Syndecan-3 Coregulates Milk Fat Metabolism and Inflammatory Reactions in Bovine Mammary Epithelial Cells through AMPK/SIRT1 Signaling Pathway
Abstract
1. Introduction
2. Results
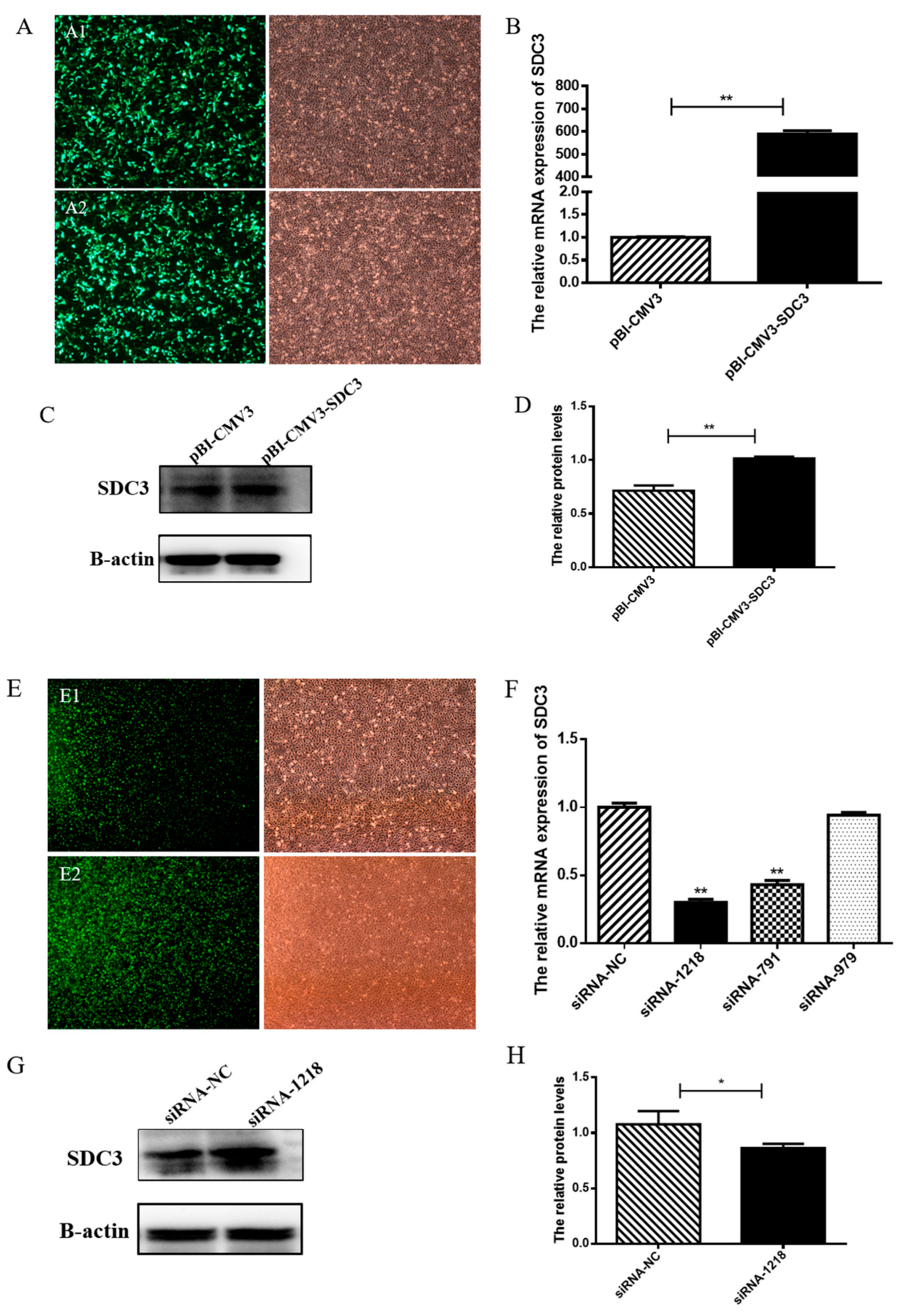
2.1. Identification of Transfection Efficiency of Plasmid and siRNAs in BMECs
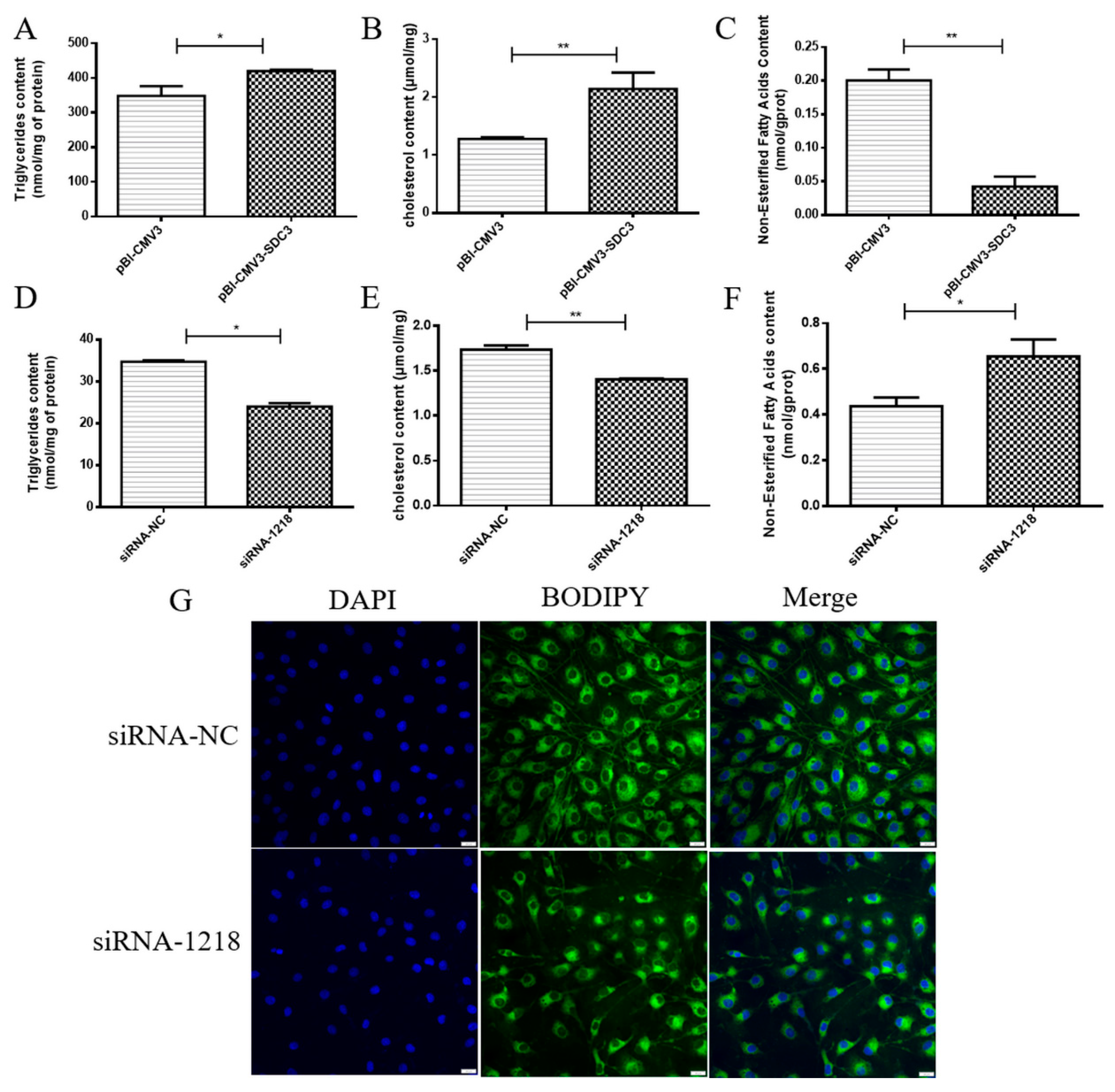
2.2. SDC3 Promotes the Synthesis of Milk Fat in BMECs
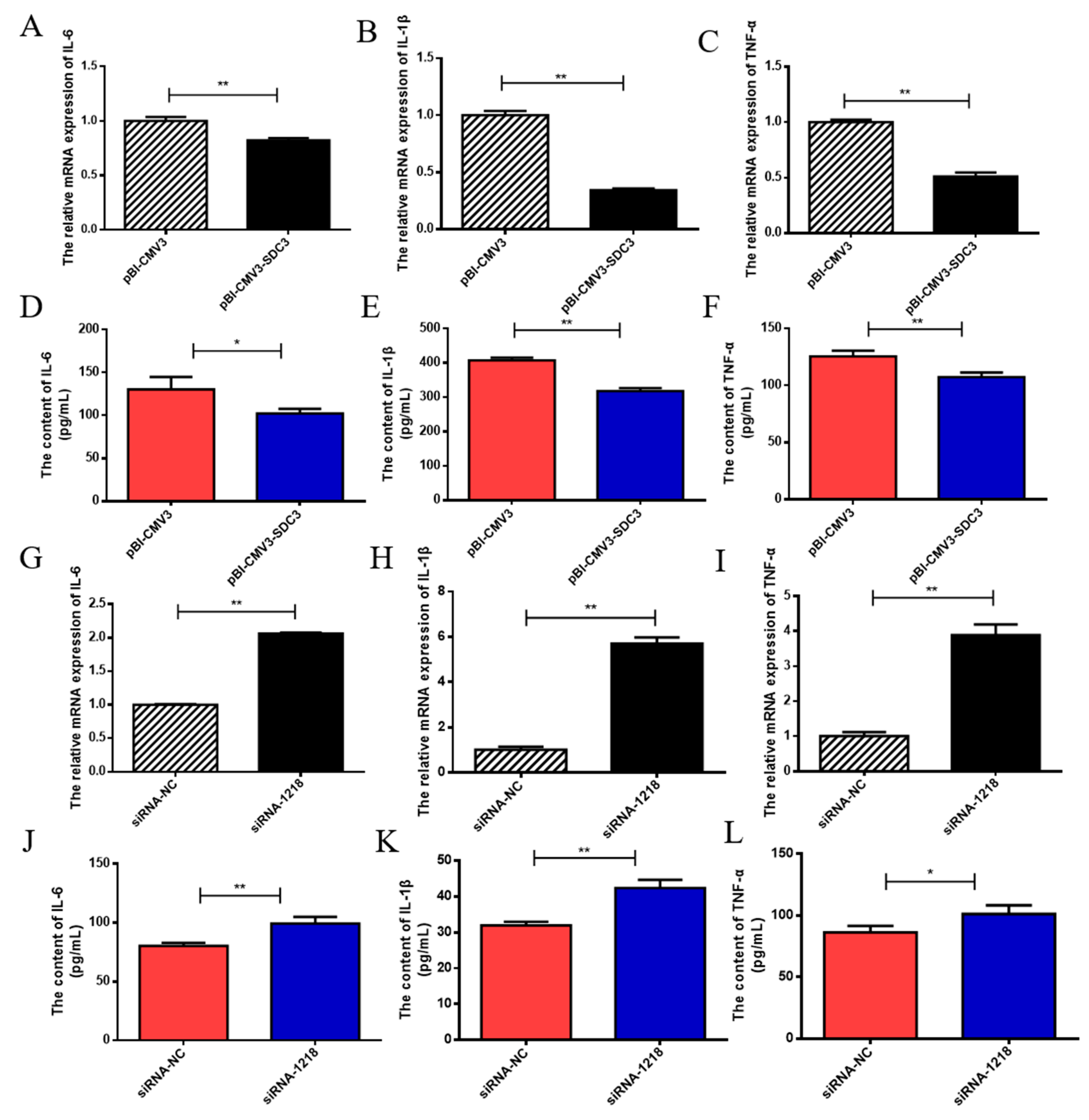
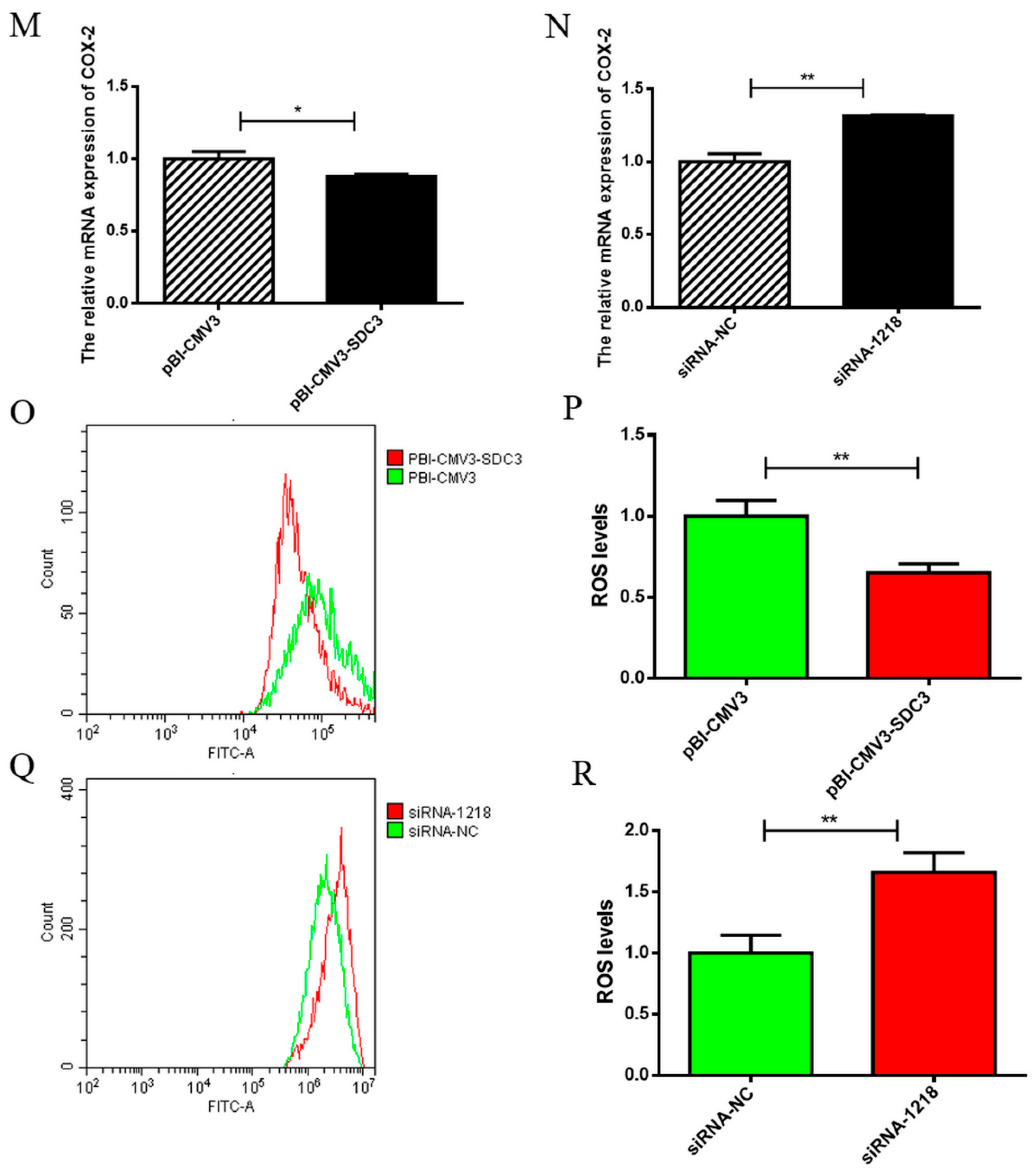
2.3. SDC3 Inhibits the Inflammatory Response of BMECs by Inhibiting the Expression of Inflammatory Factors and the Production of ROS
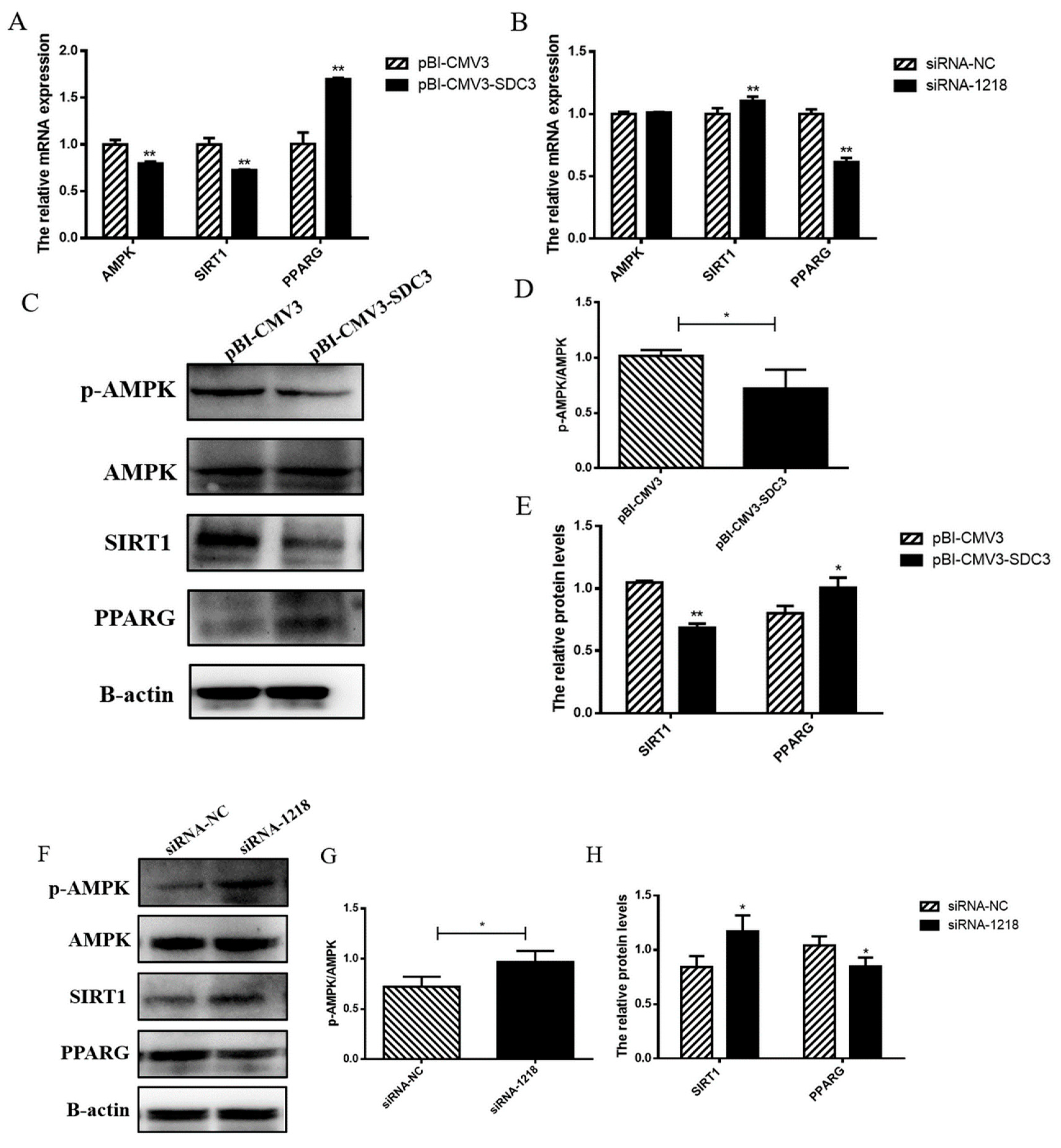
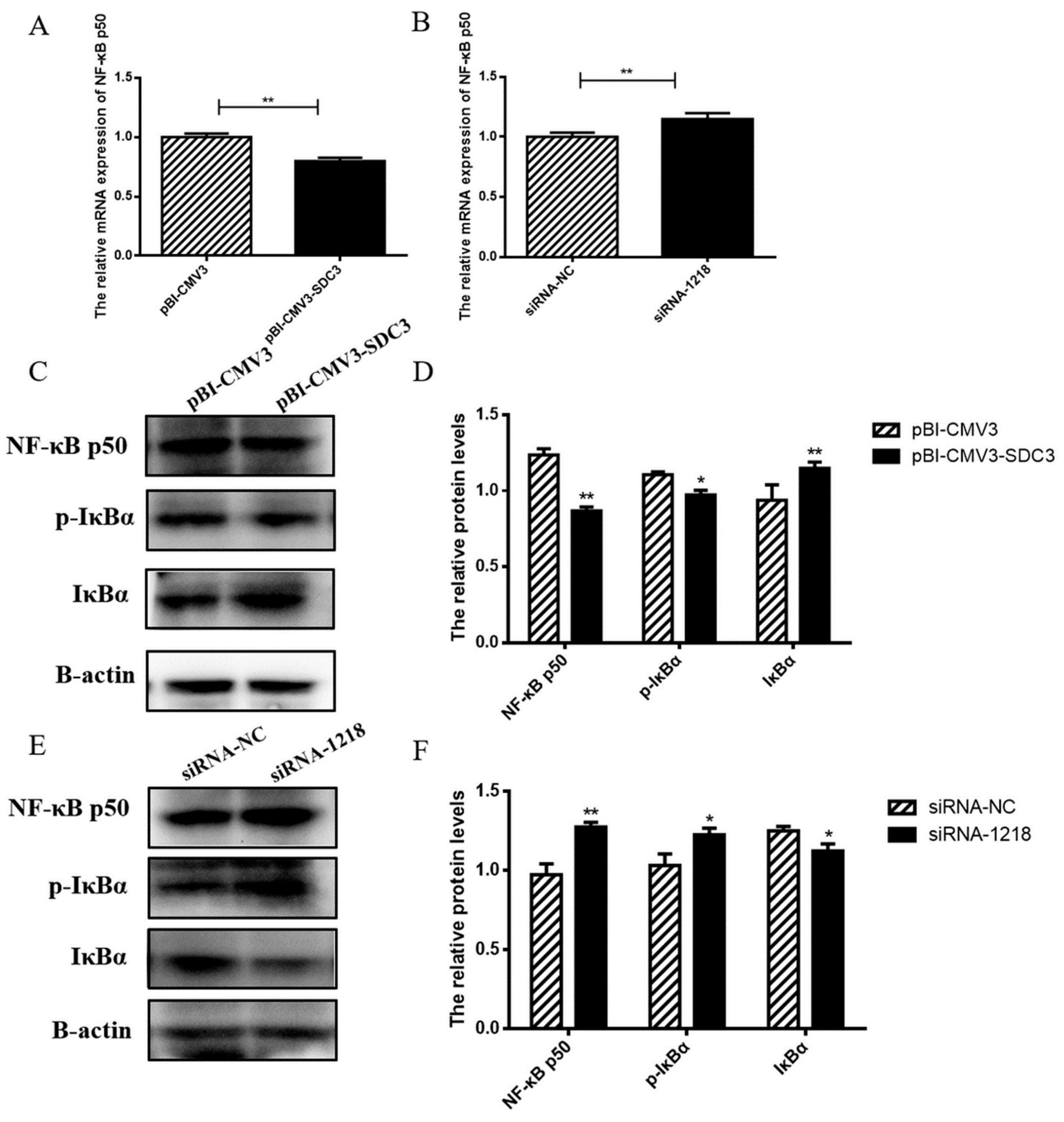
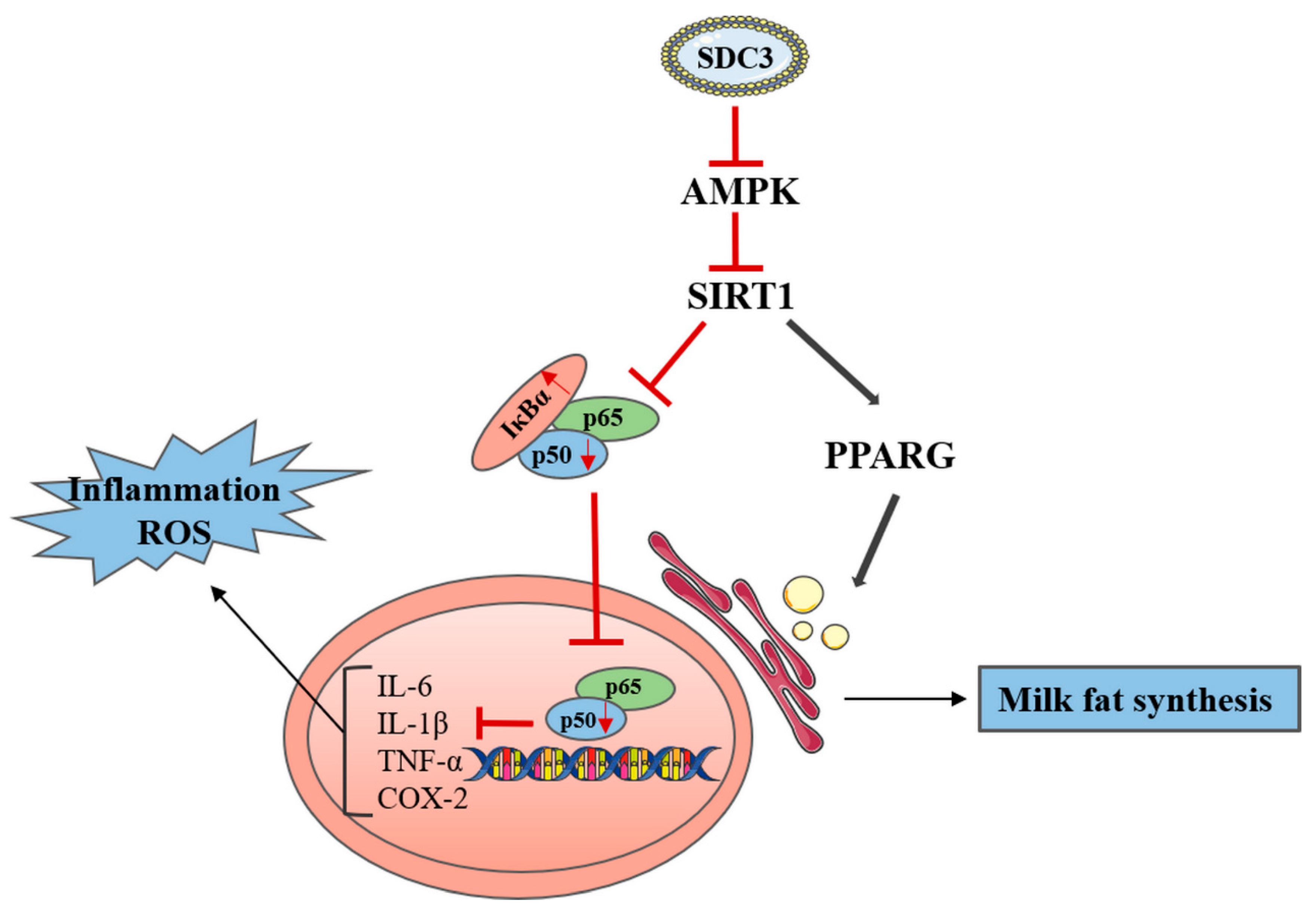
2.4. SDC3 Exerts a Cooperatively Regulatory Effect on Milk Lipid Metabolism and Inflammatory Reactions in BMECs through the AMPK/SIRT1 Signal Pathway
2.4.1. SDC3 Promotes Milk Fat Synthesis through AMPK/SIRT1/PPARG Signal Pathway
2.4.2. SDC3 Inhibits Inflammatory Reaction through the AMPK/SIRT1/NF-κB P50 Signaling Pathway
3. Discussion
4. Materials and Methods
4.1. Reagents and Antibodies
4.2. Cell Culture
4.3. Transfection
4.4. Measurement of Triglycerides and Cholesterol
4.5. Non-Esterified Fatty Acids (NEFA) Measurement
4.6. BODIPY Staining
4.7. Protein Expression Analysis
4.8. Cell ROS Analysis
4.9. Gene Expression Analysis
4.10. Western Blot
4.11. Statistical analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Randolph, H.E.; Erwin, R.E. Influence of mastitis on properties of milk. X. Fatty acid composition. J. Dairy Sci. 1974, 57, 865–868. [Google Scholar] [CrossRef]
- Pan, Z.; Zhou, Q.; Ma, H.; Gong, Q.; Wang, S.; Yao, H.; Ma, J.; Wang, K. Identification of a novel bacterial taxon associated with bovine mastitis showing a close evolutionary relationship with Elizabethkingia sp. Microbiol. Res. 2020, 236, 126443. