MYB Transcription Factor OsC1PLSr Involves the Regulation of Purple Leaf Sheath in Rice
Abstract
1. Introduction
2. Results
2.1. Observation of Leaf Sheath Phenotype
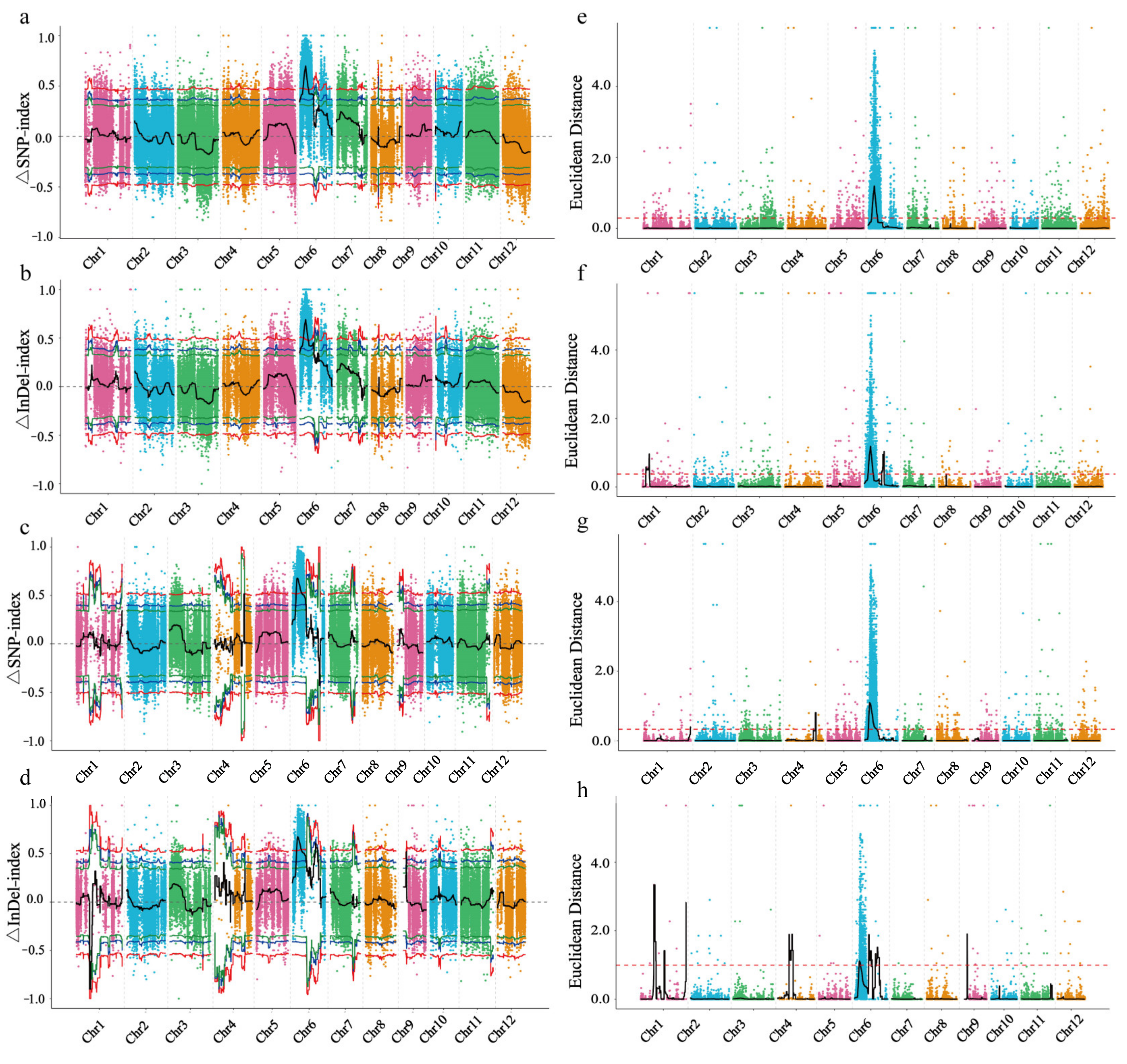
2.2. Identification of Candidate Regions for Controlling Leaf Sheath Color by BSA-Seq
2.3. Analysis of Candidate Genes
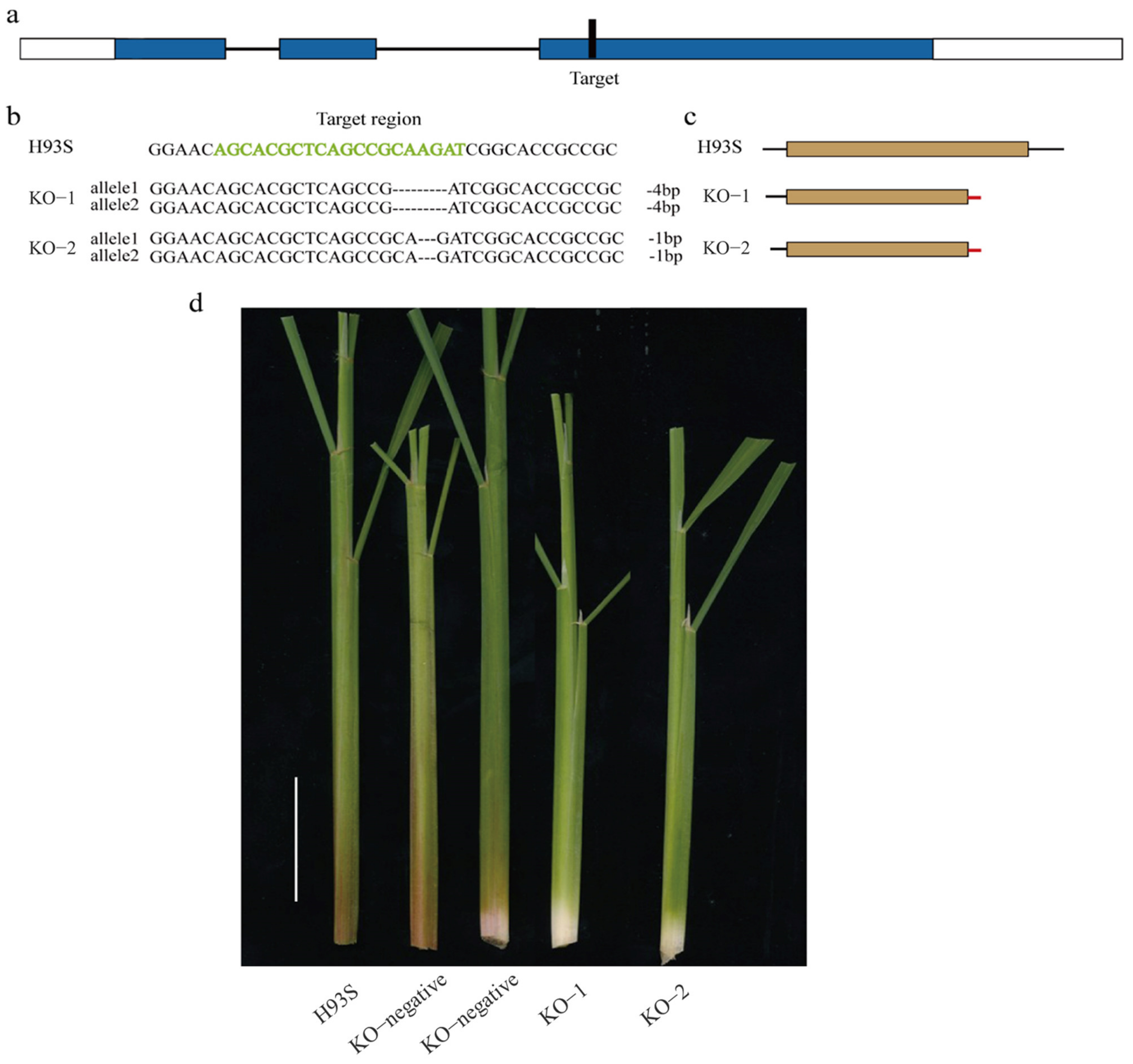
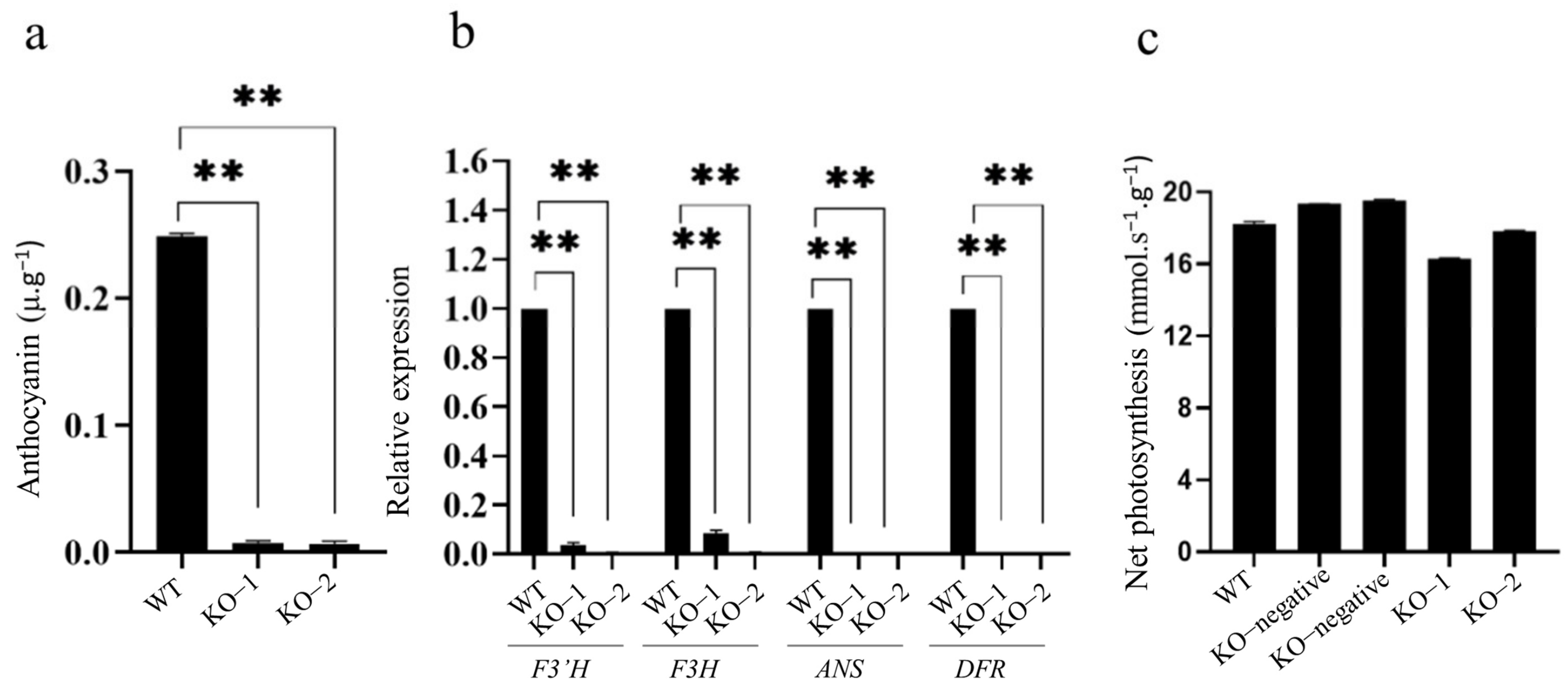
2.4. Knockout of OsC1PLSr Results in No Purple Leaf Sheath Phenotype
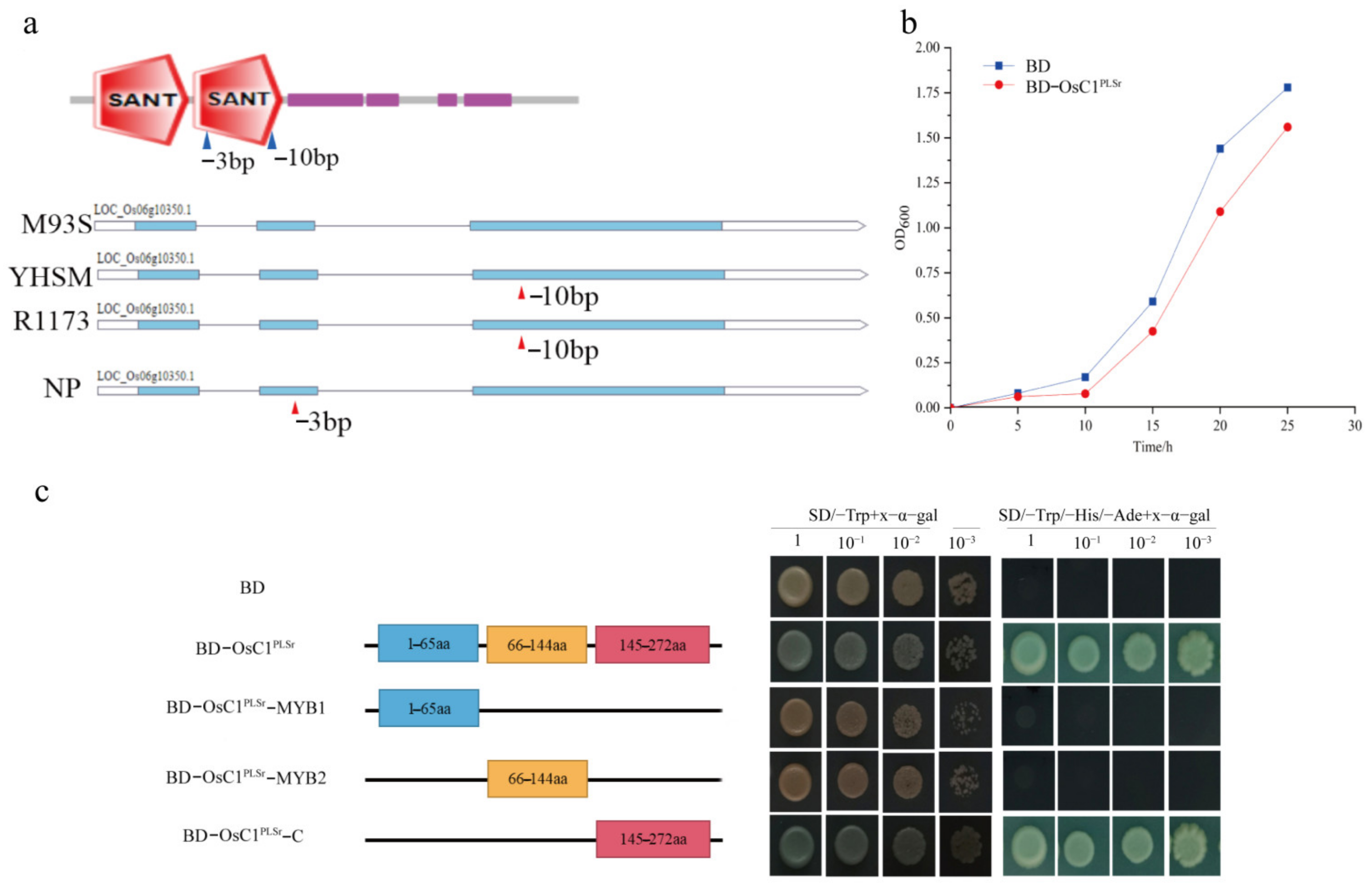
2.5. Structure Analysis, Transcription Activity Analysis, and Toxicity Detection
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Genetic Population Construction
4.2. BSA Sequencing and Association Analysis
4.3. Screening of Candidate Genes
4.4. Histological, Anthocyanin Content, and Genetic Analysis
4.5. Sequence Analysis and Structure Analysis of OsC1PLSr
4.6. CRISPR/Cas9 Generated the OsC1PLSr Mutant Plants
4.7. Transcription Activity Assay and Toxicity Detection in Yeast Report System
4.8. RNA Extraction and Quantitative qRT-PCR
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khoo, H.E.; Azlan, A.; Tang, S.T.; Lim, S.M. Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 2017, 61, 1361779. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhu, Q.; Chen, Y.; Liu, Y.-G.J.M.P. CRISPR/Cas9 platforms for genome editing in plants: Developments and applications. Mol. Plant 2016, 9, 961–974. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wang, M.H. Ultraviolet A-specific induction of anthocyanin biosynthesis and PAL expression in tomato (Solanum lycopersicum L.). Plant Growth Regul. 2010, 62, 1–8. [Google Scholar] [CrossRef]
