The Synergistic Antimicrobial Effect and Mechanism of Nisin and Oxacillin against Methicillin-Resistant Staphylococcus aureus
Abstract
1. Introduction
2. Results
2.1. The Synergy Antimicrobial Effect of OX + NIS against MRSA
2.1.1. Antimicrobial Effect against MRSA In Vitro
2.1.2. MIC and FICI of OX + NIS against MRSA
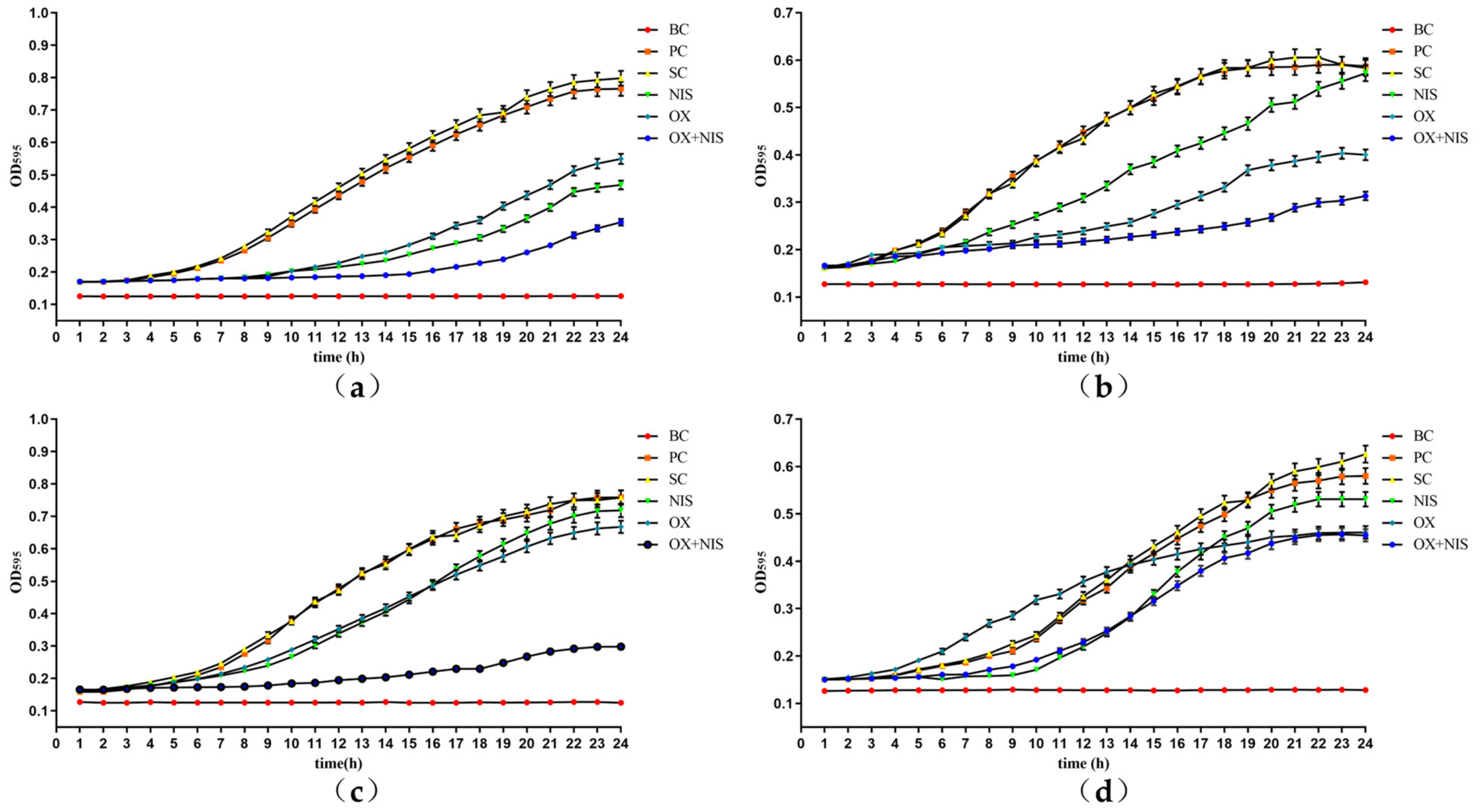
2.1.3. Effect on Growth Curve of MRSA Treated with OX + NIS
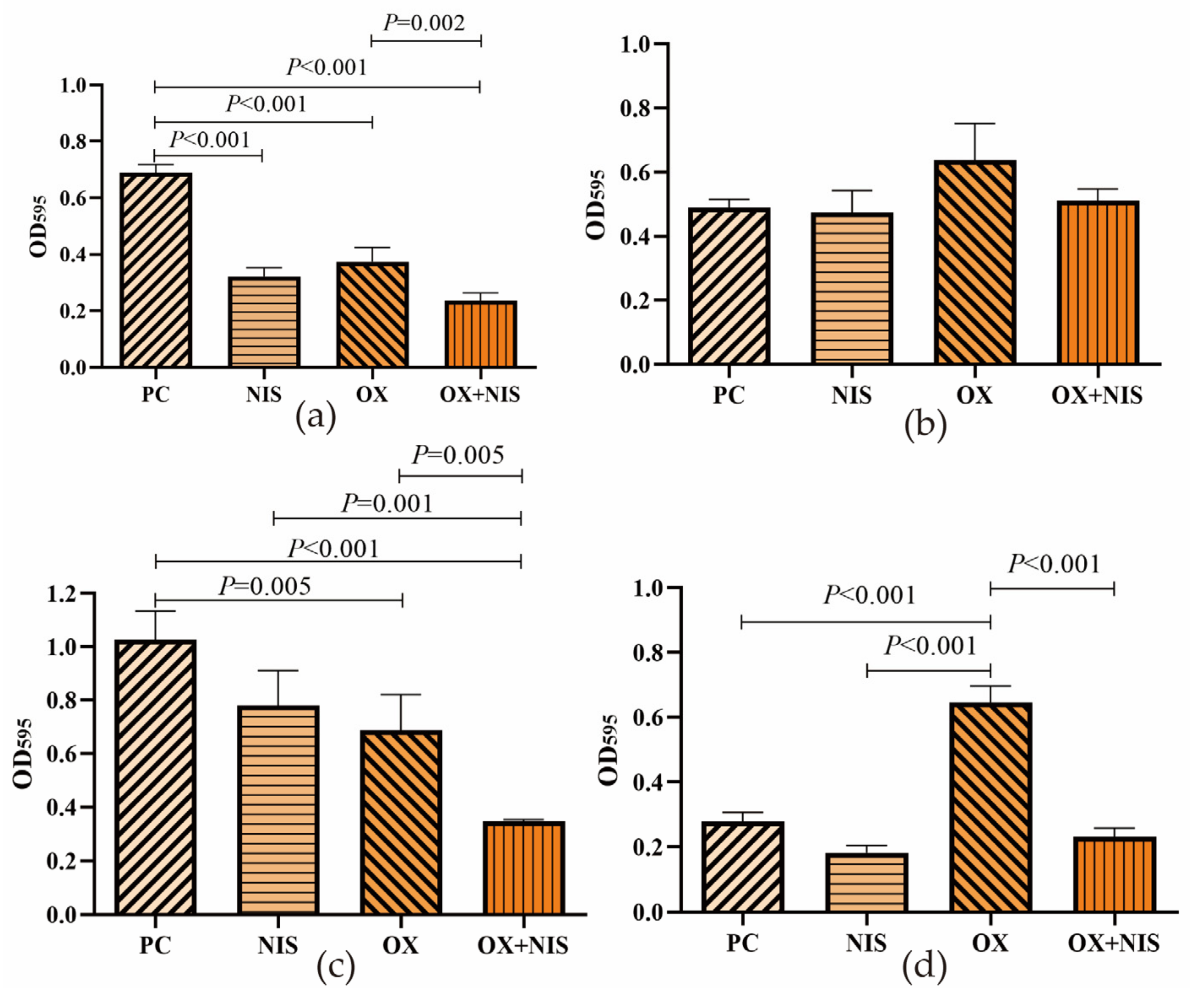
2.1.4. Effect on Biofilm Producing of MRSA Treated with OX + NIS
2.2. The Synergy Antimicrobial Mechanism of OX + NIS against MRSA
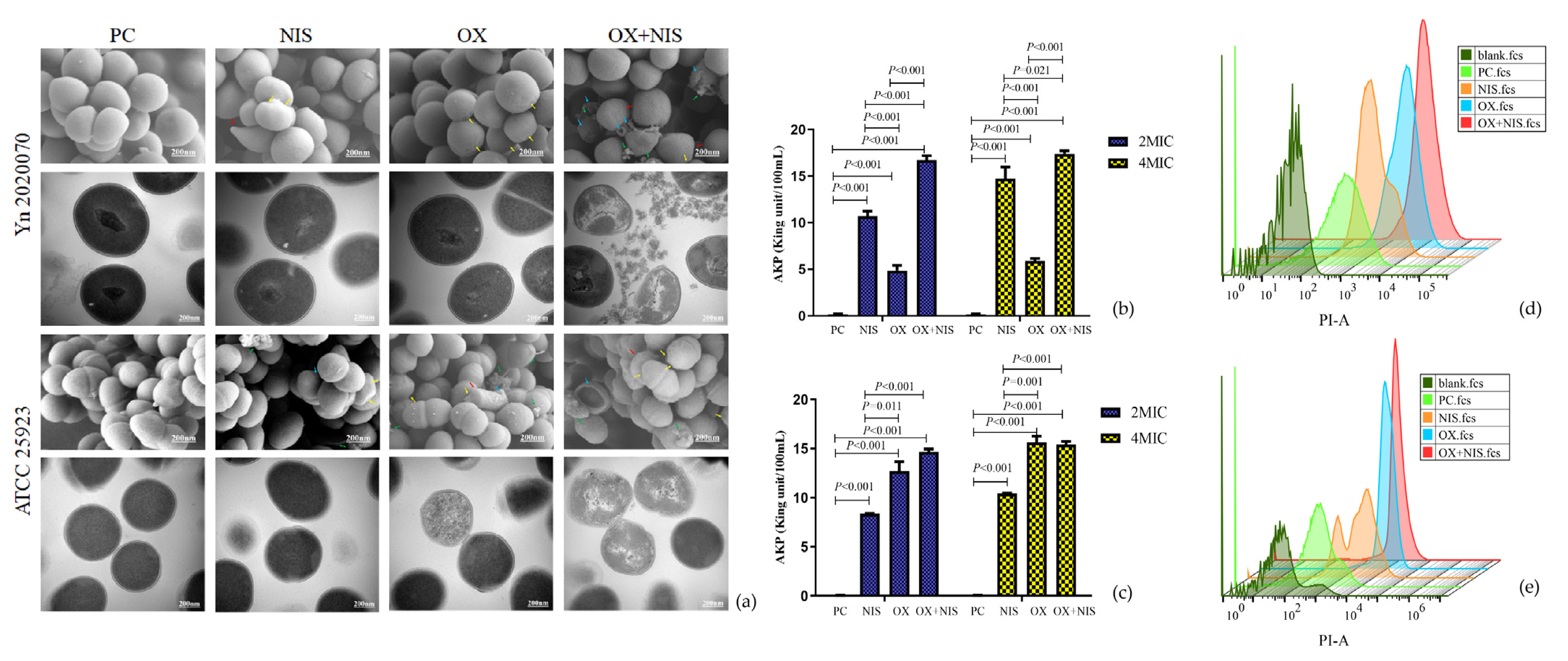
2.2.1. Morphological Changes of MRSA Cell
2.2.2. Integrity Changes of MRSA Cell Wall and Cell Membrane
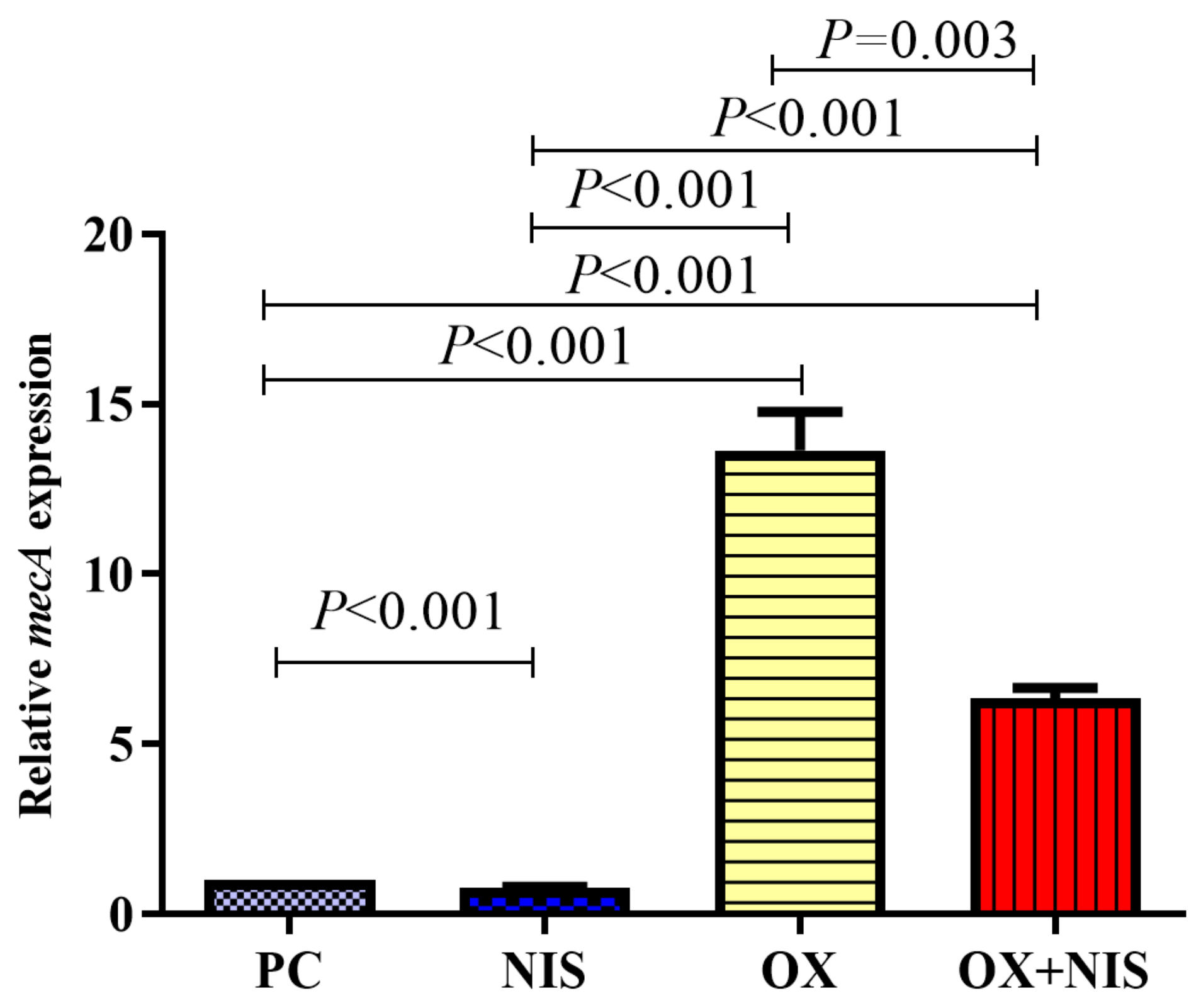
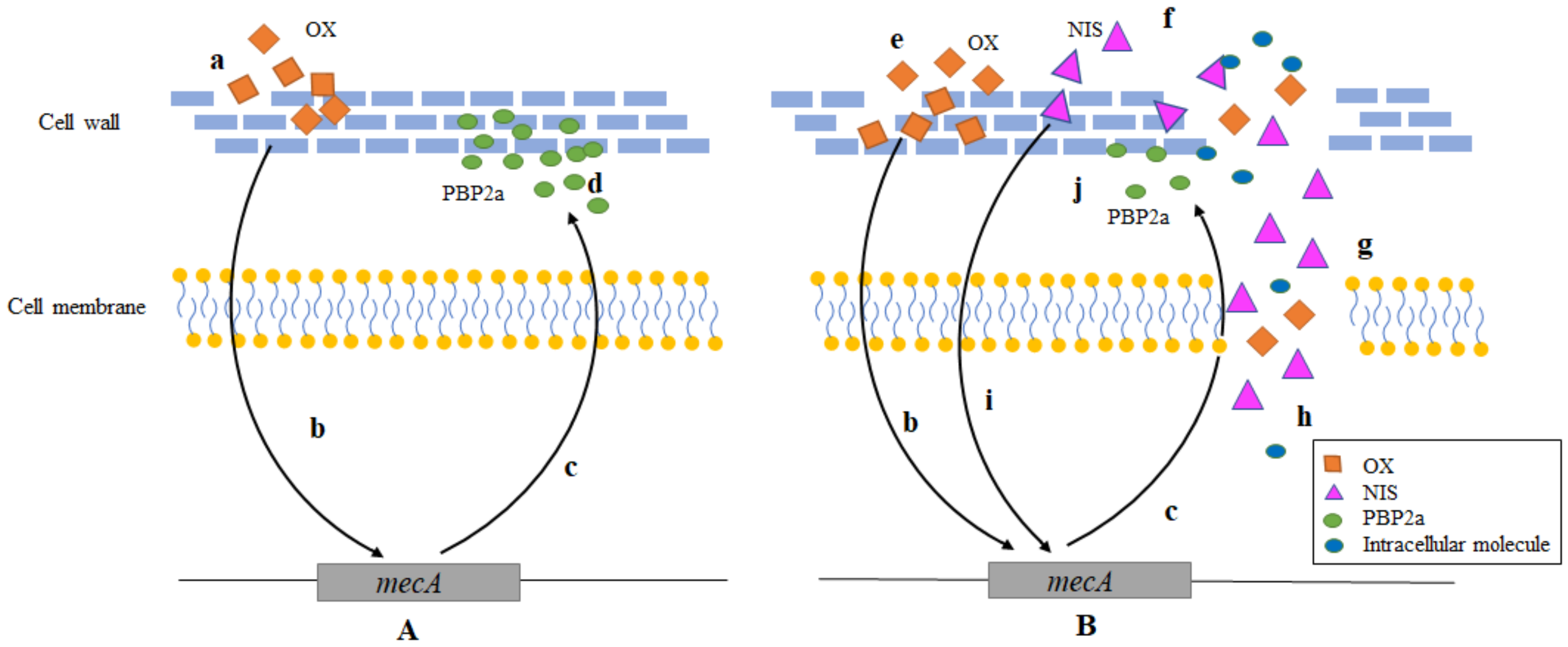
2.2.3. Transcription Change of mecA in MRSA
2.3. Efficacy of OX + NIS in the Treatment of MRSA-Induced Wound Infections in Mice Skin
2.3.1. Effect of OX + NIS Treatment on Wound Closure
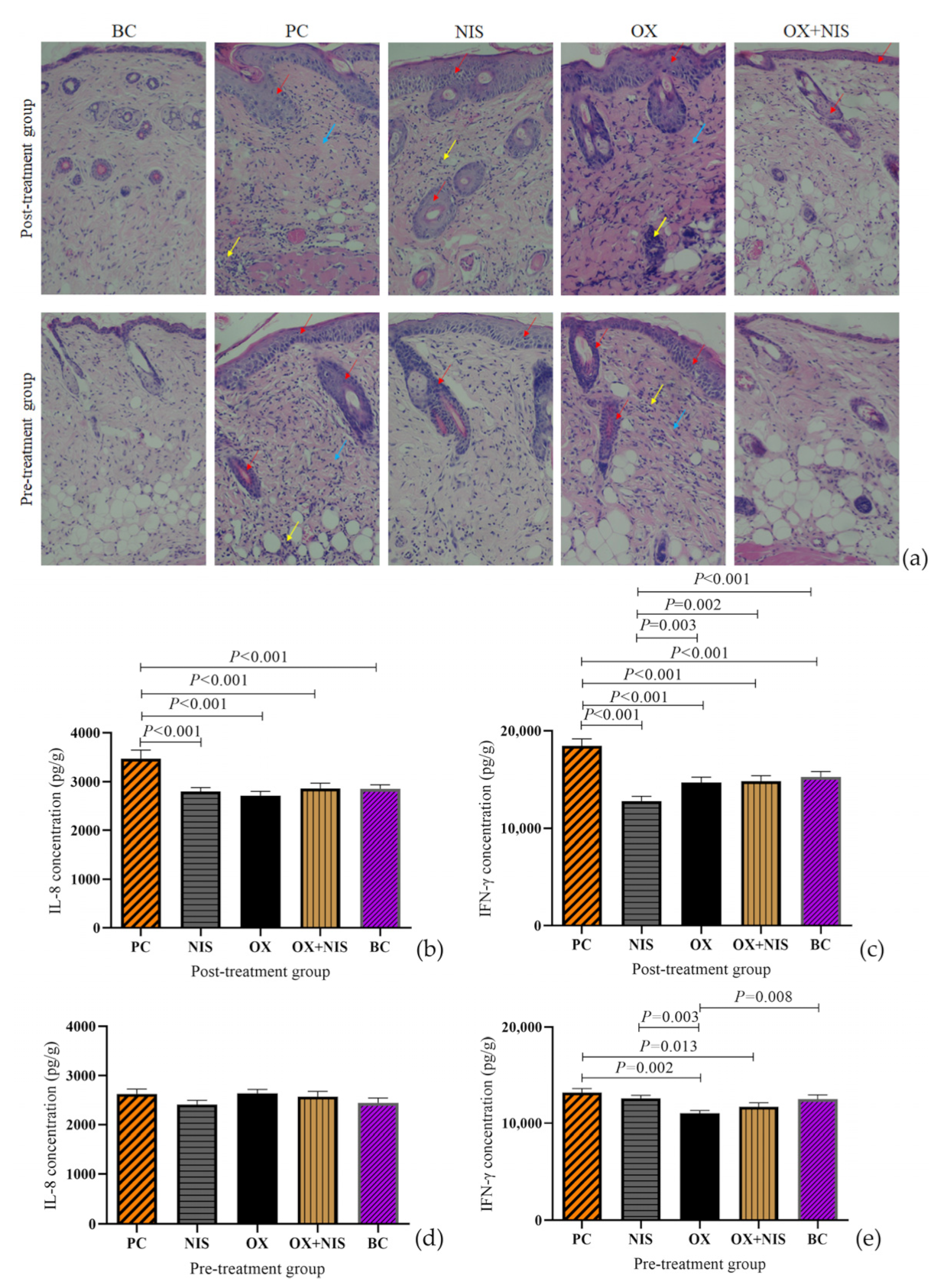
2.3.2. Histopathological Change Analysis of OX + NIS Treatment on Wound Closure
2.3.3. Inflammation Cytokines Change Analysis of OX + NIS Treatment on Wound Closure
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Antimicrobial Agents, and Culture Conditions
4.2. Minimal Inhibitory Concentration (MIC) Assay
4.3. Synergy Test of OX + NIS against MRSA