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Zhao, Z.; Yu, X.; Lu, C.; Jiang, P.; Yu, H.; Li, X.; Yu, X.; Liu, J.; Fang, X.; et al. Integrative analysis of miRNAs and mRNAs revealed regulation of lipid metabolism in dairy cattle. Funct. Integr. Genom. 2021, 21, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Carey, D.J.; Conner, K.; Asundi, V.K.; O’Mahony, D.J.; Stahl, R.C.; Showalter, L.; Cizmeci-Smith, G.; Hartman, J.; Rothblum, L.I. cDNA cloning, genomic organization, and in vivo expression of rat N-syndecan. J. Biol. Chem. 1997, 272, 2873–2879. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.Z.; Wang, Q.; Zhang, C.L.; Fang, X.T.; Song, E.L.; Chen, H. Genetic Variants in SDC3 Gene are Significantly Associated with Growth Traits in Two Chinese Beef Cattle Breeds. Anim. Biotechnol. 2016, 27, 190–198. [Google Scholar] [CrossRef]
- Reizes, O.; Lincecum, J.; Wang, Z.; Goldberger, O.; Huang, L.; Kaksonen, M.; Ahima, R.; Hinkes, M.T.; Barsh, G.S.; Rauvala, H.; et al. Transgenic expression of syndecan-1 uncovers a physiological control of feeding behavior by syndecan-3. Cell 2001, 106, 105–116. [Google Scholar] [CrossRef]
- Strader, A.D.; Reizes, O.; Woods, S.C.; Benoit, S.C.; Seeley, R.J. Mice lacking the syndecan-3 gene are resistant to diet-induced obesity. J. Clin. Investig. 2004, 114, 1354–1360. [Google Scholar] [CrossRef]
- Roh, J.J.; Kim, M.J.; Oh, D.J.; Noh, T.H.; Won, S.G.; Yang, K.Y.; Jung, D.H.; Jo, C.L.; Chung, J.H.; Kim, J.W. [P11-21] Positive association of obesity with single nucleotide polymorphisms of syndecan 3 (SDC3) in Korean population. J. Clin. Endocrinol. Metab. 2006, 91, 5095–5099. [Google Scholar]
- Kehoe, O.; Kalia, N.; King, S.; Eustace, A.; Boyes, C.; Reizes, O.; Williams, A.; Patterson, A.; Middleton, J. Syndecan-3 is selectively pro-inflammatory in the joint and contributes to antigen-induced arthritis in mice. Arthritis Res. Ther. 2014, 16, R148. [Google Scholar] [CrossRef]
- Cornelison, D.D.; Wilcox-Adelman, S.A.; Goetinck, P.F.; Rauvala, H.; Rapraeger, A.C.; Olwin, B.B. Essential and separable roles for Syndecan-3 and Syndecan-4 in skeletal muscle development and regeneration. Genes Dev. 2004, 18, 2231–2236. [Google Scholar] [CrossRef]