- Wang, Z.; Jian, X.; Zhao, Y.; Li, S.; Sui, Z.; Li, L.; Kong, L.; Luo, J. Functional characterization of cinnamate 4-hydroxylase from Helianthus annuus Linn using a fusion protein method. Gene 2020, 758, 144950. [Google Scholar] [CrossRef]
- Hemleben, V.; Dressel, A.; Epping, B.; Lukacin, R.; Martens, S.; Austin, M. Characterization and structural features of a chalcone synthase mutation in a white-flowering line of Matthiola incana R. Br. (Brassicaceae). Plant Mol. Biol. 2004, 55, 455–465. [Google Scholar] [CrossRef]
- Yin, Y.C.; Zhang, X.D.; Gao, Z.Q.; Hu, T.; Liu, Y. The research progress of chalcone isomerase (CHI) in plants. Mol. Biotechnol. 2019, 61, 32–52. [Google Scholar] [CrossRef]
- Lim, S.H.; Park, B.; Kim, D.H.; Park, S.; Yang, J.H.; Jung, J.A.; Lee, J.M.; Lee, J.Y. Cloning and functional characterization of dihydroflavonol 4-reductase gene involved in anthocyanin biosynthesis of chrysanthemum. Int. J. Mol. Sci. 2020, 21, 7960. [Google Scholar] [CrossRef]
- Kim, E.Y.; Kim, C.W.; Kim, S. Identification of two novel mutant ANS alleles responsible for inactivation of anthocyanidin synthase and failure of anthocyanin production in onion (Allium cepa L.). Euphytica 2016, 212, 427–437. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Feller, A.C. Role of the Maize Transcription Factor R in the Regulation of Anthocyanin Biosynthesis. Ph.D. Thesis, Ohio State University, Columbus, OH, USA, 2010. [Google Scholar]
- Li, S. Transcriptional control of flavonoid biosynthesis: Fine-tuning of the MYB-bHLH-WD40 (MBW) complex. Plant Signal. Behav. 2014, 9, e27522. [Google Scholar] [CrossRef]