4.4. Growth of MRSA Incubated with OX + NIS
4.5. Biofilm Formation Assay
4.6. Microscopic Observation of MRSA Cells Morphological Changes Treated with OX + NIS
4.7. Extracellular Alkaline Phosphatase Activity of MRSA Strain Treated with OX + NIS
4.8. Membrane Permeability of MRSA Strain Treated with OX + NIS
4.9. Quantification of mecA Transcription of MRSA Strain Treated with OX + NIS
4.10. Murine Skin Infection Model with MRSA
4.10.1. Animal Used
4.10.2. Skin Wound Generation and Infection with MRSA
4.10.3. Treatment and Evaluation of MRSA Wound Infections
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed]
- Jabila Mary, T.R.; Kannan, R.R.; Muthamil Iniyan, A.; Carlton Ranjith, W.A.; Nandhagopal, S.; Vishwakarma, V.; Vincent, S.G.P. β-lactamase inhibitory potential of kalafungin from marine Streptomyces in Staphylococcus aureus infected zebrafish. Microbiol. Res. 2021, 244, 126666. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.; Dai, M.; Wang, Y.; Huang, L.; Yuan, Z. Key genetic elements and regulation systems in methicillin-resistant Staphylococcus aureus. Future Microbiol. 2012, 7, 1315–1329. [Google Scholar] [CrossRef] [PubMed]
- Mahey, N.; Tambat, R.; Chandal, N.; Verma, D.K.; Thakur, K.G.; Nandanwar, H. Repurposing approved drugs as fluoroquinolone potentiators to overcome efflux pump resistance in Staphylococcus aureus. Microbiol. Spectr. 2021, 9, e0095121. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Peng, H.; Rao, X. Molecular events for promotion of vancomycin resistance in vancomycin intermediate. Front. Microbiol. 2016, 7, 1601. [Google Scholar] [CrossRef]
