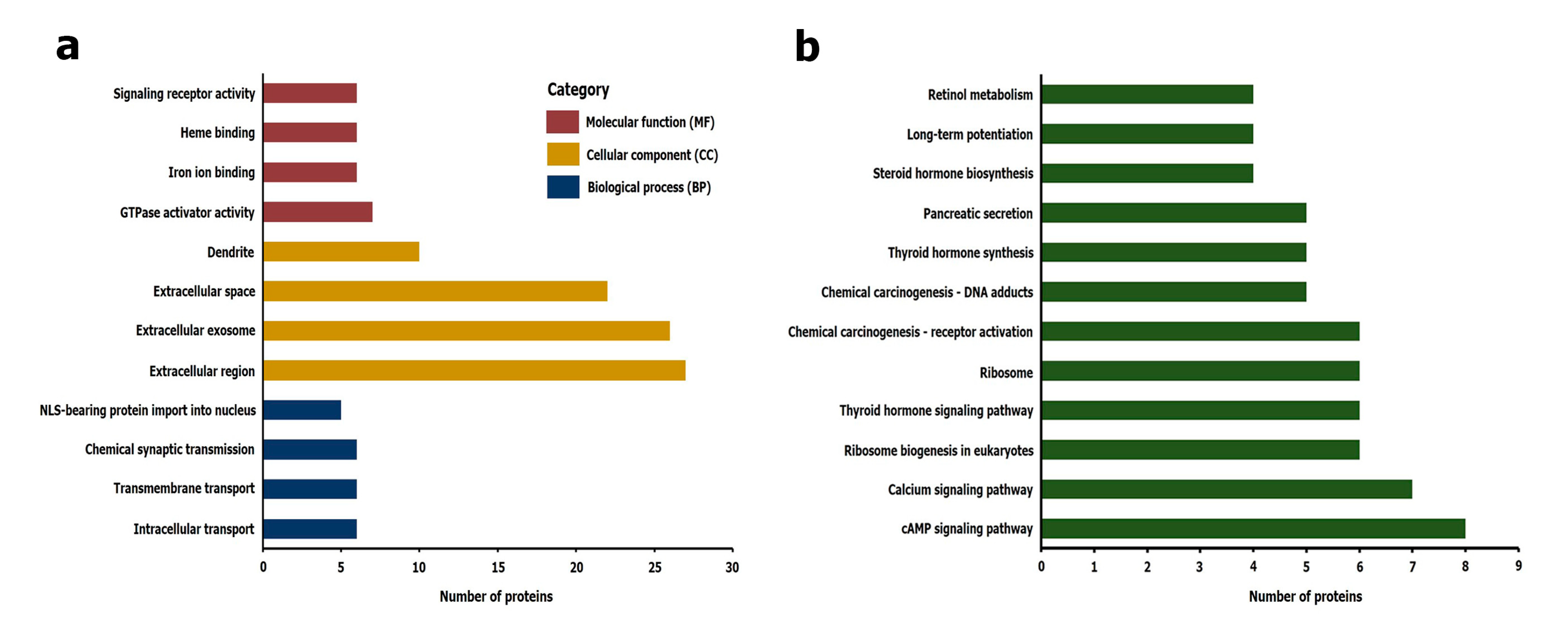
2.4. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathways
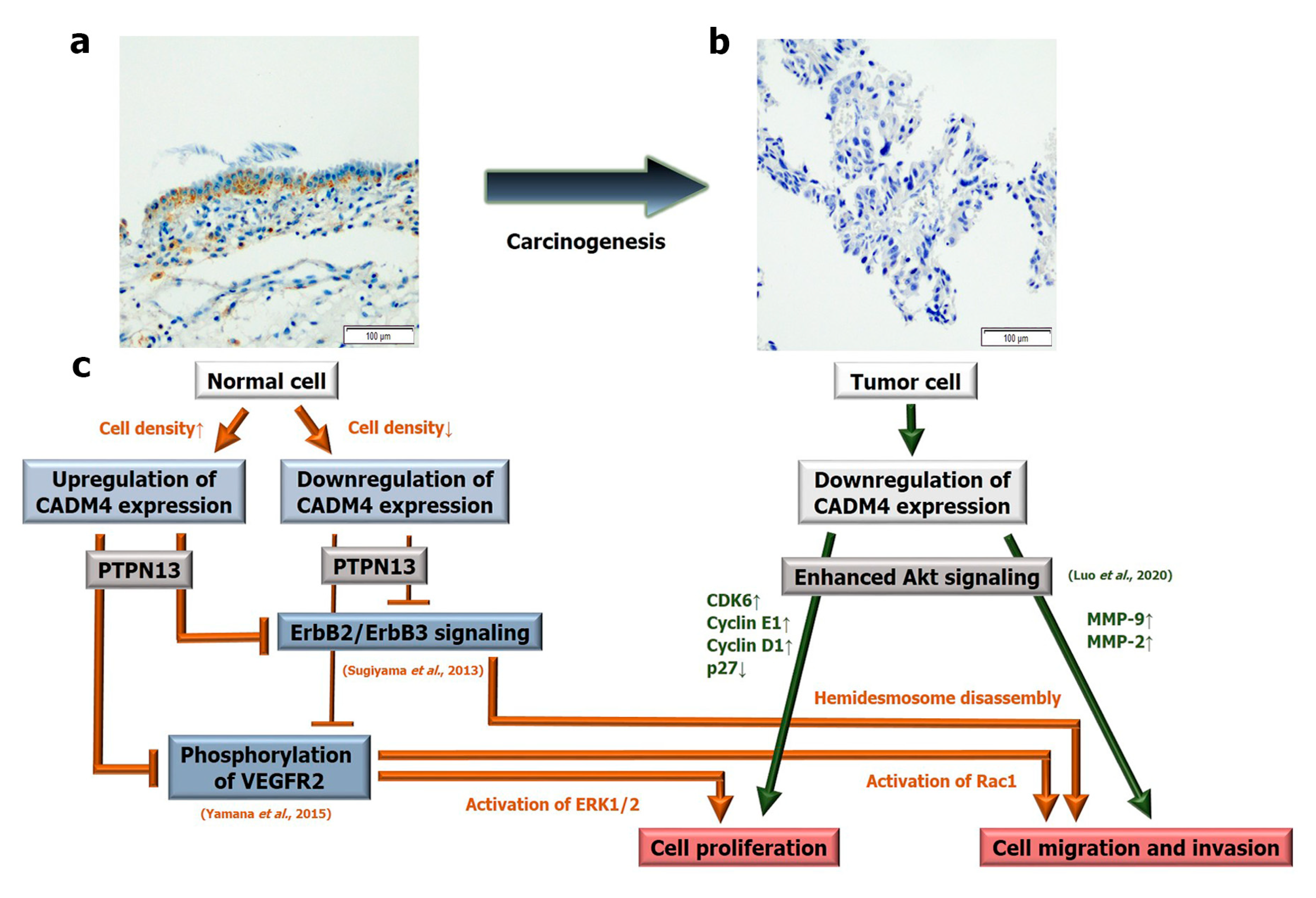
3. Discussion
4. Materials and Methods
4.1. Patients and Specimens
4.2. Collection of Clinicopathological Data
4.3. Tissue Microarray (TMA) Construction and Immunohistochemistry
4.4. Interpretation of IHC
4.5. Assessment of HER2 Status
4.6. Statistical Analyses
4.7. Pathway Enrichment Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Kanthan, R.; Senger, J.L.; Ahmed, S.; Kanthan, S.C. Gallbladder Cancer in the 21st Century. J. Oncol. 2015, 2015, 967472. [Google Scholar] [CrossRef]
- Bridgewater, J.A.; Goodman, K.A.; Kalyan, A.; Mulcahy, M.F. Biliary Tract Cancer: Epidemiology, Radiotherapy, and Molecular Profiling. Am. Soc. Clin. Oncol. Educ. Book. 2016, 35, e194–e203. [Google Scholar] [CrossRef]
- Zhu, A.X.; Hong, T.S.; Hezel, A.F.; Kooby, D.A. Current management of gallbladder carcinoma. Oncologist 2010, 15, 168–181. [Google Scholar] [CrossRef]
- Bizama, C.; Garcia, P.; Espinoza, J.A.; Weber, H.; Leal, P.; Nervi, B.; Roa, J.C. Targeting specific molecular pathways holds promise for advanced gallbladder cancer therapy. Cancer Treat. Rev. 2015, 41, 222–234. [Google Scholar] [CrossRef]
- Montalvo-Jave, E.E.; Rahnemai-Azar, A.A.; Papaconstantinou, D.; Deloiza, M.E.; Tsilimigras, D.I.; Moris, D.; Mendoza-Barrera, G.E.; Weber, S.M.; Pawlik, T.M. Molecular pathways and potential biomarkers in gallbladder cancer: A comprehensive review. Surg. Oncol. 2019, 31, 83–89. [Google Scholar] [CrossRef]
- Fukuhara, H.; Kuramochi, M.; Nobukuni, T.; Fukami, T.; Saino, M.; Maruyama, T.; Nomura, S.; Sekiya, T.; Murakami, Y. Isolation of the TSLL1 and TSLL2 genes, members of the tumor suppressor TSLC1 gene family encoding transmembrane proteins. Oncogene 2001, 20, 5401–5407. [Google Scholar] [CrossRef]
