WHIRLY1 Acts Upstream of ABA-Related Reprogramming of Drought-Induced Gene Expression in Barley and Affects Stress-Related Histone Modifications
Abstract
1. Introduction
2. Results
2.1. Overexpression of HvWHIRLY1 Delays Drought-Induced Onset of Senescence
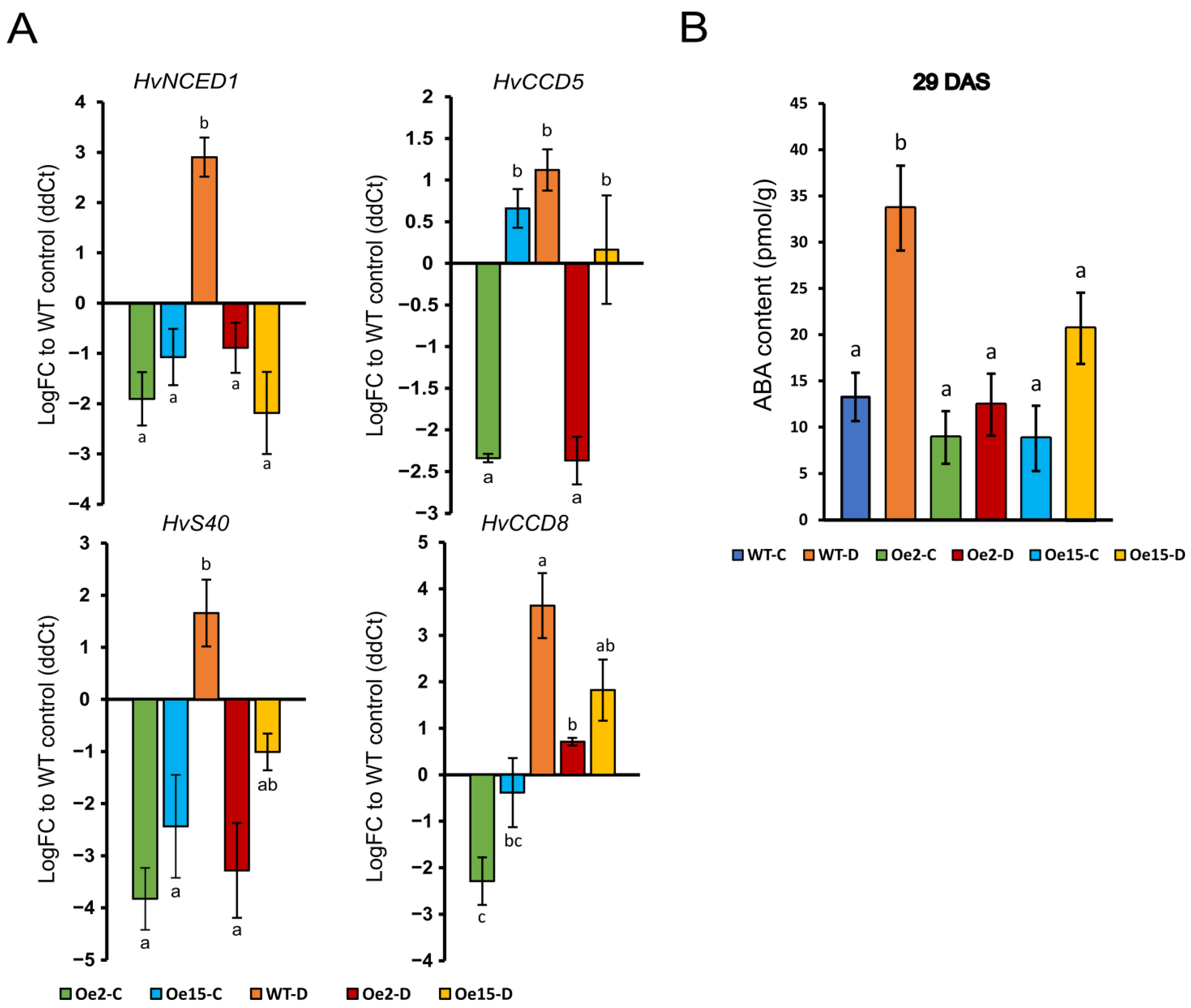
2.2. Overexpression of WHIRLY1 Suppressed Drought-Induced Expression of ABA-Related Genes and ABA Accumulation
2.3. Overexpression of WHIRLY1 Affects Reprogramming of Gene Expression during Drought Stress
2.4. Overexpression of WHIRLY1 Causes Loss of Euchromatic Marks in the ABA-Related Genes HvNCED1 and HvS40
3. Discussion
4. Material and Methods
4.1. Plant Materials, Growth Conditions and Drought Treatment
4.2. Physiological Measurements
4.2.1. Relative Chlorophyll Content
4.2.2. PS II Efficiency
4.3. Gene Expression Analysis
4.4. Transcriptome Analysis
4.5. Chromatin Immunoprecipitation (ChIP) Analysis
4.6. Quantification of Abscisic Acid (ABA)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Seppelt, R.; Klotz, S.; Peiter, E.; Volk, M. Agriculture and food security under a changing climate: An underestimated challenge. iScience 2022, 25, 105551. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, Y.; Osakabe, K.; Shinozaki, K.; Tran, L.-S. Response of plants to water stress. Front. Plant Sci. 2014, 5, 86. [Google Scholar] [CrossRef]
- Khan, I.U.; Ali, A.; Khan, H.A.; Baek, D.; Park, J.; Lim, C.J.; Zareen, S.; Jan, M.; Lee, S.Y.; Pardo, J.M.; et al. PWR/HDA9/ABI4 Complex Epigenetically Regulates ABA Dependent Drought Stress Tolerance in Arabidopsis. Front. Plant Sci. 2020, 11, 623. [Google Scholar] [CrossRef] [PubMed]
- Bouvier, F.; d’Harlingue, A.; Hugueney, P.; Marin, E.; Marion-Poll, A.; Camara, B. Xanthophyll biosynthesis. Cloning, expression, functional reconstitution, and regulation of beta-cyclohexenyl carotenoid epoxidase from pepper (Capsicum annuum). J. Biol. Chem. 1996, 271, 28861–28867. [Google Scholar] [CrossRef]
- Seiler, C.; Harshavardhan, V.T.; Rajesh, K.; Reddy, P.S.; Strickert, M.; Rolletschek, H.; Scholz, U.; Wobus, U.; Sreenivasulu, N. ABA biosynthesis and degradation contributing to ABA homeostasis during barley seed development under control and terminal drought-stress conditions. J. Exp. Bot. 2011, 62, 2615–2632. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Guo, Y.; Liu, Y.; Zhang, F.; Wang, Z.; Wang, H.; Wang, F.; Li, D.; Mao, D.; Luan, S.; et al. 9-cis-Epoxycarotenoid Dioxygenase 3 Regulates Plant Growth and Enhances Multi-Abiotic Stress Tolerance in Rice. Front. Plant Sci. 2018, 9, 162. [Google Scholar] [CrossRef]
- Biswal, B.; Joshi, P.N.; Raval, M.K.; Biswal, U.C. Photosynthesis, a global sensor of environmental stress in green plants: Stress signalling and adaptation. Curr. Sci. 2011, 101, 47–56. [Google Scholar]