- Khan, A.; Jalil, S.; Cao, H.; Chare, Y.; Hamdaoui, M.; Ziyan, C.; Shi, C.; Jin, X. The purple leaf (pl6) mutation regulates leaf color by altering the anthocyanin and chlorophyll contents in rice. Plants 2020, 9, 1477. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, T.; Maeda, H.; Oguchi, T.; Yamaguchi, T.; Tanabe, N.; Ebana, K.; Yano, M.; Ebitani, T.; Izawa, T. The birth of a black rice gene and its local spread by introgression. Plant Cell 2015, 27, 2401–2414. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wu, H.; Zhu, H.; Huang, C.; Liu, C.; Chang, Y.; Kong, Z.; Zhou, Z.; Wang, G.; Lin, Y.; et al. Determining factors, regulation system, and domestication of anthocyanin biosynthesis in rice leaves. New Phytol. 2019, 223, 705–721. [Google Scholar] [CrossRef]
- Gao, J.; Dai, G.; Zhou, W.; Liang, H.; Huang, J.; Qing, D.; Chen, W.; Wu, H.; Yang, X.; Li, D.; et al. Mapping and identifying a candidate gene Plr4, a recessive gene regulating purple leaf in rice, by using bulked segregant and transcriptome analysis with next-generation sequencing. Int. J. Mol. Sci. 2019, 20, 4335. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, Z.; Chen, C.; Wu, W.; Ren, N.; Jiang, C.; Yu, J.; Zhao, Y.; Zheng, X.; Yang, Q.; et al. The C–S–A gene system regulates hull pigmentation and reveals evolution of anthocyanin biosynthesis pathway in rice. J. Exp. Bot. 2018, 69, 1485–1498. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Zhou, T.; Han, Z.; Tan, C.; Xing, Y. Dominant complementary interaction between OsC1 and two tightly linked genes, Rb1 and Rb2, controls the purple leaf sheath in rice. Theor. Appl. Genet. 2020, 133, 2555–2566. [Google Scholar] [CrossRef]
- Chin, H.S.; Wu, Y.P.; Hour, A.L.; Hong, C.Y.; Lin, Y.R. Genetic and evolutionary analysis of purple leaf sheath in rice. Rice 2016, 9, 8. [Google Scholar] [CrossRef]