- Arias, C.A.; Murray, B.E. The rise of the Enterococcus: Beyond vancomycin resistance. Nat. Rev. Microbiol. 2012, 10, 266–278. [Google Scholar] [CrossRef]
- Shalaby, M.A.W.; Dokla, E.M.E.; Serya, R.A.T.; Abouzid, K.A.M. Penicillin binding protein 2a: An overview and a medicinal chemistry perspective. Eur. J. Med. Chem. 2020, 199, 112312. [Google Scholar] [CrossRef]
- Hall, C.W.; Mah, T.F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef]
- Cheung, G.Y.C.; Bae, J.S.; Otto, M. Pathogenicity and virulence of Staphylococcus aureus. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef]
- Thurlow, L.R.; Hanke, M.L.; Fritz, T.; Angle, A.; Aldrich, A.; Williams, S.H.; Engebretsen, I.L.; Bayles, K.W.; Horswill, A.R.; Kielian, T. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J. Immunol. 2011, 186, 6585–6596. [Google Scholar] [CrossRef]
- Mesrati, I.; Saidani, M.; Jemili, M.; Ferjeni, S.; Slim, A.; Boubaker, I.B.B. Virulence determinants, biofilm production and antimicrobial susceptibility in Staphylococcus aureus causing device-associated infections in a Tunisian hospital. Int. J. Antimicrob. Agents 2018, 52, 922–929. [Google Scholar] [CrossRef] [PubMed]