- Williams, Y.N.; Masuda, M.; Sakurai-Yageta, M.; Maruyama, T.; Shibuya, M.; Murakami, Y. Cell adhesion and prostate tumor-suppressor activity of TSLL2/IGSF4C, an immunoglobulin superfamily molecule homologous to TSLC1/IGSF4. Oncogene 2006, 25, 1446–1453. [Google Scholar] [CrossRef]
- Nagata, M.; Sakurai-Yageta, M.; Yamada, D.; Goto, A.; Ito, A.; Fukuhara, H.; Kume, H.; Morikawa, T.; Fukayama, M.; Homma, Y.; et al. Aberrations of a cell adhesion molecule CADM4 in renal clear cell carcinoma. Int. J. Cancer 2012, 130, 1329–1337. [Google Scholar] [CrossRef]
- Luo, F.; Zhao, Y.; Liu, J. Cell adhesion molecule 4 suppresses cell growth and metastasis by inhibiting the Akt signaling pathway in non-small cell lung cancer. Int. J. Biochem. Cell. Biol. 2020, 123, 105750. [Google Scholar] [CrossRef]
- Kim, K.J.; Kim, J.Y.; Hong, S.M.; Gu, M.J. Loss of CADM4 expression is associated with poor prognosis in small intestinal adenocarcinomas. APMIS 2017, 125, 437–443. [Google Scholar] [CrossRef]
- Suresh, S.; Durakoglugil, D.; Zhou, X.; Zhu, B.; Comerford, S.A.; Xing, C.; Xie, X.J.; York, B.; O'Donnell, K.A. SRC-2-mediated coactivation of anti-tumorigenic target genes suppresses MYC-induced liver cancer. PLoS Genet. 2017, 13, e1006650. [Google Scholar] [CrossRef]
- Raveh, S.; Gavert, N.; Spiegel, I.; Ben-Ze'ev, A. The cell adhesion nectin-like molecules (Necl) 1 and 4 suppress the growth and tumorigenic ability of colon cancer cells. J. Cell. Biochem. 2009, 108, 326–336. [Google Scholar] [CrossRef]
- Saito, M.; Goto, A.; Abe, N.; Saito, K.; Maeda, D.; Ohtake, T.; Murakami, Y.; Takenoshita, S. Decreased expression of CADM1 and CADM4 are associated with advanced stage breast cancer. Oncol. Lett. 2018, 15, 2401–2406. [Google Scholar] [CrossRef]
- Yamana, S.; Tokiyama, A.; Mizutani, K.; Hirata, K.; Takai, Y.; Rikitake, Y. The Cell Adhesion Molecule Necl-4/CADM4 Serves as a Novel Regulator for Contact Inhibition of Cell Movement and Proliferation. PLoS One 2015, 10, e0124259. