The Response of Triple-Negative Breast Cancer to Neoadjuvant Chemotherapy and the Epithelial–Mesenchymal Transition
Abstract
1. Introduction
2. Results
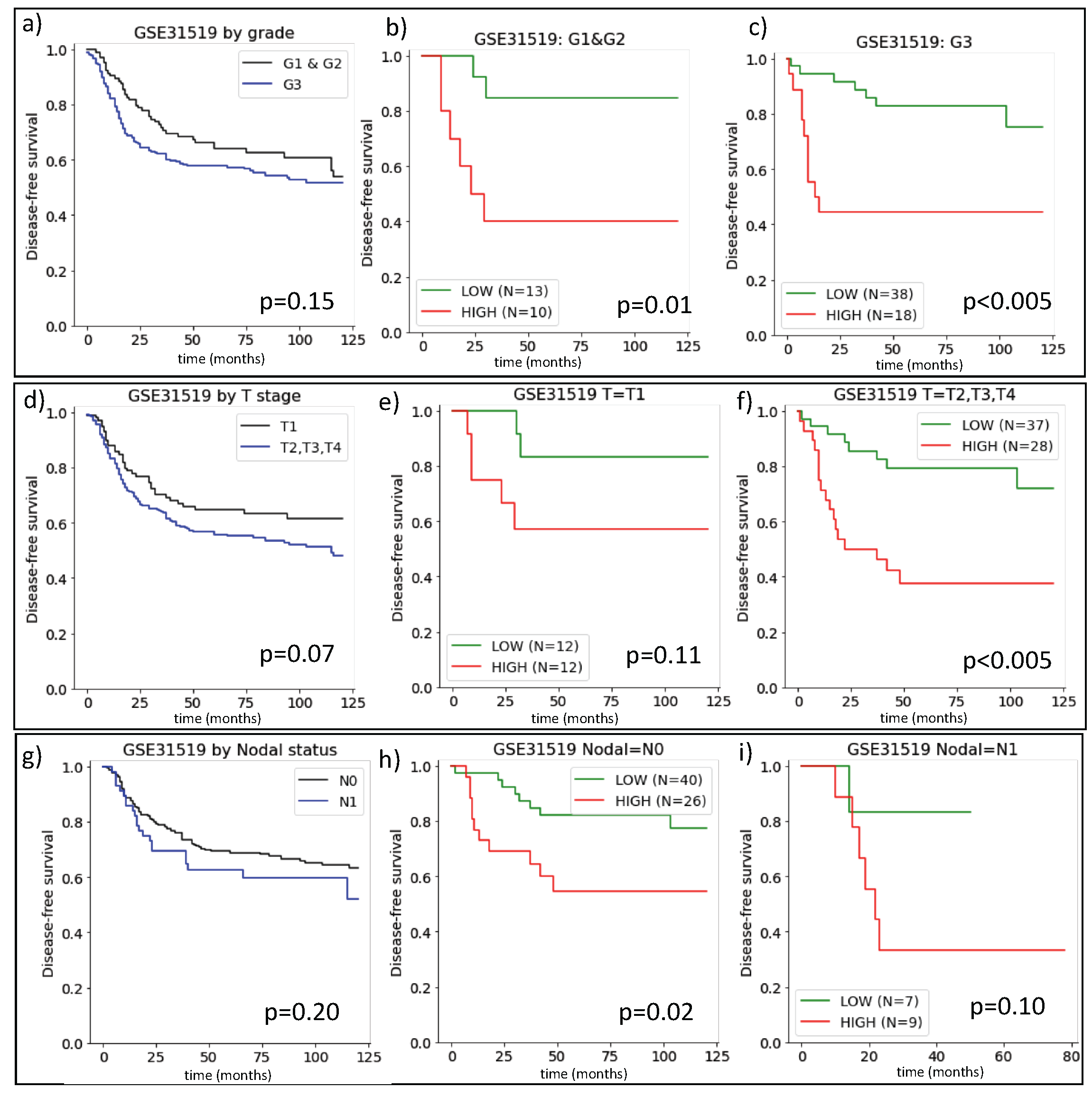