- Pokhrel, R.; Bhattarai, N.; Baral, P.; Gerstman, B.S.; Park, J.H.; Handfield, M.; Chapagain, P.P. Lipid II binding and transmembrane properties of various antimicrobial lanthipeptides. J. Chem. Theory Comput. 2022, 18, 516–525. [Google Scholar] [CrossRef] [PubMed]
- Jahangiri, A.; Neshani, A.; Mirhosseini, S.A.; Ghazvini, K.; Zare, H.; Sedighian, H. Synergistic effect of two antimicrobial peptides, Nisin and P10 with conventional antibiotics against extensively drug-resistant Acinetobacter baumannii and colistin-resistant Pseudomonas aeruginosa isolates. Microb. Pathog. 2021, 150, 104700. [Google Scholar] [CrossRef] [PubMed]
- Dosler, S.; Gerceker, A.A. In vitro activities of nisin alone or in combination with vancomycin and ciprofloxacin against methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. Chemotherapy 2011, 57, 511–516. [Google Scholar] [CrossRef]
- Li, J.; Shi, L.; Liu, J.; Wang, J.; Da, R.; Han, B. Inhibitory effects of nisin combined with meropenem on methicillin resistant Staphylococcus aureus. Natl. Prod. Res. Dev. 2022, 34, 003. [Google Scholar] [CrossRef]
- Shin, J.M.; Gwak, J.W.; Kamarajan, P.; Fenno, J.C.; Rickard, A.H.; Kapila, Y.L. Biomedical applications of nisin. J. Appl. Microbiol. 2016, 120, 1449–1465. [Google Scholar] [CrossRef]
- Vishwakarma, A.; Dang, F.; Ferrell, A.; Barton, H.A.; Joy, A. Peptidomimetic polyurethanes inhibit bacterial biofilm formation and disrupt surface established biofilms. J. Am. Chem. Soc. 2021, 143, 9440–9449. [Google Scholar] [CrossRef]