- Arokiasamy, S.; Balderstone, M.J.M.; De Rossi, G.; Whiteford, J.R. Syndecan-3 in Inflammation and Angiogenesis. Front. Immunol. 2019, 10, 3031. [Google Scholar] [CrossRef]
- Kempf, A.; Boda, E.; Kwok, J.C.F.; Fritz, R.; Grande, V.; Kaelin, A.M.; Ristic, Z.; Schmandke, A.; Schmandke, A.; Tews, B.; et al. Control of Cell Shape, Neurite Outgrowth, and Migration by a Nogo-A/HSPG Interaction. Dev. Cell 2017, 43, 24–34.e25. [Google Scholar] [CrossRef]
- Pisconti, A.; Cornelison, D.D.; Olguín, H.C.; Antwine, T.L.; Olwin, B.B. Syndecan-3 and Notch cooperate in regulating adult myogenesis. J. Cell Biol. 2010, 190, 427–441. [Google Scholar] [CrossRef]
- Yamada, Y.; Arai, T.; Kojima, S.; Sugawara, S.; Kato, M.; Okato, A.; Yamazaki, K.; Naya, Y.; Ichikawa, T.; Seki, N. Regulation of antitumor miR-144-5p targets oncogenes: Direct regulation of syndecan-3 and its clinical significance. Cancer Sci. 2018, 109, 2919–2936. [Google Scholar] [CrossRef] [PubMed]
- Hardie, D.G.; Ross, F.A.; Hawley, S.A. AMPK: A nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 2012, 13, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Mihaylova, M.M.; Shaw, R.J. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 2011, 13, 1016–1023. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Zhang, Y.; Ge, Y.; Li, W.; Cao, Y.; Qu, Y.; Liu, S.; Guo, Y.; Fu, S.; Liu, J. Sodium butyrate promotes milk fat synthesis in bovine mammary epithelial cells via GPR41 and its downstream signalling pathways. Life Sci 2020, 259, 118375. [Google Scholar] [CrossRef]
- Zhang, T.Y.; Huang, J.T.; Tian, H.B.; Ma, Y.; Chen, Z.; Wang, J.J.; Shi, H.P.; Luo, J. trans-10,cis-12 conjugated linoleic acid alters lipid metabolism of goat mammary epithelial cells by regulation of de novo synthesis and the AMPK signaling pathway. J. Dairy Sci. 2018, 101, 5571–5581. [Google Scholar] [CrossRef]