- Liebers, M.; Cozzi, C.; Uecker, F.; Chambon, L.; Blanvillain, R.; Pfannschmidt, T. Biogenic signals from plastids and their role in chloroplast development. J. Exp. Bot. 2022, 73, 7105–7125. [Google Scholar] [CrossRef]
- Endo, A.; Sawada, Y.; Takahashi, H.; Okamoto, M.; Ikegami, K.; Koiwai, H.; Seo, M.; Toyomasu, T.; Mitsuhashi, W.; Shinozaki, K.; et al. Drought induction of Arabidopsis 9-cis-epoxycarotenoid dioxygenase occurs in vascular parenchyma cells. Plant Physiol. 2008, 147, 1984–1993. [Google Scholar] [CrossRef]
- Kim, J.M.; Sasaki, T.; Ueda, M.; Sako, K.; Seki, M. Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front. Plant Sci. 2015, 6, 114. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; He, Z. Small DNA Methylation, Big Player in Plant Abiotic Stress Responses and Memory. Front. Plant Sci. 2020, 11, 595603. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.K. Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 2009, 12, 133–139. [Google Scholar] [CrossRef]
- Janack, B.; Sosoi, P.; Krupinska, K.; Humbeck, K. Knockdown of WHIRLY1 Affects Drought Stress-Induced Leaf Senescence and Histone Modifications of the Senescence-Associated Gene HvS40. Plants 2016, 5, 37. [Google Scholar] [CrossRef] [PubMed]
- Vendramin, S.; Huang, J.; Crisp, P.A.; Madzima, T.F.; McGinnis, K.M. Epigenetic Regulation of ABA-Induced Transcriptional Responses in Maize. G3 Genes Genomes Genet. 2020, 10, 1727–1743. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.E.; West, C.E.; Foyer, C.H. WHIRLY protein functions in plants. Food Energy Secur. 2023, 12, e379. [Google Scholar] [CrossRef]
- Krupinska, K.; Desel, C.; Frank, S.; Hensel, G. WHIRLIES Are Multifunctional DNA-Binding Proteins With Impact on Plant Development and Stress Resistance. Front. Plant Sci. 2022, 13, 880423. [Google Scholar] [CrossRef] [PubMed]
- Desveaux, D.; Subramaniam, R.; Després, C.; Mess, J.N.; Lévesque, C.; Fobert, P.R.; Dangl, J.L.; Brisson, N. A “Whirly” transcription factor is required for salicylic acid-dependent disease resistance in Arabidopsis. Dev. Cell 2004, 6, 229–240. [Google Scholar] [CrossRef]
- Desveaux, D.; Després, C.; Joyeux, A.; Subramaniam, R.; Brisson, N. PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10a gene activation in potato. Plant Cell 2000, 12, 1477–1489. [Google Scholar] [CrossRef]
- Akbudak, M.A.; Filiz, E. Whirly (Why) transcription factors in tomato (Solanum lycopersicum L.): Genome-wide identification and transcriptional profiling under drought and salt stresses. Mol. Biol. Rep. 2019, 46, 4139–4150. [Google Scholar] [CrossRef]