- Lister, J.L.; Horswill, A.R. Staphylococcus aureus biofilms: Recent developments in biofilm dispersal. Front. Cell. Infect. Microbiol. 2014, 4, 178. [Google Scholar] [CrossRef]
- Wu, B.C.; Blimkie, T.M.; Haney, E.F.; Falsafi, R.; Akhoundsadegh, N.; Hancock, R.E.W. Host response of human epidermis to methicillin-resistant Staphylococcus aureus biofilm infection and synthetic antibiofilm peptide treatment. Cells 2022, 11, 3459. [Google Scholar] [CrossRef]
- Arêde, P.; Milheiriço, C.; de Lencastre, H.; Oliveira, D.C. The anti-repressor MecR2 promotes the proteolysis of the mecA repressor and enables optimal expression of β-lactam resistance in MRSA. PLoS Pathog. 2012, 8, e1002816. [Google Scholar] [CrossRef]
- Parsaeimehr, M.; Basti, A.A.; Radmehr, B.; Misaghi, A.; Abbasifar, A.; Karim, G.; Rokni, N.; Motlagh, M.S.; Gandomi, H.; Noori, N.; et al. Effect of Zataria multiflora Boiss. essential oil, nisin, and their combination on the production of enterotoxin C and alpha-hemolysin by Staphylococcus aureus. Foodborne Pathog. Dis. 2010, 7, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Han, J.Z.; Ma, Z.; Gao, P.; Lu, Z.X.; Liu, H.X.; Gao, L.; Lu, W.J.; Ju, X.Y.; Lv, F.X.; Zhao, H.Z.; et al. The antibacterial activity of LI-F type peptide against methicillin-resistant Staphylococcus aureus (MRSA) in vitro and inhibition of infections in murine scalded epidermis. Appl. Microbiol. Biotechnol. 