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, H.; Mizutani, K.; Kurita, S.; Okimoto, N.; Shimono, Y.; Takai, Y. Interaction of Necl-4/CADM4 with ErbB3 and integrin alpha6 beta4 and inhibition of ErbB2/ErbB3 signaling and hemidesmosome disassembly. Genes. Cells 2013, 18, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Roa, I.; de Toro, G.; Schalper, K.; de Aretxabala, X.; Churi, C.; Javle, M. Overexpression of the HER2/neu Gene: A New Therapeutic Possibility for Patients with Advanced Gallbladder Cancer. Gastrointest. Cancer Res. 2014, 7, 42–48. [Google Scholar] [PubMed]
- Hryciuk, B.; Peksa, R.; Bienkowski, M.; Szymanowski, B.; Radecka, B.; Winnik, K.; Zok, J.; Cichowska, N.; Iliszko, M.; Duchnowska, R. Expression of Female Sex Hormone Receptors, Connective Tissue Growth Factor and HER2 in Gallbladder Cancer. Sci. Rep. 2020, 10, 1871. [Google Scholar] [CrossRef]
- Sung, Y.N.; Kim, S.J.; Jun, S.Y.; Yoo, C.; Kim, K.P.; Lee, J.H.; Hwang, D.W.; Hwang, S.; Lee, S.S.; Hong, S.M. Expression of HER2 and Mismatch Repair Proteins in Surgically Resected Gallbladder Adenocarcinoma. Front. Oncol. 2021, 11, 658564. [Google Scholar] [CrossRef]
- Bang, S.; Jee, S.; Son, H.; Cha, H.; Sim, J.; Kim, Y.; Park, H.; Myung, J.; Shin, S.J.; Kim, H.; et al. Decreased Expression of Cell Adhesion Molecule 4 in Gastric Adenocarcinoma and Its Prognostic Implications. Diagnostics 2022, 12. [Google Scholar] [CrossRef]
- Lee, S.E.; Kim, K.S.; Kim, W.B.; Kim, I.G.; Nah, Y.W.; Ryu, D.H.; Park, J.S.; Yoon, M.H.; Cho, J.Y.; Hong, T.H.; et al. Practical guidelines for the surgical treatment of gallbladder cancer. J. Korean Med. Sci. 2014, 29, 1333–1340. [Google Scholar] [CrossRef] [PubMed]
- Ilyas, M.; Grabsch, H.; Ellis, I.O.; Womack, C.; Brown, R.; Berney, D.; Fennell, D.; Salto-Tellez, M.; Jenkins, M.; Landberg, G.; et al. Guidelines and considerations for conducting experiments using tissue microarrays. Histopathology 2013, 62, 827–839. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Kir, J.; Liu, D.; Bryant, D.; Guo, Y.; Stephens, R.; Baseler, M.W.; Lane, H.C.; et al. DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007, 35, W169–W175. [Google Scholar] [CrossRef] [PubMed]