2.1. The Prognostic Value of ARIADNE for TNBC in Comparison with Grade, Stage and Nodal Status
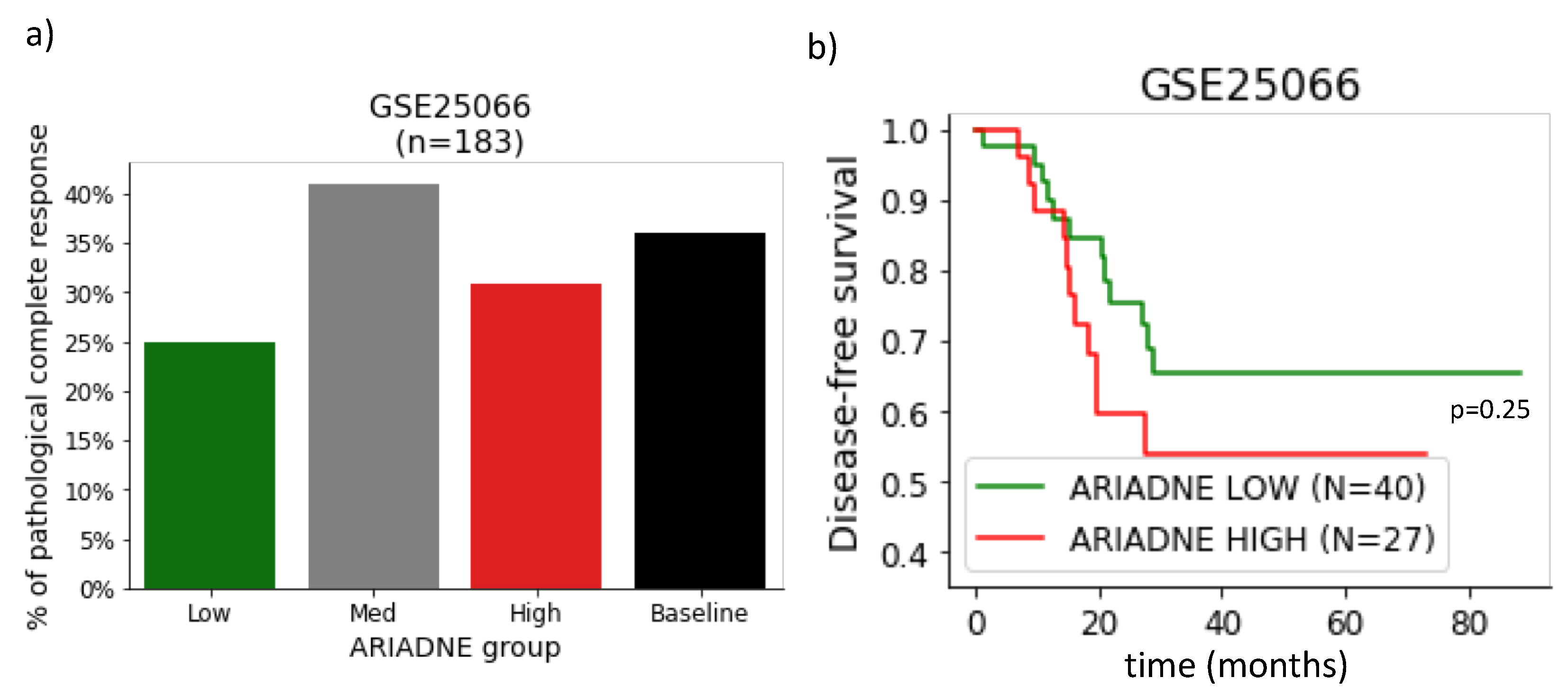
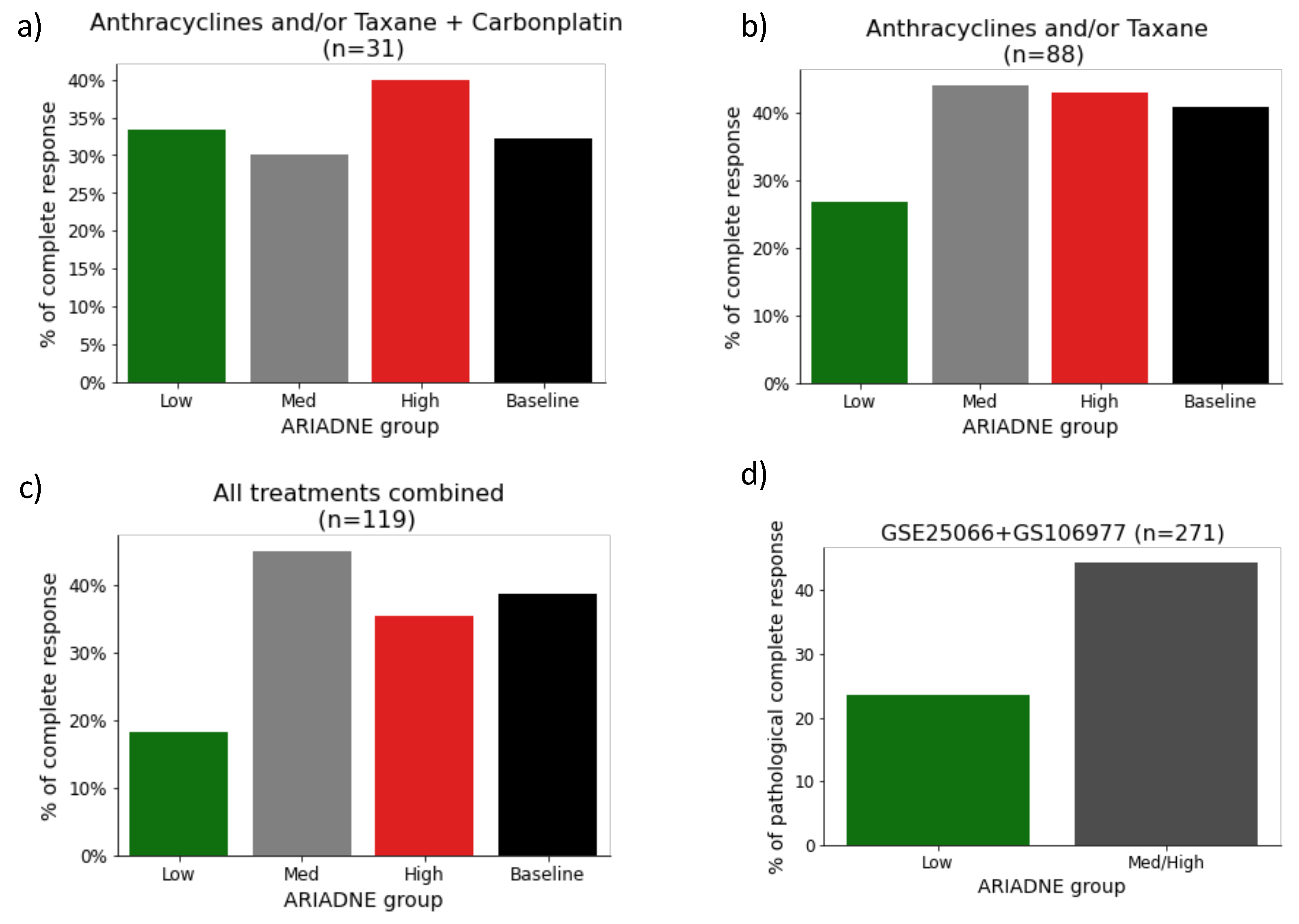
2.2. Evaluation of the Predictive Value of ARIADNE for the Response to Neoadjuvant Chemotherapy
3. Discussion
4. Materials and Methods
4.1. Gene Expression Data
4.2. Data Normalization and Computation of ARIADNE Scores
- 1.