2018, 102, 2301–2311. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, G.J.; Riddle, G.; Elborn, J.S.; Ennis, M.; Skibinski, G. Staphylococcus aureus enterotoxins induce IL-8 secretion by human nasal epithelial cells. Respir. Res. 2006, 7, 115. [Google Scholar] [CrossRef]
- Verma, A.K.; Bauer, C.; Palani, S.; Metzger, D.W.; Sun, K. IFN-γ drives TNF-α hyperproduction and lethal lung inflammation during antibiotic treatment of postinfluenza Staphylococcus aureus pneumonia. J. Immunol. 2021, 207, 1371–1376. [Google Scholar] [CrossRef] [PubMed]
- Kussmann, M.; Obermueller, M.; Karer, M.; Kriz, R.; Chen, R.Y.; Hohl, L.; Schneider, L.; Burgmann, H.; Traby, L.; Vossen, M.G. Synergistic effect of cefazolin plus fosfomycin against Staphylococcus aureus in vitro and in vivo in an experimental Galleria mellonella model. Front. Pharm. 2021, 12, 685807. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Da, R.; Tuo, X.H.; Cheng, Y.; Wei, J.; Jiang, K.C.; Lv, J.; Adediji, O.M.; Han, B. Probiotic and safety properties screening of Enterococcus faecalis from healthy Chinese infants. Probiotics Antimicrob. 2020, 12, 1115–1125. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Lázaro, D.; Alonso-Calleja, C.; Oniciuc, E.A.; Capita, R.; Gallego, D.; González-Machado, C.; Wagner, M.; Barbu, V.; Eiros-Bouza, J.M.; Nicolau, A.I.; et al. Characterization of biofilms formed by foodborne methicillin-resistant Staphylococcus aureus. Front. Microbiol. 