- Zhuang, K.; Kong, F.; Zhang, S.; Meng, C.; Yang, M.; Liu, Z.; Wang, Y.; Ma, N.; Meng, Q. Whirly1 enhances tolerance to chilling stress in tomato via protection of photosystem II and regulation of starch degradation. New Phytol. 2019, 221, 1998–2012. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, K.; Wang, J.; Jiao, B.; Chen, C.; Zhang, J.; Ma, N.; Meng, Q. WHIRLY1 maintains leaf photosynthetic capacity in tomato by regulating the expression of RbcS1 under chilling stress. J. Exp. Bot. 2020, 71, 3653–3663. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Liu, W.; Wei, Y.; Shi, H. MeCIPK23 interacts with Whirly transcription factors to activate abscisic acid biosynthesis and regulate drought resistance in cassava. Plant Biotechnol. J. 2020, 18, 1504–1506. [Google Scholar] [CrossRef]
- Grabowski, E.; Miao, Y.; Mulisch, M.; Krupinska, K. Single-stranded DNA-binding protein Whirly1 in barley leaves is located in plastids and the nucleus of the same cell. Plant Physiol. 2008, 147, 1800–1804. [Google Scholar] [CrossRef] [PubMed]
- Isemer, R.; Mulisch, M.; Schäfer, A.; Kirchner, S.; Koop, H.U.; Krupinska, K. Recombinant Whirly1 translocates from transplastomic chloroplasts to the nucleus. FEBS Lett. 2012, 586, 85–88. [Google Scholar] [CrossRef]
- Pfalz, J.; Liere, K.; Kandlbinder, A.; Dietz, K.J.; Oelmüller, R. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 2006, 18, 176–197. [Google Scholar] [CrossRef] [PubMed]
- Melonek, J.; Mulisch, M.; Schmitz-Linneweber, C.; Grabowski, E.; Hensel, G.; Krupinska, K. Whirly1 in chloroplasts associates with intron containing RNAs and rarely co-localizes with nucleoids. Planta 2010, 232, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Majeran, W.; Friso, G.; Asakura, Y.; Qu, X.; Huang, M.; Ponnala, L.; Watkins, K.P.; Barkan, A.; van Wijk, K.J. Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: A new conceptual framework for nucleoid functions. Plant Physiol. 2012, 158, 156–189. [Google Scholar] [CrossRef]
- Krupinska, K.; Dähnhardt, D.; Fischer-Kilbienski, I.; Kucharewicz, W.; Scharrenberg, C.; Trösch, M.; Buck, F. Identification of WHIRLY1 as a Factor Binding to the Promoter of the Stress- and Senescence-Associated Gene HvS40. J. Plant Growth Regul. 2014, 33, 91–105. [Google Scholar] [CrossRef]
- Nia, M.S.; Frank, S.; Schäfer, A.; Desel, C.; Mulisch, M.; Voigt, U.; Nowara, D.; Tandron Moya, Y.A.; Bilger, W.; Wiren, N.v.; et al. The balance between growth and resistance is shifted to the latter by over-accumulation of chloroplast-nucleus located WHIRLY1 in barley. bioRxiv 2023. [Google Scholar] [CrossRef]
- Bucchini, F.; Del Cortona, A.; Kreft, Ł.; Botzki, A.; Van Bel, M.; Vandepoele, K. TRAPID 2.0: A web application for taxonomic and functional analysis of de novo transcriptomes. Nucleic Acids Res. 2021, 49, e101. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Fang, L.; Li, X.; Liu, J.; Liu, F. Endogenous ABA level modulates the effects of CO2 elevation and soil water deficit on growth, water and nitrogen use efficiencies in barley and tomato plants. Agric. Water Manag. 2021, 249, 106808. [Google Scholar] [CrossRef]