- log2 transformation of MAS5 values;
- 2.
- Median centering of arrays;
- 3.
- Magnitude normalization of arrays.
4.3. Computation of Survival Curves
4.4. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Morris, G.J.; Naidu, S.; Topham, A.K.; Guiles, F.; Xu, Y.; McCue, P.; Schwartz, G.F.; Park, P.K.; Rosenberg, A.L.; Brill, K.; et al. Differences in breast carcinoma characteristics in newly diagnosed African–American and Caucasian patients: A single-institution compilation compared with the National Cancer Institute’s Surveillance, Epidemiology, and end results database. Cancer Interdiscip. Int. J. Am. Cancer Soc. 2007, 110, 876–884. [Google Scholar] [CrossRef]
- Bareche, Y.; Venet, D.; Ignatiadis, M.; Aftimos, P.; Piccart, M.; Rothe, F.; Sotiriou, C. Unravelling triple-negative breast cancer molecular heterogeneity using an integrative multiomic analysis. Ann. Oncol. 2018, 29, 895–902. [Google Scholar] [CrossRef]
- Jiang, Y.Z.; Ma, D.; Suo, C.; Shi, J.; Xue, M.; Hu, X.; Xiao, Y.; Yu, K.D.; Liu, Y.R.; Yu, Y.; et al. Genomic and Transcriptomic Landscape of Triple-Negative Breast Cancers: Subtypes and Treatment Strategies. Cancer Cell 2019, 35, 428–440.e5. [Google Scholar] [CrossRef]
- Bianchini, G.; Balko, J.M.; Mayer, I.A.; Sanders, M.E.; Gianni, L. Triple-negative breast cancer: Challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 2016, 13, 674–690. [Google Scholar] [CrossRef]
- Garrido-Castro, A.C.; Lin, N.U.; Polyak, K. Insights into Molecular Classifications of Triple-Negative Breast Cancer: Improving Patient Selection for Treatment. Cancer Discov. 2019, 9, 176–198. [Google Scholar] [CrossRef]
- Won, K.A.; Spruck, C. Triple-negative breast cancer therapy: Current and future perspectives (Review). Int. J. Oncol. 2020, 57, 1245–1261. [Google Scholar] [CrossRef] [PubMed]
- Bergin, A.R.T.; Loi, S. Triple-negative breast cancer: Recent treatment advances. F1000Res 2019, 8. [Google Scholar] [CrossRef]
- Lebert, J.M.; Lester, R.; Powell, E.; Seal, M.; McCarthy, J. Advances in the systemic treatment of triple-negative breast cancer. Curr. Oncol. 2018, 25, S142–S150. [Google Scholar] [CrossRef] [PubMed]
- Von Minckwitz, G.; Untch, M.; Blohmer, J.U.; Costa, S.D.; Eidtmann, H.; Fasching, P.A.; Gerber, B.; Eiermann, W.; Hilfrich, J.; Huober, J.; et al. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J. Clin. Oncol. 2012, 30, 1796–1804. [Google Scholar] [CrossRef]
- Bidard, F.C.; Matthieu, M.C.; Chollet, P.; Raoefils, I.; Abrial, C.; Dômont, J.; Spielmann, M.; Delaloge, S.; André, F.; Penault-Llorca, F. p53 status and efficacy of primary anthracyclines/alkylating agent-based regimen according to breast cancer molecular classes. Ann. Oncol. 2008, 19, 1261–1265. [Google Scholar] [CrossRef]
- Carey, L.A.; Dees, E.C.; Sawyer, L.; Gatti, L.; Moore, D.T.; Collichio, F.; Ollila, D.W.; Sartor, C.I.; Graham, M.L.; Perou, C.M. The triple negative paradox: Primary tumor chemosensitivity of breast cancer subtypes. Clin. Cancer Res. 2007, 13, 2329–2334. [Google Scholar] [CrossRef]