2018, 9, 3004. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Shi, L.; Wang, J.; Yuan, J.; Liu, J.; Liu, L.J.; Da, R.; Cheng, Y.; Han, B. The probiotic potential analysis and safety evaluation of Enterococcus durans A8-1 isolated from healthy Chinese infant. Front. Microbiol. 2022, 12, 799173. [Google Scholar] [CrossRef]
- Perrine-Walker, F.; Le, K. Propidium iodide enabled live imaging of Pasteuria sp.-Pratylenchus zeae infection studies under fluorescence microscopy. Protoplasma 2021, 258, 279–287. [Google Scholar] [CrossRef]
- Wang, S.; Kang, O.H.; Kwon, D.Y. Trans-cinnamaldehyde exhibits synergy with conventional antibiotic against Methicillin-Resistant Staphylococcus aureus. Int. J. Mol. Sci. 2021, 22, 2752. [Google Scholar] [CrossRef]
- Ovchinnikov, K.V.; Kranjec, C.; Thorstensen, T.; Carlsen, H.; Diep, D.B. Successful development of bacteriocins into therapeutic formulation for treatment of MRSA skin infection in a murine model. Antimicrob. Agents. Chemother. 2020, 64, e00829-20. [Google Scholar] [CrossRef] [PubMed]


| Strain | MIC (Alone) | MIC (Combination) | FICI | Inhibitory Effect | ||
|---|---|---|---|---|---|---|
| OX (μg/mL) | NIS (μg/mL) | OX (μg/mL) | NIS (μg/mL) | |||
| Yn2020043 | 32 | 12,800 | 8 | 3200 | 0.500 | synergy |
| Yn2020051 | 64 | 12,800 | 16 | 3200 | 0.500 | synergy |
| Yn2020070 | 16 | 12,800 | 4 | 1600 | 0.375 | synergy |