| Characteristics | n (%) |
|---|---|
| Age, median (range, years) | 65 (28–90) |
| Sex | |
| Female | 51 (51.0%) |
| Male | 49 (49.0%) |
| Tumor size, mean (range, cm) * | 4.5 (0.2–10.0) |
| Location | |
| Neck | 12 (12.0%) |
| Body | 48 (48.0%) |
| Fundus | 22 (22.0%) |
| Neck to body | 5 (5.0%) |
| Body to fundus | 5 (5.0%) |
| Neck to fundus | 4 (4.0%) |
| Whole gallbladder | 4 (4.0%) |
| Histological type | |
| Adenocarcinoma | 89 (89.0%) |
| Adenocarcinoma with neuroendocrine differentiation | 2 (2.0%) |
| Adenocarcinoma with sarcomatoid differentiation | 3 (3.0%) |
| Mixed adenocarcinoma | 2 (2.0%) |
| Adenosquamous carcinoma | 4 (4.0%) |
| Histological grade | |
| G1 (well differentiated) | 22 (22.0%) |
| G2 (moderately differentiated) | 57 (57.0%) |
| G3 (poorly differentiated) | 21 (21.0%) |
| Lymphovascular invasion | |
| Present | 50 (50.0%) |
| Not identified | 50 (50.0%) |
| Perineural invasion | |
| Present | 32 (32.0%) |
| Not identified | 68 (68.0%) |
| T category | |
| pT1a | 8 (8.0%) |
| pT1b | 9 (9.0%) |
| pT2 | 49 (49.0%) |
| pT3 | 31 (31.0%) |
| pT4 | 3 (3.0 %) |
| N category | |
| pNx | 15 (15.0%) |
| pN0 | 49 (49.0%) |
| pN1 | 33 (33.0%) |
| pN2 | 3 (3.0%) |
| TNM stage (AJCC 8th edition) | |
| I | 17 (17.0%) |
| II | 34 (34.0%) |
| IIIA | 13 (13.0%) |
| IIIB | 31 (31.0%) |
| IVA | 2 (2.0%) |
| IVB | 3 (3.0%) |
| HER2 status | |
| No amplification | 91 (91.0%) |
| Amplification | 9 (9.0%) |
| Variables | CADM4 Expression | p-Value | |
|---|---|---|---|
| Low Expression (%) (n = 28) | High Expression (%) (n = 72) | ||
| Age | 0.914 | ||
| <65 years | 12 (28.6%) | 30 (71.4%) | |
| ≥65 years | 16 (27.6%) | 42 (72.4%) | |
| Sex | 0.748 | ||
| Female | 15 (29.4%) | 36 (70.6%) | |
| Male | 13 (26.5%) | 36 (73.5%) | |
| Histological grade | 0.948 | ||
| Grade 1 or 2 | 22 (27.8%) | 57 (72.2%) | |
| Grade 3 | 6 (28.6%) | 15 (71.4%) | |
| Lymphovascular invasion | 1.000 | ||
