Influence of MMR, MGMT Promotor Methylation and Protein Expression on Overall and Progression-Free Survival in Primary Glioblastoma Patients Treated with Temozolomide
Abstract
1. Introduction
2. Results
2.1. Patient Characteristics
2.2. Overall Survival, Sex, Age and Resection Association
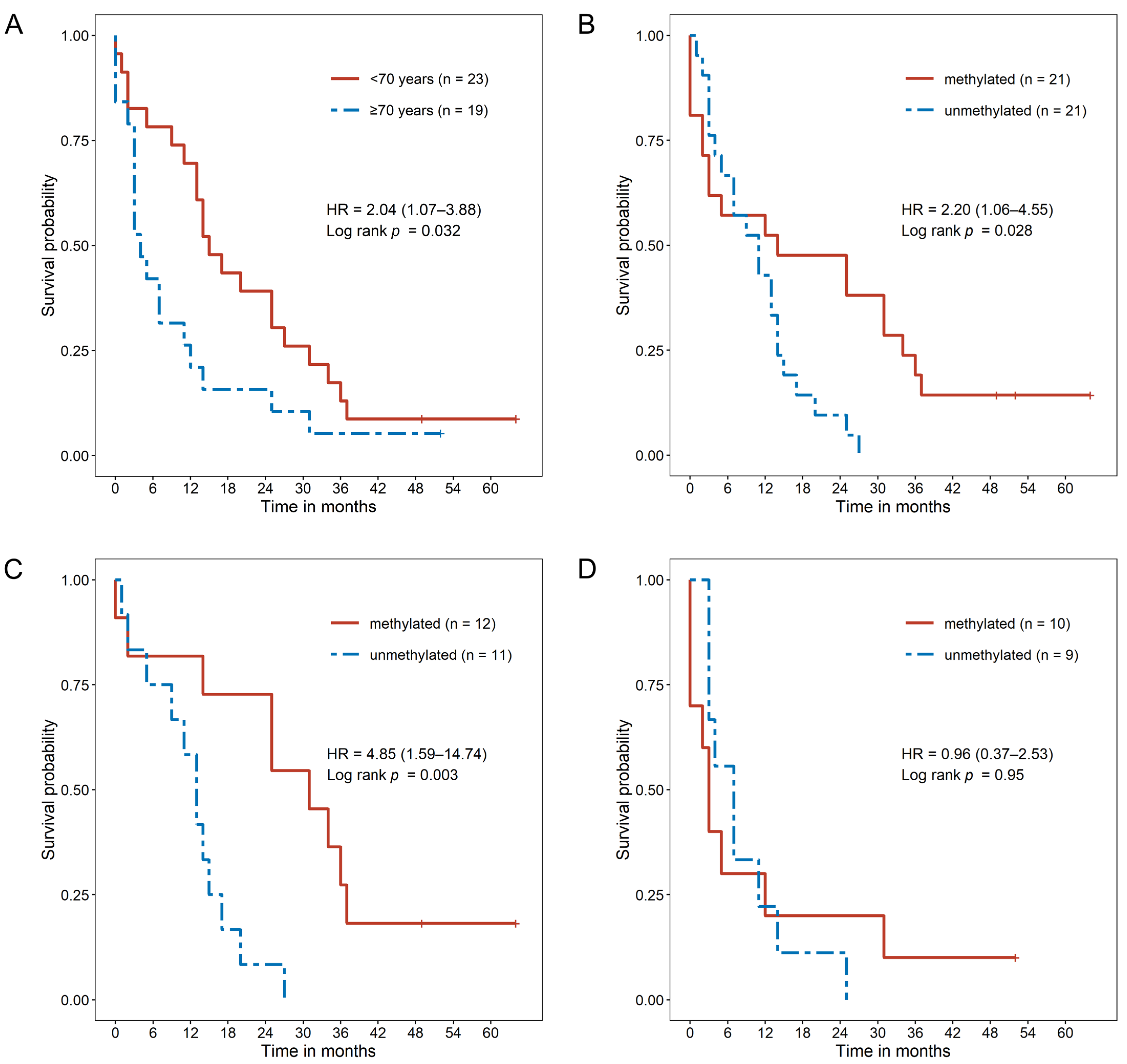
2.3. MGMT Promoter Methylation Causes a Survival Benefit in Younger GBM Patients
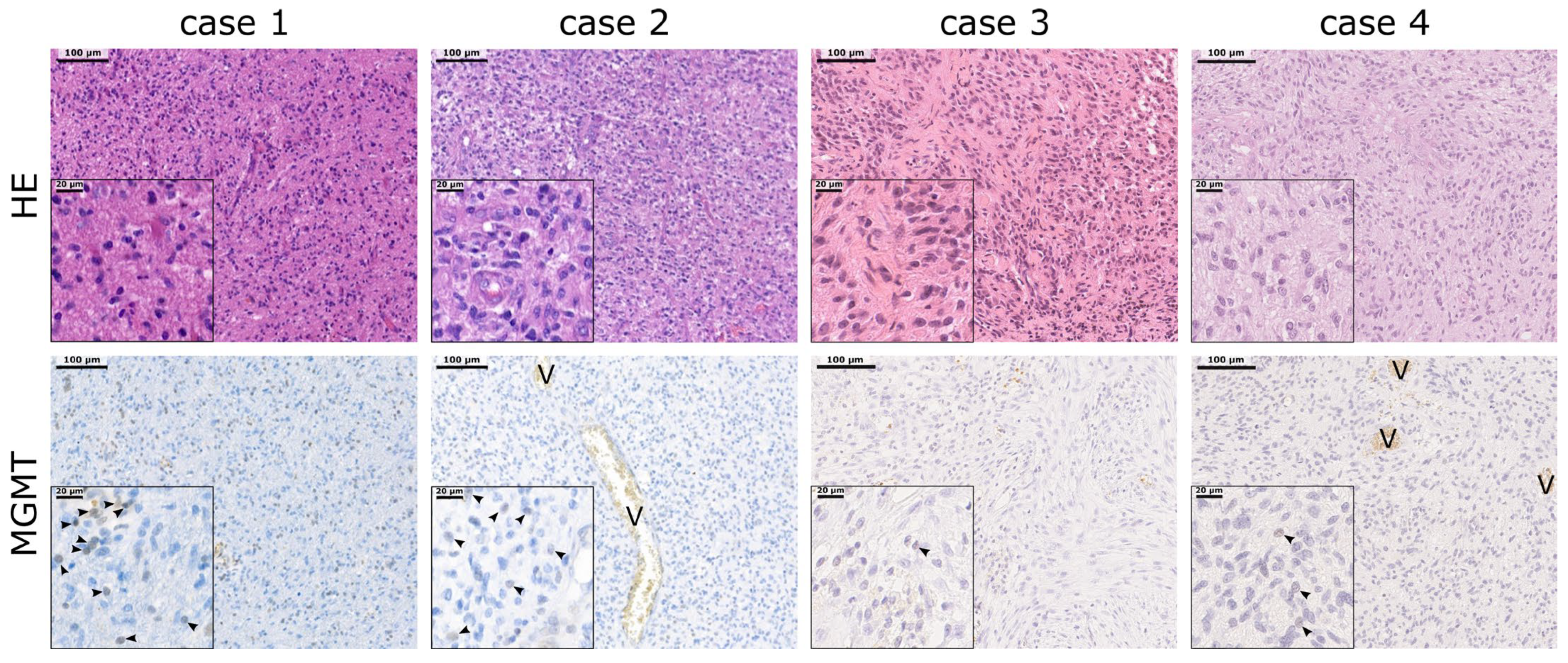
2.4. MGMT Promoter Methylation Does Not Correlate with MGMT Protein Expression
2.5. MGMT Promoter Methylation Status as a Prognostic Factor Dependent on Patient Age
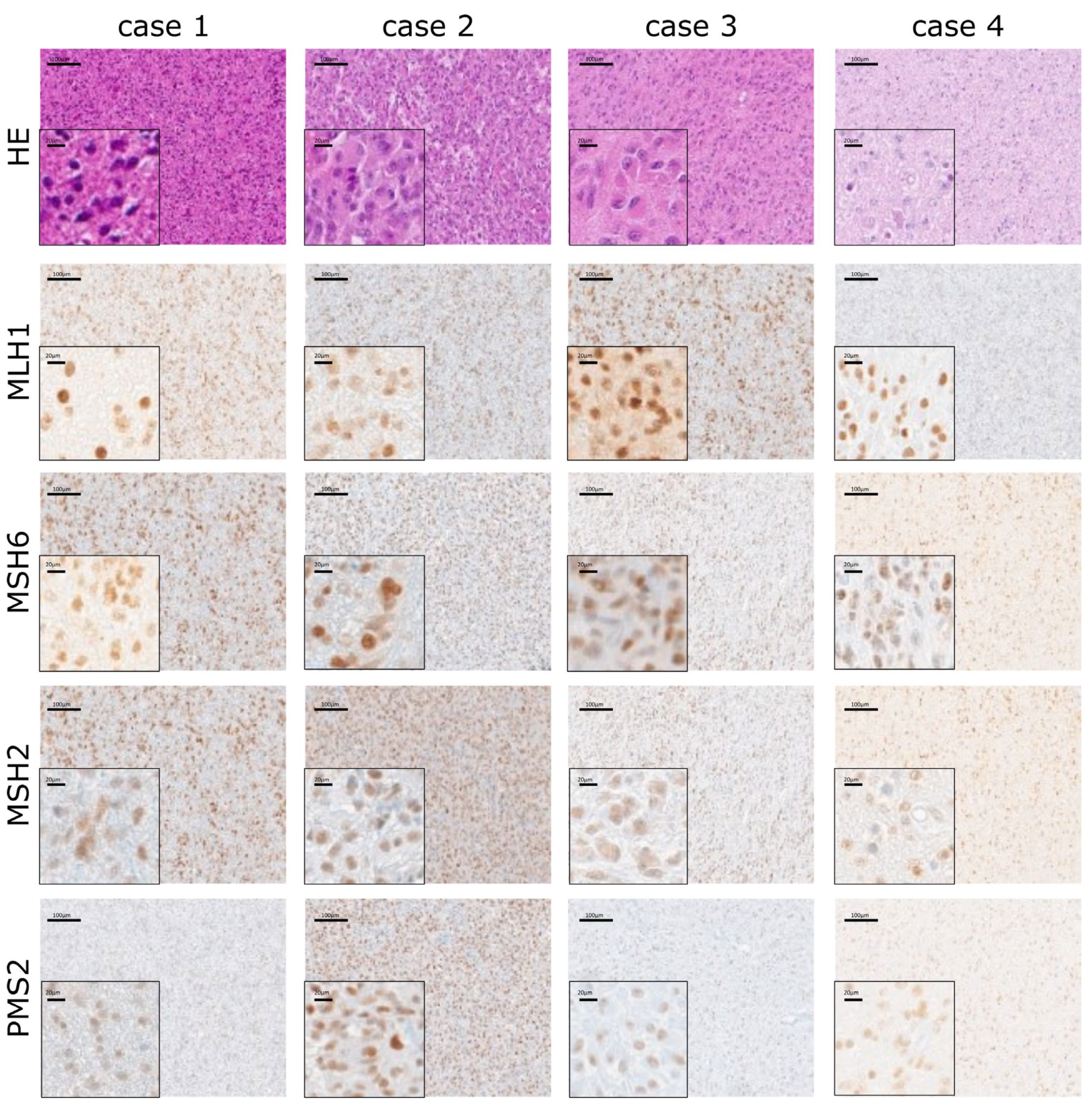
2.6. Stable Expression of MMR Proteins Independent of MGMT Status
3. Discussion
4. Materials and Methods
4.1. Patient Selection
4.2. Tissue Sampling
4.3. Methylation Analysis
4.4. Immunohistochemical Analysis
4.5. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abd-Elghany, A.A.; Naji, A.A.; Alonazi, B.; Aldosary, H.; Alsufayan, M.A.; Alnasser, M.; Mohammad, E.A.; Mahmoud, M.Z. Radiological Characteristics of Glioblastoma Multiforme Using CT and MRI Examination. J. Radiat. Res. Appl. Sci. 2019, 12, 289–293. [Google Scholar] [CrossRef]
- Magazzù, A.; Marcuello, C. Investigation of Soft Matter Nanomechanics by Atomic Force Microscopy and Optical Tweezers: A Comprehensive Review. Nanomaterials 2023, 13, 963. [Google Scholar] [CrossRef]
- Tsitlakidis, A.; Tsingotjidou, A.S.; Kritis, A.; Cheva, A.; Selviaridis, P.; Aifantis, E.C.; Foroglou, N. Atomic Force Microscope Nanoindentation Analysis of Diffuse Astrocytic Tumor Elasticity: Relation with Tumor Histopathology. Cancers 2021, 13, 4539. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Hegi, M.E.; Mason, W.P.; van den Bent, M.J.; Taphoorn, M.J.B.; Janzer, R.C.; Ludwin, S.K.; Allgeier, A.; Fisher, B.; Belanger, K.; et al. Effects of Radiotherapy with Concomitant and Adjuvant Temozolomide versus Radiotherapy Alone on Survival in Glioblastoma in a Randomised Phase III Study: 5-Year Analysis of the EORTC-NCIC Trial. Lancet Oncol. 2009, 10, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Hegi, M.E.; Liu, L.; Herman, J.G.; Stupp, R.; Wick, W.; Weller, M.; Mehta, M.P.; Gilbert, M.R. Correlation of O6-Methylguanine Methyltransferase (MGMT) Promoter Methylation with Clinical Outcomes in Glioblastoma and Clinical Strategies to Modulate MGMT Activity. J. Clin. Oncol. 2008, 26, 4189–4199. [Google Scholar] [CrossRef] [PubMed]
- Hegi, M.E.; Diserens, A.-C.; Gorlia, T.; Hamou, M.-F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT Gene Silencing and Benefit from Temozolomide in Glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M.; Herman, J.G. Generating Mutations but Providing Chemosensitivity: The Role of O6-Methylguanine DNA Methyltransferase in Human Cancer. Oncogene 2004, 23, 1–8. [Google Scholar] [CrossRef]
- Spiegl-Kreinecker, S.; Pirker, C.; Filipits, M.; Lötsch, D.; Buchroithner, J.; Pichler, J.; Silye, R.; Weis, S.; Micksche, M.; Fischer, J.; et al. O6-Methylguanine DNA Methyltransferase Protein Expression in Tumor Cells Predicts Outcome of Temozolomide Therapy in Glioblastoma Patients. Neuro Oncol. 2010, 12, 28–36. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Bauchet, L.; Davis, F.G.; Deltour, I.; Fisher, J.L.; Langer, C.E.; Pekmezci, M.; Schwartzbaum, J.A.; Turner, M.C.; Walsh, K.M.; et al. The Epidemiology of Glioma in Adults: A “State of the Science” Review. Neuro Oncol. 2014, 16, 896–913. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, X.; Zhou, B.; Zhang, L. The Prognostic Value of MGMT Promoter Methylation in Glioblastoma Multiforme: A Meta-Analysis. Fam. Cancer 2013, 12, 449–458. [Google Scholar] [CrossRef]