| ATCC25923 | 8 | 12,800 | 4 | 6400 | 1.000 | addition |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Ma, X.; Li, J.; Shi, L.; Liu, L.; Hou, X.; Jiang, S.; Li, P.; Lv, J.; Han, L.; et al. The Synergistic Antimicrobial Effect and Mechanism of Nisin and Oxacillin against Methicillin-Resistant Staphylococcus aureus. Int. J. Mol. Sci. 2023, 24, 6697. https://doi.org/10.3390/ijms24076697
Wang J, Ma X, Li J, Shi L, Liu L, Hou X, Jiang S, Li P, Lv J, Han L, et al. The Synergistic Antimicrobial Effect and Mechanism of Nisin and Oxacillin against Methicillin-Resistant Staphylococcus aureus. International Journal of Molecular Sciences. 2023; 24(7):6697. https://doi.org/10.3390/ijms24076697
Chicago/Turabian StyleWang, Jun, Xinxin Ma, Jing Li, Lu Shi, Lijuan Liu, Xinyao Hou, Sijin Jiang, Pu Li, Jia Lv, Lei Han, and et al. 2023. "The Synergistic Antimicrobial Effect and Mechanism of Nisin and Oxacillin against Methicillin-Resistant Staphylococcus aureus" International Journal of Molecular Sciences 24, no. 7: 6697. https://doi.org/10.3390/ijms24076697
APA StyleWang, J., Ma, X., Li, J., Shi, L., Liu, L., Hou, X., Jiang, S., Li, P., Lv, J., Han, L., Cheng, Y., & Han, B. (2023). The Synergistic Antimicrobial Effect and Mechanism of Nisin and Oxacillin against Methicillin-Resistant Staphylococcus aureus. International Journal of Molecular Sciences, 24(7), 6697. https://doi.org/10.3390/ijms24076697