- Hardie, D.G.; Schaffer, B.E.; Brunet, A. AMPK: An Energy-Sensing Pathway with Multiple Inputs and Outputs. Trends Cell Biol. 2016, 26, 190–201. [Google Scholar] [CrossRef]
- Quentin, T.; Kitz, J.; Steinmetz, M.; Poppe, A.; Bär, K.; Krätzner, R. Different expression of the catalytic alpha subunits of the AMP activated protein kinase--an immunohistochemical study in human tissue. Histol. Histopathol. 2011, 26, 589–596. [Google Scholar] [CrossRef]
- Ji, S.; Zheng, Z.; Liu, S.; Ren, G.; Gao, J.; Zhang, Y.; Li, G. Resveratrol promotes oxidative stress to drive DLC1 mediated cellular senescence in cancer cells. Exp. Cell Res. 2018, 370, 292–302. [Google Scholar] [CrossRef]
- Picard, F.; Kurtev, M.; Chung, N.; Topark-Ngarm, A.; Senawong, T.; Machado De Oliveira, R.; Leid, M.; McBurney, M.W.; Guarente, L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature 2004, 429, 771–776. [Google Scholar] [CrossRef]
- Zheng, Z.; Bian, Y.; Zhang, Y.; Ren, G.; Li, G. Metformin activates AMPK/SIRT1/NF-κB pathway and induces mitochondrial dysfunction to drive caspase3/GSDME-mediated cancer cell pyroptosis. Cell Cycle 2020, 19, 1089–1104. [Google Scholar] [CrossRef]
- Ansari, N.; Khodagholi, F.; Amini, M.; Shaerzadeh, F. Attenuation of LPS-induced apoptosis in NGF-differentiated PC12 cells via NF-κB pathway and regulation of cellular redox status by an oxazine derivative. Biochimie 2011, 93, 899–908. [Google Scholar] [CrossRef]
- Hayden, M.S.; Ghosh, S. Shared principles in NF-kappaB signaling. Cell 2008, 132, 344–362. [Google Scholar] [CrossRef]
- Rawat, C.; Kukal, S.; Dahiya, U.R.; Kukreti, R. Cyclooxygenase-2 (COX-2) inhibitors: Future therapeutic strategies for epilepsy management. J. Neuroinflammation 2019, 16, 197. [Google Scholar] [CrossRef]
- Bagul, P.K.; Deepthi, N.; Sultana, R.; Banerjee, S.K. Resveratrol ameliorates cardiac oxidative stress in diabetes through deacetylation of NFkB-p65 and histone 3. J. Nutr. Biochem. 2015, 26, 1298–1307. [Google Scholar] [CrossRef]