- Hegi, M.E.; Diserens, A.-C.; Godard, S.; Dietrich, P.-Y.; Regli, L.; Ostermann, S.; Otten, P.; Van Melle, G.; de Tribolet, N.; Stupp, R. Clinical Trial Substantiates the Predictive Value of O-6-Methylguanine-DNA Methyltransferase Promoter Methylation in Glioblastoma Patients Treated with Temozolomide. Clin. Cancer Res. 2004, 10, 1871–1874. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M.; Toyota, M.; Sanchez-Cespedes, M.; Capella, G.; Peinado, M.A.; Watkins, D.N.; Issa, J.P.; Sidransky, D.; Baylin, S.B.; Herman, J.G. Inactivation of the DNA Repair Gene O6-Methylguanine-DNA Methyltransferase by Promoter Hypermethylation Is Associated with G to A Mutations in K-Ras in Colorectal Tumorigenesis. Cancer Res. 2000, 60, 2368–2371. [Google Scholar] [PubMed]
- Uno, M.; Oba-Shinjo, S.M.; Camargo, A.A.; Moura, R.P.; de Aguiar, P.H.; Cabrera, H.N.; Begnami, M.; Rosemberg, S.; Teixeira, M.J.; Marie, S.K.N. Correlation of MGMT Promoter Methylation Status with Gene and Protein Expression Levels in Glioblastoma. Clinics 2011, 66, 1747–1755. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Taillibert, S.; Kanner, A.A.; Kesari, S.; Steinberg, D.M.; Toms, S.A.; Taylor, L.P.; Lieberman, F.; Silvani, A.; Fink, K.L.; et al. Maintenance Therapy with Tumor-Treating Fields plus Temozolomide vs. Temozolomide Alone for Glioblastoma: A Randomized Clinical Trial. JAMA 2015, 314, 2535–2543. [Google Scholar] [CrossRef]
- Wick, W.; Platten, M.; Meisner, C.; Felsberg, J.; Tabatabai, G.; Simon, M.; Nikkhah, G.; Papsdorf, K.; Steinbach, J.P.; Sabel, M.; et al. Temozolomide Chemotherapy Alone versus Radiotherapy Alone for Malignant Astrocytoma in the Elderly: The NOA-08 Randomised, Phase 3 Trial. Lancet Oncol. 