- Crupi, P.; Pichierri, A.; Basile, T.; Antonacci, D. Postharvest stilbenes and flavonoids enrichment of table grape cv redglobe (Vitis vinifera L.) as affected by interactive UV-C exposure and storage conditions. Food Chem. 2013, 141, 802–808. [Google Scholar] [CrossRef]
- Heppel, S.C.; Jaffé, F.W.; Takos, A.M.; Schellmann, S.; Rausch, T.; Walker, A.R.; Bogs, J. Identification of key amino acids for the evolution of promoter target specificity of anthocyanin and proanthocyanidin regulating MYB factors. Plant Mol. Biol. 2013, 82, 457–471. [Google Scholar] [CrossRef]
- Oshima, M.; Taniguchi, Y.; Akasaka, M.; Abe, K.; Ichikawa, H.; Tabei, Y.; Tanaka, J. Development of a visible marker trait based on leaf sheath–specific anthocyanin pigmentation applicable to various genotypes in rice. Breed. Sci. 2019, 69, 244–254. [Google Scholar] [CrossRef]
- Leon, R.; Lightbourn, L.; Lopez-Meyer, M.; Amarillas, L.; Heredia, J.; Martínez-Bastidas, T.; Villicaña, C.; León-Félix, J. Differential gene expression of anthocyanin biosynthetic genes under low temperature and ultraviolet-B radiation in bell pepper (Capsicum annuum). Int. J. Agric. Biol. 2020, 23, 531–538. [Google Scholar] [CrossRef]
- Nakatsuka, A.; Mizuta, D.; Kii, Y.; Miyajima, I.; Kobayashi, N. Isolation and expression analysis of flavonoid biosynthesis genes in evergreen azalea. Sci. Hortic. 2008, 118, 314–320. [Google Scholar] [CrossRef]
- Hiei, Y.; Ohta, S.; Komari, T.; Kumashiro, T.J.T.P.J. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994, 6, 271–282. [Google Scholar] [CrossRef] [PubMed]


| Population Name | Tested Plants | Purple Plants | Green Plants | Mendelian Expectations | χ2 | χ2(0.05, 1) |
|---|---|---|---|---|---|---|