- Lubary, M.; Hofland, G.W.; ter Horst, J.H. The potential of milk fat for the synthesis of valuable derivatives. Eur. Food Res. Technol. 2011, 232, 1–8. [Google Scholar] [CrossRef]
- Cantó, C.; Gerhart-Hines, Z.; Feige, J.N.; Lagouge, M.; Noriega, L.; Milne, J.C.; Elliott, P.J.; Puigserver, P.; Auwerx, J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 2009, 458, 1056–1060. [Google Scholar] [CrossRef]
- Chen, X.Y.; Cai, C.Z.; Yu, M.L.; Feng, Z.M.; Zhang, Y.W.; Liu, P.H.; Zeng, H.; Yu, C.H. LB100 ameliorates nonalcoholic fatty liver disease via the AMPK/Sirt1 pathway. World J. Gastroenterol. 2019, 25, 6607–6618. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, J.; Bruckbauer, A.; Zemel, M.B. Activation of the AMPK/Sirt1 pathway by a leucine-metformin combination increases insulin sensitivity in skeletal muscle, and stimulates glucose and lipid metabolism and increases life span in Caenorhabditis elegans. Metab. Clin. Exp. 2016, 65, 1679–1691. [Google Scholar] [CrossRef] [PubMed]
- Itoh, T.; Fairall, L.; Amin, K.; Inaba, Y.; Szanto, A.; Balint, B.L.; Nagy, L.; Yamamoto, K.; Schwabe, J.W. Structural basis for the activation of PPARgamma by oxidized fatty acids. Nat. Struct. Mol. Biol. 2008, 15, 924–931. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.D.; Sarraf, P.; Troy, A.E.; Bradwin, G.; Moore, K.; Milstone, D.S.; Spiegelman, B.M.; Mortensen, R.M. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell 1999, 4, 611–617. [Google Scholar] [CrossRef]
- Matsusue, K.; Aibara, D.; Hayafuchi, R.; Matsuo, K.; Takiguchi, S.; Gonzalez, F.J.; Yamano, S. Hepatic PPARγ and LXRα independently regulate lipid accumulation in the livers of genetically obese mice. FEBS Lett. 2014, 588, 2277–2281. [Google Scholar] [CrossRef]
- Lee, H.S.; Lim, S.M.; Jung, J.I.; Kim, S.M.; Lee, J.K.; Kim, Y.H.; Cha, K.M.; Oh, T.K.; Moon, J.M.; Kim, T.Y.; et al. Gynostemma Pentaphyllum Extract Ameliorates High-Fat Diet-Induced Obesity in C57BL/6N Mice by Upregulating SIRT1. Nutrients 2019, 11, 2475. [Google Scholar] [CrossRef]