2012, 13, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Malmström, A.; Grønberg, B.H.; Marosi, C.; Stupp, R.; Frappaz, D.; Schultz, H.; Abacioglu, U.; Tavelin, B.; Lhermitte, B.; Hegi, M.E.; et al. Temozolomide versus Standard 6-Week Radiotherapy versus Hypofractionated Radiotherapy in Patients Older than 60 Years with Glioblastoma: The Nordic Randomised, Phase 3 Trial. Lancet Oncol. 2012, 13, 916–926. [Google Scholar] [CrossRef] [PubMed]
- Jiapaer, S.; Furuta, T.; Tanaka, S.; Kitabayashi, T.; Nakada, M. Potential Strategies Overcoming the Temozolomide Resistance for Glioblastoma. Neurol. Med. Chir. 2018, 58, 405–421. [Google Scholar] [CrossRef]
- Li, G.-M. Mechanisms and Functions of DNA Mismatch Repair. Cell Res. 2008, 18, 85–98. [Google Scholar] [CrossRef]
- Pegg, A.E.; Dolan, M.E.; Moschel, R.C. Structure, Function, and Inhibition of O6-Alkylguanine-DNA Alkyltransferase. Prog. Nucleic Acid Res. Mol. Biol. 1995, 51, 167–223. [Google Scholar] [CrossRef]
- Koukourakis, G.V.; Kouloulias, V.; Zacharias, G.; Papadimitriou, C.; Pantelakos, P.; Maravelis, G.; Fotineas, A.; Beli, I.; Chaldeopoulos, D.; Kouvaris, J. Temozolomide with Radiation Therapy in High Grade Brain Gliomas: Pharmaceuticals Considerations and Efficacy; a Review Article. Molecules 2009, 14, 1561–1577. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, R.N.; Almeida, K.H.; Fornsaglio, J.L.; Schamus, S.; Sobol, R.W. The Role of Base Excision Repair in the Sensitivity and Resistance to Temozolomide-Mediated Cell Death. Cancer Res. 2005, 65, 6394–6400. [Google Scholar] [CrossRef] [PubMed]
- Helleday, T.; Petermann, E.; Lundin, C.; Hodgson, B.; Sharma, R.A. DNA Repair Pathways as Targets for Cancer Therapy. Nat. Rev. Cancer 2008, 8, 193–204. [Google Scholar] [CrossRef]
- Maxwell, J.A.; Johnson, S.P.; McLendon, R.E.; Lister, D.W.; Horne, K.S.; Rasheed, A.; Quinn, J.A.; Ali-Osman, F.; Friedman, A.H.; Modrich, P.L.; et al. Mismatch Repair Deficiency Does Not Mediate Clinical Resistance to Temozolomide in Malignant Glioma. Clin. Cancer Res. 2008, 14, 4859–4868. [Google Scholar] [CrossRef]