| F2-1 (H93S/R1173) | 500 | 390 | 110 | 3:1 | 2.40 | 3.84 |
| F2-2 (H93S/YHSM) | 500 | 388 | 112 | 3:1 | 1.80 | 3.84 |
| Samples | Filtered Dates | Q30 | GC | Mapped | Ave_Depth | Coverage |
|---|---|---|---|---|---|---|
| R1173 | 42,356,107 | 90.60% | 44.45% | 98.01% | 28× | 94.95% |
| F2-1 (P) | 55,322,041 | 90.32% | 44.40% | 97.96% | 37× | 97.25% |
| F2-1 (G) | 60,695,930 | 90.94% | 44.77% | 98.49% | 40× | 97.23% |
| H93S | 36,790,415 | 91.30% | 44.33% | 98.56% | 25× | 94.85% |
| F2-2 (P) | 55,367,274 | 91.27% | 44.54% | 98.58% | 37× | 97.33% |
| F2-2 (G) | 56,536,080 | 91.08% | 44.40% | 98.50% | 38× | 96.76% |
| YHSM | 43,800,417 | 91.33% | 44.36% | 98.56% | 30× | 95.09% |
| Population | FT | TL | MG | RSL4 | CGMP | RPNP | CHQS/I |
|---|---|---|---|---|---|---|---|
| F2-1 | SNPs | 1,432,222 | 1961 | 21,971 | 443,822 | 288,956 | 675,512 |
| InDels | 312,912 | 8875 | 10,217 | 97,239 | 56,207 | 140,374 | |
| F2-2 | SNPs | 1,251,158 | 1362 | 25,629 | 478,438 | 278,933 | 466,796 |
| InDels | 274,708 | 6863 | 10,764 | 105,401 | 54,942 | 96,738 |
| Population | CRAT | Chr. | Start Position | End Position | Size (Mb) | Genes |
|---|---|---|---|---|---|---|
| F2-1 | SNP AA | 6 | 3,330,000 | 8,720,000 | 5.39 | 1415 |
| InDel AA | 6 | 3,730,000 | 8,550,000 | 4.82 | 1274 | |
| F2-2 | SNP AA | 6 | 4,030,000 | 9,060,000 | 5.03 | 1275 |
| InDel AA | 6 | 4,610,000 | 6,030,000 | 1.42 | 415 |
| Genes | MSU_Locus | Annotation |
|---|---|---|
| OsR498G0611776200.01 | LOC_Os06g09540 | SAC domain containing protein, putative, expressed |
| OsR498G0611780600.01 | LOC_Os06g09620 | expressed protein |
| OsR498G0611782900.01 | LOC_Os06g09679 | chaperonin, putative, expressed |
| OsR498G0611789000.01 | LOC_Os06g09850 | 25.3 kDa vesicle transport protein, putative, expressed |