| Not identified | 14 (28.0%) | 36 (72.0%) | |
| Present | 14 (28.0%) | 36 (72.0%) | |
| Perineural invasion | 0.620 | ||
| Not identified | 18 (26.5%) | 50 (73.5%) | |
| Present | 10 (31.3%) | 22 (68.8%) | |
| T category | 0.010 | ||
| pT1 or pT2 | 13 (19.7%) | 53 (80.3%) | |
| pT3 or pT4 | 15 (44.1%) | 19 (55.9%) | |
| N category | 0.373 | ||
| pNx or pN0 | 16 (25.0%) | 48 (75.0%) | |
| pN1 or pN2 | 12 (33.3%) | 24 (66.7%) | |
| TNM stage (AJCC 8th edition) | 0.019 | ||
| I or II | 9 (17.6%) | 42 (82.4%) | |
| III or IV | 19 (38.8%) | 30 (61.2%) | |
| HER2 status | 0.262 * | ||
| No amplification | 24 (26.4%) | 67 (73.6%) | |
| Amplification | 4 (44.4%) | 5 (55.6%) | |
| OS | ||||||
|---|---|---|---|---|---|---|
| Variables | Univariate Analysis | Multivariate Analysis | ||||
| HR | 95% CI | p-Value | HR | 95% CI | p-Value | |
| CADM4 expression (high vs. low) | 2.654 | 1.440–4.892 | 0.002 | 2.199 | 1.182–4.091 | 0.013 |
| Age group (<65 years vs. ≥65 years) | 1.505 | 0.805–2.816 | 0.201 | |||
| Histological grade (grade 1 or 2 vs. grade 3) | 1.604 | 0.820–3.134 | 0.167 | |||
| T category (pT1 or pT2 vs. pT3 or pT4) | 2.889 | 1.570–5.313 | 0.001 | |||
| N category (pNx or pN0 vs. pN1 or pN2) | 2.628 | 1.427–4.838 | 0.002 | |||
| TNM stage (AJCC 8th edition) (I or II vs. III or IV) | 3.475 | 1.775–6.803 | < 0.001 | 2.267 | 1.040–4.944 | 0.040 |
| Lymphovascular invasion (not identified vs. present) | 2.507 | 1.337–4.700 | 0.004 | 1.424 | 0.682–2.976 | 0.347 |
| Perineural invasion (not identified vs. present) | 2.493 | 1.348–4.613 | 0.004 | 1.419 | 0.688–2.925 | 0.343 |
| HER2 status (no amplification or amplification) | 0.996 | 0.391–2.539 | 0.994 | |||
| RFS | ||||||
| Variables | Univariate Analysis | Multivariate Analysis | ||||
| HR | 95% CI | p-Value | HR | 95% CI | p-Value | |
| CADM4 expression (high vs. low) | 2.181 | 1.114–4.271 | 0.023 | 1.923 | 0.978–3.782 | 0.058 |
| Age group (<65 years vs. ≥65 years) | 0.691 | 0.359–1.331 | 0.269 | |||