- Han, Y.; Watanabe, S.; Shimada, H.; Sakamoto, A. Dynamics of the leaf endoplasmic reticulum modulate β-glucosidase-mediated stress-activated ABA production from its glucosyl ester. J. Exp. Bot. 2020, 71, 2058–2071. [Google Scholar] [CrossRef] [PubMed]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef]
- Wu, J.; Kamanga, B.M.; Zhang, W.; Xu, Y.; Xu, L. Research progress of aldehyde oxidases in plants. PeerJ 2022, 10, e13119. [Google Scholar] [CrossRef]
- Saeed, F.; Chaudhry, U.K.; Bakhsh, A.; Raza, A.; Saeed, Y.; Bohra, A.; Varshney, R.K. Moving Beyond DNA Sequence to Improve Plant Stress Responses. Front. Genet. 2022, 13, 874648. [Google Scholar] [CrossRef]
- Chang, Y.N.; Zhu, C.; Jiang, J.; Zhang, H.; Zhu, J.K.; Duan, C.G. Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant Biol. 2020, 62, 563–580. [Google Scholar] [CrossRef]
- Comadira, G.; Rasool, B.; Kaprinska, B.; García, B.M.; Morris, J.; Verrall, S.R.; Bayer, M.; Hedley, P.E.; Hancock, R.D.; Foyer, C.H. WHIRLY1 Functions in the Control of Responses to Nitrogen Deficiency But Not Aphid Infestation in Barley. Plant Physiol. 2015, 168, 1140–1151. [Google Scholar] [CrossRef]
- Miao, Y.; Jiang, J.; Ren, Y.; Zhao, Z. The Single-Stranded DNA-Binding Protein WHIRLY1 Represses WRKY53 Expression and Delays Leaf Senescence in a Developmental Stage-Dependent Manner in Arabidopsis. Plant Physiol. 2013, 163, 746–756. [Google Scholar] [CrossRef]
- Rothman-Denes, L.B.; Dai, X.; Davydova, E.; Carter, R.; Kazmierczak, K. Transcriptional regulation by DNA structural transitions and single-stranded DNA-binding proteins. Cold Spring Harb. Symp. Quant. Biol. 1998, 63, 63–73. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, Y.; Huang, D.; Liu, H.; Justin, N.; Zhao, W.; Liu, J.; Peng, Y. Structural features of the single-stranded DNA-binding protein MoSub1 from Magnaporthe oryzae. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Niehrs, C.; Luke, B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat. Rev. Mol. Cell Biol. 2020, 21, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.; Wang, G.G. R-loop and its functions at the regulatory interfaces between transcription and (epi)genome. Biochim. Biophys. Acta Gene Regul. Mech. 2021, 1864, 194750. [Google Scholar] [CrossRef]
- Guo, J.T.; Malik, F. Single-Stranded DNA Binding Proteins and Their Identification Using Machine Learning-Based Approaches. Biomolecules 2022, 12, 1187. [Google Scholar] [CrossRef]
- Kobayashi, K.; Endo, K.; Wada, H. Multiple Impacts of Loss of Plastidic Phosphatidylglycerol Biosynthesis on Photosynthesis during Seedling Growth of Arabidopsis. Front. Plant Sci. 2016, 7, 336. [Google Scholar] [CrossRef] [PubMed]
- Melonek, J.; Oetke, S.; Krupinska, K. Multifunctionality of plastid nucleoids as revealed by proteome analyses. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2016, 1864, 1016–1038. [Google Scholar] [CrossRef]
- Powikrowska, M.; Oetke, S.; Jensen, P.E.; Krupinska, K. Dynamic composition, shaping and organization of plastid nucleoids. Front. Plant Sci. 2014, 5, 424. [Google Scholar] [CrossRef]