| OsR498G0611790200.01 | LOC_Os06g09890 | smr domain containing protein, expressed |
| OsR498G0611790800.01 | LOC_Os06g09900 | expressed protein |
| OsR498G0611792500.01 | LOC_Os06g09930 | G protein coupled receptor, putative, expressed |
| OsR498G0611812900.01 | LOC_Os06g10340 | autophagy-related protein 12, putative, expressed |
| OsR498G0611814100.01 | LOC_Os06g10350 | MYB family transcription factor, putative, expressed |
| OsR498G0611822700.01 | LOC_Os06g10520 | pantothenate kinase, putative, expressed |
| OsR498G0611830400.01 | LOC_Os06g10620 | transcription elongation factor SPT5 homolog 1, putative, expressed |
| OsR498G0611870100.01 | LOC_Os06g11620 | RNA recognition motif containing protein, putative, expressed |
| OsR498G0611870800.01 | LOC_Os06g11640 | serine/threonine-protein phosphatase 2A activator 2, putative, expressed |
| OsR498G0611835200.01 | LOC_Os06g10690 | PHD-finger domain containing protein, putative, expressed |
| OsR498G0611836500.01 | LOC_Os06g10750 | integral membrane protein DUF6 containing protein, expressed |
| OsR498G0611837500.01 | LOC_Os06g10790 | lectin-like receptor kinase, putative, expressed |
| OsR498G0611842100.01 | LOC_Os06g10900 | DELLA protein RGL3, putative, expressed |
| OsR498G0611842900.01 | LOC_Os06g10910 | xyloglucan fucosyltransferase, putative, expressed |
| OsR498G0611843200.01 | LOC_Os06g10920 | xyloglucan fucosyltransferase, putative, expressed |
| OsR498G0611848000.01 | LOC_Os06g11020 | tic22-like family domain containing protein, expressed |
| OsR498G0611849000.01 | LOC_Os06g11040 | expressed protein |
| OsR498G0611856800.01 | LOC_Os06g11270 | Anthocyanidin 3-O-glucosyltransferase, putative, expressed |