- Keam, B.; Im, S.A.; Kim, H.J.; Oh, D.Y.; Kim, J.H.; Lee, S.H.; Chie, E.K.; Han, W.; Kim, D.W.; Moon, W.K.; et al. Prognostic impact of clinicopathologic parameters in stage II/III breast cancer treated with neoadjuvant docetaxel and doxorubicin chemotherapy: Paradoxical features of the triple negative breast cancer. BMC Cancer 2007, 7, 203. [Google Scholar] [CrossRef] [PubMed]
- Huober, J.; von Minckwitz, G.; Denkert, C.; Tesch, H.; Weiss, E.; Zahm, D.M.; Belau, A.; Khandan, F.; Hauschild, M.; Thomssen, C.; et al. Effect of neoadjuvant anthracycline-taxane-based chemotherapy in different biological breast cancer phenotypes: Overall results from the GeparTrio study. Breast Cancer Res. Treat. 2010, 124, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Castrellon, A.B.; Pidhorecky, I.; Valero, V.; Raez, L.E. The role of carboplatin in the neoadjuvant chemotherapy treatment of triple negative breast cancer. Oncol. Rev. 2017, 11, 324. [Google Scholar] [CrossRef] [PubMed]
- Vidra, R.; Nemes, A.; Vidrean, A.; Pintea, S.; Tintari, S.; Deac, A.; Ciuleanu, T. Pathological complete response following cisplatin or carboplatin-based neoadjuvant chemotherapy for triple-negative breast cancer: A systematic review and meta-analysis. Exp. Ther. Med. 2022, 23, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Schmid, P.; Park, Y.H.; Munoz-Couselo, E.; Kim, S.B.; Sohn, J.; Im, S.A.; Holgado, E.; Wang, Y.; Dang, T.; Aktan, G.; et al. Pembrolizumab (pembro)+ chemotherapy (chemo) as neoadjuvant treatment for triple negative breast cancer (TNBC): Preliminary results from KEYNOTE-173. J. Clin. Oncol. 2017, 35, 556. [Google Scholar] [CrossRef]
- Schmid, P.; Park, Y.; Muñoz-Couselo, E.; Kim, S.; Sohn, J.; Im, S.; Holgado, E.; Foukakis, T.; Kuemmel, S.; Dent, R.; et al. Abstract PD5-01: KEYNOTE-173: Phase 1b multicohort study of pembrolizumab (Pembro) in combination with chemotherapy as neoadjuvant treatment for triple-negative breast cancer (TNBC). Cancer Res. 2019, 79, PD5-01. [Google Scholar] [CrossRef]
- Debien, V.; De Caluwé, A.; Wang, X.; Piccart-Gebhart, M.; Tuohy, V.K.; Romano, E.; Buisseret, L. Immunotherapy in breast cancer: An overview of current strategies and perspectives. NPJ Breast Cancer 2023, 9, 7. [Google Scholar] [CrossRef]
- Van den Ende, N.S.; Nguyen, A.H.; Jager, A.; Kok, M.; Debets, R.; van Deurzen, C.H. Triple-Negative Breast Cancer and Predictive Markers of Response to Neoadjuvant Chemotherapy: A Systematic Review. Int. J. Mol. Sci. 2023, 24, 2969. [Google Scholar] [CrossRef]
- Petrosyan, V.; Dobrolecki, L.E.; Thistlethwaite, L.; Lewis, A.N.; Sallas, C.; Srinivasan, R.R.; Lei, J.T.; Kovacevic, V.; Obradovic, P.; Ellis, M.J.; et al. Identifying biomarkers of differential chemotherapy response in TNBC patient-derived xenografts with a CTD/WGCNA approach. Iscience 2023, 26, 105799. [Google Scholar] [CrossRef]
- Paik, S.; Shak, S.; Tang, G.; Kim, C.; Baker, J.; Cronin, M.; Baehner, F.L.; Walker, M.G.; Watson, D.; Park, T.; et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 2004, 351, 2817–2826. [Google Scholar] [CrossRef] [PubMed]