- Berens, M.L.; Wolinska, K.W.; Spaepen, S.; Ziegler, J.; Nobori, T.; Nair, A.; Krüler, V.; Winkelmüller, T.M.; Wang, Y.; Mine, A.; et al. Balancing trade-offs between biotic and abiotic stress responses through leaf age-dependent variation in stress hormone cross-talk. Proc. Natl. Acad. Sci. USA 2019, 116, 2364–2373. [Google Scholar] [CrossRef]
- Parasyri, A.; Barth, O.; Zschiesche, W.; Humbeck, K. The Barley Heavy Metal Associated Isoprenylated Plant Protein HvFP1 Is Involved in a Crosstalk between the Leaf Development and Abscisic Acid-Related Drought Stress Responses. Plants 2022, 11, 2851. [Google Scholar] [CrossRef]
- Desveaux, D.; Maréchal, A.; Brisson, N. Whirly transcription factors: Defense gene regulation and beyond. Trends Plant Sci. 2005, 10, 95–102. [Google Scholar] [CrossRef]
- Witzel, F.; Maddison, L.; Blüthgen, N. How scaffolds shape MAPK signaling: What we know and opportunities for systems approaches. Front. Physiol. 2012, 3, 475. [Google Scholar] [CrossRef] [PubMed]
- Casar, B.; Crespo, P. ERK Signals: Scaffolding Scaffolds? Front. Cell Dev. Biol. 2016, 4, 49. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Pan, C.Q.; Li, B.; Tucker-Kellogg, L.; Tidor, B.; Chen, Y.; Low, B.C. Simulating EGFR-ERK Signaling Control by Scaffold Proteins KSR and MP1 Reveals Differential Ligand-Sensitivity Co-Regulated by Cbl-CIN85 and Endophilin. PLoS ONE 2011, 6, e22933. [Google Scholar] [CrossRef] [PubMed]
- Levchenko, A.; Bruck, J.; Sternberg, P.W. Scaffold proteins may biphasically affect the levels of mitogen-activated protein kinase signaling and reduce its threshold properties. Proc. Natl. Acad. Sci. USA 2000, 97, 5818–5823. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Lan, W.; Ma, W.; Huang, R.; Lin, W.; Li, M.; Chen, C.Y.; Wu, K.; Miao, Y. WHIRLY1 recruits the histone deacetylase HDA15 repressing leaf senescence and flowering in Arabidopsis. J. Integr. Plant Biol. 2022, 64, 1411–1429. [Google Scholar] [CrossRef]
- Liu, S.; Li, M.; Su, L.; Ge, K.; Li, L.; Li, X.; Liu, X.; Li, L. Negative feedback regulation of ABA biosynthesis in peanut (Arachis hypogaea): A transcription factor complex inhibits AhNCED1 expression during water stress. Sci. Rep. 2016, 6, 37943. [Google Scholar] [CrossRef]
- Jamsheer, K.M.; Jindal, S.; Sharma, M.; Awasthi, P.; Sreejath, S.; Sharma, M.; Mannully, C.T.; Laxmi, A. A negative feedback loop of TOR signaling balances growth and stress-response trade-offs in plants. Cell Rep. 2022, 39, 110631. [Google Scholar] [CrossRef]
- Wehner, G.; Balko, C.; Ordon, F. Experimental Design to Determine Drought Stress Response and Early Leaf Senescence in Barley (Hordeum vulgare L.). Bio-Protocol 2016, 6, e1749. [Google Scholar] [CrossRef]
- Zhu, J.; Tremblay, N.; Liang, Y. Comparing SPAD and atLEAF values for chlorophyll assessment in crop species. Can. J. Soil Sci. 2012, 92, 645–648. [Google Scholar] [CrossRef]