| OsR498G0611859800.01 | LOC_Os06g11330 | OsMADS55—MADS-box family gene with MIKCc type-box, expressed |
| OsR498G0611860600.01 | LOC_Os06g11370 | mediator of RNA polymerase II transcription subunit 6, putative, expressed |
| OsR498G0611862900.01 | LOC_Os06g11410 | cyclin, putative, expressed |
| OsR498G0611871200.01 | LOC_Os06g11660 | phosphate-induced protein 1 conserved region domain containing protein, expressed |
| OsR498G0611864700.01 | LOC_Os06g11430 | GRAM domain containing protein, expressed |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zou, T.; Wang, X.; Sun, T.; Rong, H.; Wu, L.; Deng, J.; Guo, T.; Wang, H.; Wang, J.; Huang, M. MYB Transcription Factor OsC1PLSr Involves the Regulation of Purple Leaf Sheath in Rice. Int. J. Mol. Sci. 2023, 24, 6655. https://doi.org/10.3390/ijms24076655
Zou T, Wang X, Sun T, Rong H, Wu L, Deng J, Guo T, Wang H, Wang J, Huang M. MYB Transcription Factor OsC1PLSr Involves the Regulation of Purple Leaf Sheath in Rice. International Journal of Molecular Sciences. 2023; 24(7):6655. https://doi.org/10.3390/ijms24076655
Chicago/Turabian StyleZou, Ting, Xinyi Wang, Tong Sun, Huazhen Rong, Linxuan Wu, Jing Deng, Tao Guo, Hui Wang, Jiafeng Wang, and Ming Huang. 2023. "MYB Transcription Factor OsC1PLSr Involves the Regulation of Purple Leaf Sheath in Rice" International Journal of Molecular Sciences 24, no. 7: 6655. https://doi.org/10.3390/ijms24076655
APA StyleZou, T., Wang, X., Sun, T., Rong, H., Wu, L., Deng, J., Guo, T., Wang, H., Wang, J., & Huang, M. (2023). MYB Transcription Factor OsC1PLSr Involves the Regulation of Purple Leaf Sheath in Rice. International Journal of Molecular Sciences, 24(7), 6655. https://doi.org/10.3390/ijms24076655