- Paik, S.; Tang, G.; Shak, S.; Kim, C.; Baker, J.; Kim, W.; Cronin, M.; Baehner, F.L.; Watson, D.; Bryant, J.; et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J. Clin. Oncol. 2006, 24, 3726–3734. [Google Scholar] [CrossRef] [PubMed]
- Van’t Veer, L.J.; Dai, H.; Van De Vijver, M.J.; He, Y.D.; Hart, A.A.; Mao, M.; Peterse, H.L.; Van Der Kooy, K.; Marton, M.J.; Witteveen, A.T.; et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002, 415, 530–536. [Google Scholar] [CrossRef] [PubMed]
- Buyse, M.; Loi, S.; Van’t Veer, L.; Viale, G.; Delorenzi, M.; Glas, A.M.; Saghatchian d’Assignies, M.; Bergh, J.; Lidereau, R.; Ellis, P.; et al. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J. Natl. Cancer Inst. 2006, 98, 1183–1192. [Google Scholar] [CrossRef]
- Ein-Dor, L.; Kela, I.; Getz, G.; Givol, D.; Domany, E. Outcome signature genes in breast cancer: Is there a unique set? Bioinformatics 2005, 21, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Drier, Y.; Domany, E. Do two machine-learning based prognostic signatures for breast cancer capture the same biological processes? PLoS ONE 2011, 6, e17795. [Google Scholar] [CrossRef]
- La Porta, C.A.; Zapperi, S. Artificial intelligence in breast cancer diagnostics. Cell Rep. Med. 2022, 3, 100851. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef]
- Burstein, M.D.; Tsimelzon, A.; Poage, G.M.; Covington, K.R.; Contreras, A.; Fuqua, S.A.; Savage, M.I.; Osborne, C.K.; Hilsenbeck, S.G.; Chang, J.C.; et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin. Cancer Res. 2015, 21, 1688–1698. [Google Scholar] [CrossRef]
- Yu, G.; Li, X.; He, T.F.; Gruosso, T.; Zuo, D.; Souleimanova, M.; Ramos, V.M.; Omeroglu, A.; Meterissian, S.; Guiot, M.C.; et al. Predicting Relapse in Patients With Triple Negative Breast Cancer (TNBC) Using a Deep-Learning Approach. Front. Physiol. 2020, 11, 511071. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Jovanović, B.; Chen, X.; Estrada, M.V.; Johnson, K.N.; Shyr, Y.; Moses, H.L.; Sanders, M.E.; Pietenpol, J.A. Refinement of triple-negative breast cancer molecular subtypes: Implications for neoadjuvant chemotherapy selection. PLoS ONE 2016, 11, e0157368. [Google Scholar] [CrossRef] [PubMed]
- Font-Clos, F.; Zapperi, S.; La Porta, C.A.M. Classification of triple-negative breast cancers through a Boolean network model of the epithelial-mesenchymal transition. Cell Syst. 2021, 12, 457–462.e4. [Google Scholar] [CrossRef]
- Huber, M.A.; Kraut, N.; Beug, H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr. Opin. Cell Biol. 2005, 17, 548–558. [Google Scholar] [CrossRef]
- Rhim, A.D.; Mirek, E.T.; Aiello, N.M.; Maitra, A.; Bailey, J.M.; McAllister, F.; Reichert, M.; Beatty, G.L.; Rustgi, A.K.; Vonderheide, R.H.; et al. EMT and dissemination precede pancreatic tumor formation. Cell 2012, 148, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Sarrió, D.; Rodriguez-Pinilla, S.M.; Hardisson, D.; Cano, A.; Moreno-Bueno, G.; Palacios, J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008, 68, 989–997. [Google Scholar] [CrossRef] [PubMed]