- Genty, B.; Briantais, J.-M.; Baker, N.R. The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim. Biophys. Acta (BBA)-Gen. Subj. 1989, 990, 87–92. [Google Scholar] [CrossRef]
- Chomczynski, P.; Mackey, K. Short technical reports. Modification of the TRI reagent procedure for isolation of RNA from polysaccharide- and proteoglycan-rich sources. BioTechniques 1995, 19, 942–945. [Google Scholar] [PubMed]
- Mascher, M. Pseudomolecules and annotation of the second version of the reference genome sequence assembly of barley cv. Morex [Morex V2]. 2019. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013, 41, e108. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Gendrel, A.V.; Lippman, Z.; Yordan, C.; Colot, V.; Martienssen, R.A. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 2002, 297, 1871–1873. [Google Scholar] [CrossRef]
- Ay, N.; Irmler, K.; Fischer, A.; Uhlemann, R.; Reuter, G.; Humbeck, K. Epigenetic programming via histone methylation at WRKY53 controls leaf senescence in Arabidopsis thaliana. Plant J. 2009, 58, 333–346. [Google Scholar] [CrossRef]
- Johnson, L.; Cao, X.; Jacobsen, S. Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 2002, 12, 1360–1367. [Google Scholar] [CrossRef]
- Balcke, G.U.; Handrick, V.; Bergau, N.; Fichtner, M.; Henning, A.; Stellmach, H.; Tissier, A.; Hause, B.; Frolov, A. An UPLC-MS/MS method for highly sensitive high-throughput analysis of phytohormones in plant tissues. Plant Methods 2012, 8, 47. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manh, M.B.; Ost, C.; Peiter, E.; Hause, B.; Krupinska, K.; Humbeck, K. WHIRLY1 Acts Upstream of ABA-Related Reprogramming of Drought-Induced Gene Expression in Barley and Affects Stress-Related Histone Modifications. Int. J. Mol. Sci. 2023, 24, 6326. https://doi.org/10.3390/ijms24076326
Manh MB, Ost C, Peiter E, Hause B, Krupinska K, Humbeck K. WHIRLY1 Acts Upstream of ABA-Related Reprogramming of Drought-Induced Gene Expression in Barley and Affects Stress-Related Histone Modifications. International Journal of Molecular Sciences. 2023; 24(7):6326. https://doi.org/10.3390/ijms24076326
Chicago/Turabian StyleManh, Minh Bui, Charlotte Ost, Edgar Peiter, Bettina Hause, Karin Krupinska, and Klaus Humbeck. 2023. "WHIRLY1 Acts Upstream of ABA-Related Reprogramming of Drought-Induced Gene Expression in Barley and Affects Stress-Related Histone Modifications" International Journal of Molecular Sciences 24, no. 7: 6326. https://doi.org/10.3390/ijms24076326
APA StyleManh, M. B., Ost, C., Peiter, E., Hause, B., Krupinska, K., & Humbeck, K. (2023). WHIRLY1 Acts Upstream of ABA-Related Reprogramming of Drought-Induced Gene Expression in Barley and Affects Stress-Related Histone Modifications. International Journal of Molecular Sciences, 24(7), 6326. https://doi.org/10.3390/ijms24076326