- Guo, W.; Liu, B.; Hu, G.; Kan, X.; Li, Y.; Gong, Q.; Xu, D.; Ma, H.; Cao, Y.; Huang, B.; et al. Vanillin protects the blood-milk barrier and inhibits the inflammatory response in LPS-induced mastitis in mice. Toxicol. Appl. Pharmacol. 2019, 365, 9–18. [Google Scholar] [CrossRef]
- Alluwaimi, A.M. The cytokines of bovine mammary gland: Prospects for diagnosis and therapy. Res. Vet. Sci. 2004, 77, 211–222. [Google Scholar] [CrossRef]
- Günther, J.; Liu, S.; Esch, K.; Schuberth, H.J.; Seyfert, H.M. Stimulated expression of TNF-alpha and IL-8, but not of lingual antimicrobial peptide reflects the concentration of pathogens contacting bovine mammary epithelial cells. Vet. Immunol. Immunopathol. 2010, 135, 152–157. [Google Scholar] [CrossRef]
- Kehoe, O.; Cartwright, A.; Askari, A.; El Haj, A.J.; Middleton, J. Intra-articular injection of mesenchymal stem cells leads to reduced inflammation and cartilage damage in murine antigen-induced arthritis. J. Transl. Med. 2014, 12, 157. [Google Scholar] [CrossRef]
- Eustace, A.D.; McNaughton, E.F.; King, S.; Kehoe, O.; Kungl, A.; Mattey, D.; Nobbs, A.H.; Williams, N.; Middleton, J. Soluble syndecan-3 binds chemokines, reduces leukocyte migration in vitro and ameliorates disease severity in models of rheumatoid arthritis. Arthritis Res. Ther. 2019, 21, 172. [Google Scholar] [CrossRef]
- Reizes, O.; Clegg, D.J.; Strader, A.D.; Benoit, S.C. A role for syndecan-3 in the melanocortin regulation of energy balance. Peptides 2006, 27, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, C.A.; Selleck, S.B. Heparan sulfate proteoglycans at a glance. J. Cell Sci. 2007, 120, 1829–1832. [Google Scholar] [CrossRef] [PubMed]
- Sarrazin, S.; Lamanna, W.C.; Esko, J.D. Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 2011, 3, a004952. [Google Scholar] [CrossRef] [PubMed]