- Aleskandarany, M.A.; Negm, O.H.; Green, A.R.; Ahmed, M.A.H.; Nolan, C.C.; Tighe, P.J.; Ellis, I.O.; Rakha, E.A. Epithelial mesenchymal transition in early invasive breast cancer: An immunohistochemical and reverse phase protein array study. Breast Cancer Res. Treat. 2014, 145, 339–348. [Google Scholar] [CrossRef]
- Grosse-Wilde, A.; Fouquier d’Hérouël, A.; McIntosh, E.; Ertaylan, G.; Skupin, A.; Kuestner, R.E.; del Sol, A.; Walters, K.A.; Huang, S. Stemness of the hybrid Epithelial/Mesenchymal State in Breast Cancer and Its Association with Poor Survival. PLoS ONE 2015, 10, e0126522. [Google Scholar] [CrossRef]
- Bitterman, P.; Chun, B.; Kurman, R.J. The significance of epithelial differentiation in mixed mesodermal tumors of the uterus. A clinicopathologic and immunohistochemical study. Am. J. Surg. Pathol. 1990, 14, 317–328. [Google Scholar] [CrossRef]
- Haraguchi, S.; Fukuda, Y.; Sugisaki, Y.; Yamanaka, N. Pulmonary carcinosarcoma: Immunohistochemical and ultrastructural studies. Pathol. Int. 1999, 49, 903–908. [Google Scholar] [CrossRef]
- Paniz Mondolfi, A.E.; Jour, G.; Johnson, M.; Reidy, J.; Cason, R.C.; Barkoh, B.A.; Benaim, G.; Singh, R.; Luthra, R. Primary cutaneous carcinosarcoma: Insights into its clonal origin and mutational pattern expression analysis through next-generation sequencing. Hum. Pathol. 2013, 44, 2853–2860. [Google Scholar] [CrossRef]
- Revenu, C.; Gilmour, D. EMT 2.0: Shaping epithelia through collective migration. Curr. Opin. Genet. Dev. 2009, 19, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Bardia, A.; Wittner, B.S.; Stott, S.L.; Smas, M.E.; Ting, D.T.; Isakoff, S.J.; Ciciliano, J.C.; Wells, M.N.; Shah, A.M.; et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Jolly, M.K.; Tripathi, S.C.; Jia, D.; Mooney, S.M.; Celiktas, M.; Hanash, S.M.; Mani, S.A.; Pienta, K.J.; Ben-Jacob, E.; Levine, H. Stability of the hybrid epithelial/mesenchymal phenotype. Oncotarget 2016, 7, 27067–27084. [Google Scholar] [CrossRef] [PubMed]
- George, J.T.; Jolly, M.K.; Xu, S.; Somarelli, J.A.; Levine, H. Survival Outcomes in Cancer Patients Predicted by a Partial EMT Gene Expression Scoring Metric. Cancer Res. 2017, 77, 6415–6428. [Google Scholar] [CrossRef] [PubMed]
- Pastushenko, I.; Brisebarre, A.; Sifrim, A.; Fioramonti, M.; Revenco, T.; Boumahdi, S.; Van Keymeulen, A.; Brown, D.; Moers, V.; Lemaire, S.; et al. Identification of the tumour transition states occurring during EMT. Nature 2018, 556, 463–468. [Google Scholar] [CrossRef]
- Chakraborty, P.; George, J.T.; Tripathi, S.; Levine, H.; Jolly, M.K. Comparative Study of Transcriptomics-Based Scoring Metrics for the Epithelial-Hybrid-Mesenchymal Spectrum. Front. Bioeng. Biotechnol. 2020, 8, 220. [Google Scholar] [CrossRef]
- Font-Clos, F.; Zapperi, S.; La Porta, C.A.M. Topography of epithelial-mesenchymal plasticity. Proc. Natl. Acad. Sci. USA 2018, 115, 5902–5907. [Google Scholar] [CrossRef]