| Histological grade (grade 1 or 2 vs. grade 3) | 1.451 | 0.682–3.088 | 0.334 | |||
| T category (pT1 or pT2 vs. pT3 or pT4) | 5.401 | 2.690–10.845 | < 0.001 | |||
| N category (pNx or pN0 vs. pN1 or pN2) | 3.141 | 1.613–6.116 | 0.001 | |||
| TNM stage (AJCC 8th edition) (I or II vs. III or IV) | 9.230 | 3.577–23.813 | < 0.001 | 5.726 | 2.077–15.788 | 0.001 |
| Lymphovascular invasion (not identified vs. present) | 4.945 | 2.240–10.919 | < 0.001 | 2.398 | 0.977–5.882 | 0.056 |
| Perineural invasion (not identified vs. present) | 3.059 | 1.580–5.923 | 0.001 | 1.234 | 0.589–2.586 | 0.577 |
| HER2 status (no amplification or amplification) | 0.466 | 0.112–1.942 | 0.294 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bang, S.; Jee, S.; Son, H.; Cha, H.; Song, K.; Park, H.; Myung, J.; Kim, H.; Paik, S. Clinicopathological Significance of Cell Adhesion Molecule 4 Expression in Gallbladder Cancer and Its Prognostic Role. Int. J. Mol. Sci. 2023, 24, 6898. https://doi.org/10.3390/ijms24086898
Bang S, Jee S, Son H, Cha H, Song K, Park H, Myung J, Kim H, Paik S. Clinicopathological Significance of Cell Adhesion Molecule 4 Expression in Gallbladder Cancer and Its Prognostic Role. International Journal of Molecular Sciences. 2023; 24(8):6898. https://doi.org/10.3390/ijms24086898
Chicago/Turabian StyleBang, Seongsik, Seungyun Jee, Hwangkyu Son, Hyebin Cha, Kihyuk Song, Hosub Park, Jaekyung Myung, Hyunsung Kim, and Seungsam Paik. 2023. "Clinicopathological Significance of Cell Adhesion Molecule 4 Expression in Gallbladder Cancer and Its Prognostic Role" International Journal of Molecular Sciences 24, no. 8: 6898. https://doi.org/10.3390/ijms24086898
APA StyleBang, S., Jee, S., Son, H., Cha, H., Song, K., Park, H., Myung, J., Kim, H., & Paik, S. (2023). Clinicopathological Significance of Cell Adhesion Molecule 4 Expression in Gallbladder Cancer and Its Prognostic Role. International Journal of Molecular Sciences, 24(8), 6898. https://doi.org/10.3390/ijms24086898