- Karin, M.; Lin, A. NF-kappaB at the crossroads of life and death. Nat. Immunol. 2002, 3, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Sen, R.; Baltimore, D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell 1986, 46, 705–716. [Google Scholar] [CrossRef]
- Kong, L.M.; Deng, X.; Zuo, Z.L.; Sun, H.D.; Zhao, Q.S.; Li, Y. Identification and validation of p50 as the cellular target of eriocalyxin B. Oncotarget 2014, 5, 11354–11364. [Google Scholar] [CrossRef]
- Yi, H.; Peng, R.; Zhang, L.Y.; Sun, Y.; Peng, H.M.; Liu, H.D.; Yu, L.J.; Li, A.L.; Zhang, Y.J.; Jiang, W.H.; et al. LincRNA-Gm4419 knockdown ameliorates NF-κB/NLRP3 inflammasome-mediated inflammation in diabetic nephropathy. Cell Death Dis. 2017, 8, e2583. [Google Scholar] [CrossRef]
- Jiang, P.; Xia, L.; Jin, Z.; Ali, S.; Wang, M.; Li, X.; Yang, R.; Fang, X.; Zhao, Z. New function of the CD44 gene: Lipid metabolism regulation in bovine mammary epithelial cells. J. Dairy Sci. 2020, 103, 6661–6671. [Google Scholar] [CrossRef]
- Jiang, P.; Zhao, Z.; Li, X.; Wang, M.; Xia, L.; Cao, Y.; Yang, R.; Fang, X. RNA Interference Mediated Knockdown of ATP Binding Cassette Subfamily A Member 1 Decreases the Triglyceride Content of Bovine Mammary Epithelial Cells. Pak. J. Zool 2020, 52, 239–245. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-DDCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Jiang, P.; Iqbal, A.; Wang, M.; Li, X.; Fang, X.; Yu, H.; Zhao, Z. Transcriptomic Analysis of Short/Branched-Chain Acyl-Coenzyme a Dehydrogenase Knocked Out BMECs Revealed Its Reg-ulatory Effect on Lipid Metabolism. Front. Vet. Sci. 2021, 8, 744287. [Google Scholar] [CrossRef]
| Gene | Sequence Number | Primer Sequence (5′ to 3′) | Product Size (bp) |
|---|---|---|---|
| AMPKα1 | XM_005221557.3 | F: CCCGTATTATTTGCGTGTTCG | 160 |
| R: CTGTGGCGTAGCAGTCCCT | |||
| SIRT1 | NM_001192980.3 | F: TGAAGAATGCTGTCTCCAAT | 271 |
| R: GCGTTTACTAATCTGCTCCT | |||
| PPARG | NM_181024.2 | F: TGACCCGATGGTTGCAGATTAT | 100 |
| R: ATGAGGGAGTTGGAAGGCTCT | |||
| NF-κB p50 | NM_001076409.1 | F: AACATCCACCTGCATGCACAC | 155 |