- Font-Clos, F.; Zapperi, S.; La Porta, C.A. Classification of triple negative breast cancer by epithelial mesenchymal transition and the tumor immune microenvironment. Sci. Rep. 2022, 12, 9651. [Google Scholar] [CrossRef]
- Gruosso, T.; Gigoux, M.; Manem, V.S.K.; Bertos, N.; Zuo, D.; Perlitch, I.; Saleh, S.M.I.; Zhao, H.; Souleimanova, M.; Johnson, R.M.; et al. Spatially distinct tumor immune microenvironments stratify triple-negative breast cancers. J. Clin. Investig. 2019, 129, 1785–1800. [Google Scholar] [CrossRef]
- Jia, X.; Wang, K.; Xu, L.; Li, N.; Zhao, Z.; Li, M. A systematic review and meta-analysis of BRCA1/2 mutation for predicting the effect of platinum-based chemotherapy in triple-negative breast cancer. Breast 2022, 66, 31–39. [Google Scholar] [CrossRef]
- Hatzis, C.; Pusztai, L.; Valero, V.; Booser, D.J.; Esserman, L.; Lluch, A.; Vidaurre, T.; Holmes, F.; Souchon, E.; Wang, H.; et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA 2011, 305, 1873–1881. [Google Scholar] [CrossRef] [PubMed]
- Santonja, A.; Sánchez-Muñoz, A.; Lluch, A.; Chica-Parrado, M.R.; Albanell, J.; Chacón, J.I.; Antolín, S.; Jerez, J.M.; de la Haba, J.; de Luque, V.; et al. Triple negative breast cancer subtypes and pathologic complete response rate to neoadjuvant chemotherapy. Oncotarget 2018, 9, 26406. [Google Scholar] [CrossRef] [PubMed]
- Karn, T.; Rody, A.; Müller, V.; Schmidt, M.; Becker, S.; Holtrich, U.; Pusztai, L. Control of dataset bias in combined Affymetrix cohorts of triple negative breast cancer. Genom. Data 2014, 2, 354–356. [Google Scholar] [CrossRef] [PubMed]

| Dataset | No. Patients | Treatment | Analysis Performed |
|---|---|---|---|
| GSE31519 | 383 | Not available. | Statification, according to GNT and survival. |
| GSE25066 | 183 | Anthracycline/taxane. | Drug response and survival. |
| GSE10677 | 119 | Anthracycline/taxane and carboplatin. | Drug response. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zapperi, S.; La Porta, C.A.M. The Response of Triple-Negative Breast Cancer to Neoadjuvant Chemotherapy and the Epithelial–Mesenchymal Transition. Int. J. Mol. Sci. 2023, 24, 6422. https://doi.org/10.3390/ijms24076422
Zapperi S, La Porta CAM. The Response of Triple-Negative Breast Cancer to Neoadjuvant Chemotherapy and the Epithelial–Mesenchymal Transition. International Journal of Molecular Sciences. 2023; 24(7):6422. https://doi.org/10.3390/ijms24076422
Chicago/Turabian StyleZapperi, Stefano, and Caterina A. M. La Porta. 2023. "The Response of Triple-Negative Breast Cancer to Neoadjuvant Chemotherapy and the Epithelial–Mesenchymal Transition" International Journal of Molecular Sciences 24, no. 7: 6422. https://doi.org/10.3390/ijms24076422
APA StyleZapperi, S., & La Porta, C. A. M. (2023). The Response of Triple-Negative Breast Cancer to Neoadjuvant Chemotherapy and the Epithelial–Mesenchymal Transition. International Journal of Molecular Sciences, 24(7), 6422. https://doi.org/10.3390/ijms24076422