| R: GGCATCTGTCATTCGTGCTTC | |||
| IL-6 | NM_173923.2 | F: GATGCTTCCAATCTGGGTTCA | 115 |
| R: TCCTGATTTCCCTCATACTCG | |||
| IL-1β | NM_174093.1 | F: GATGGCTTACTACAGTGACGA | 254 |
| R: AGATGAATGAAAGGATGCTC | |||
| TNF-α | XM_005223596.4 | F: ACGGGCTTTACCTCATCTACTC | 132 |
| R: TGGCAGACAGGATGTTGACC | |||
| COX-2 | YP_209208.1 | F: CCAGAGCTCTTCCTCCTGTG | 213 |
| R: AAGCTGGTCCTCGTTCAAAA | |||
| SDC3 | NM_001206531.2 | F: CGCCCTCTTTGCTGCCTT | 103 |
| R: TGTGACGCTCGCCTGCTT | |||
| ACTB | NM_173979.3 | F:AGAGCAAGAGAGGCATCC | 133 |
| R:TCGTTGTAGAAGGTGTGGT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, J.; Zhao, Z.; Wu, H.; Fang, X.; Miao, F.; Chen, X.; Jiang, X.; Li, J.; Jiang, P.; Yu, H. Syndecan-3 Coregulates Milk Fat Metabolism and Inflammatory Reactions in Bovine Mammary Epithelial Cells through AMPK/SIRT1 Signaling Pathway. Int. J. Mol. Sci. 2023, 24, 6657. https://doi.org/10.3390/ijms24076657
Fan J, Zhao Z, Wu H, Fang X, Miao F, Chen X, Jiang X, Li J, Jiang P, Yu H. Syndecan-3 Coregulates Milk Fat Metabolism and Inflammatory Reactions in Bovine Mammary Epithelial Cells through AMPK/SIRT1 Signaling Pathway. International Journal of Molecular Sciences. 2023; 24(7):6657. https://doi.org/10.3390/ijms24076657
Chicago/Turabian StyleFan, Jing, Zhihui Zhao, Haochen Wu, Xibi Fang, Fengshuai Miao, Xuanxu Chen, Xinyi Jiang, Jing Li, Ping Jiang, and Haibin Yu. 2023. "Syndecan-3 Coregulates Milk Fat Metabolism and Inflammatory Reactions in Bovine Mammary Epithelial Cells through AMPK/SIRT1 Signaling Pathway" International Journal of Molecular Sciences 24, no. 7: 6657. https://doi.org/10.3390/ijms24076657
APA StyleFan, J., Zhao, Z., Wu, H., Fang, X., Miao, F., Chen, X., Jiang, X., Li, J., Jiang, P., & Yu, H. (2023). Syndecan-3 Coregulates Milk Fat Metabolism and Inflammatory Reactions in Bovine Mammary Epithelial Cells through AMPK/SIRT1 Signaling Pathway. International Journal of Molecular Sciences, 24(7), 6657. https://doi.org/10.3390/ijms24076657