- Perazzoli, G.; Prados, J.; Ortiz, R.; Caba, O.; Cabeza, L.; Berdasco, M.; Gónzalez, B.; Melguizo, C. Temozolomide Resistance in Glioblastoma Cell Lines: Implication of MGMT, MMR, P-Glycoprotein and CD133 Expression. PLoS ONE 2015, 10, e0140131. [Google Scholar] [CrossRef]
- Quiros, S.; Roos, W.P.; Kaina, B. Processing of O6-Methylguanine into DNA Double-Strand Breaks Requires Two Rounds of Replication Whereas Apoptosis Is Also Induced in Subsequent Cell Cycles. Cell Cycle 2010, 9, 168–178. [Google Scholar] [CrossRef]
- Novo, N.; Romero-Tamayo, S.; Marcuello, C.; Boneta, S.; Blasco-Machin, I.; Velázquez-Campoy, A.; Villanueva, R.; Moreno-Loshuertos, R.; Lostao, A.; Medina, M.; et al. Beyond a Platform Protein for the Degradosome Assembly: The Apoptosis-Inducing Factor as an Efficient Nuclease Involved in Chromatinolysis. PNAS Nexus 2022, 2, pgac312. [Google Scholar] [CrossRef] [PubMed]
- Artus, C.; Boujrad, H.; Bouharrour, A.; Brunelle, M.N.; Hoos, S.; Yuste, V.J.; Lenormand, P.; Rousselle, J.C.; Namane, A.; England, P.; et al. AIF Promotes Chromatinolysis and Caspase-Independent Programmed Necrosis by Interacting with Histone H2AX. EMBO J. 2010, 29, 1585–1599. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.-P.; Zhang, Y.-L.; Zhou, D.-Y.; Gao, C.-F.; Lai, Z.-S. Microsatellite Instability, MMR Gene Expression and Proliferation Kinetics in Colorectal Cancer with Famillial Predisposition. World J. Gastroenterol. 2000, 6, 902–905. [Google Scholar] [CrossRef]
- McConechy, M.K.; Talhouk, A.; Li-Chang, H.H.; Leung, S.; Huntsman, D.G.; Gilks, C.B.; McAlpine, J.N. Detection of DNA Mismatch Repair (MMR) Deficiencies by Immunohistochemistry Can Effectively Diagnose the Microsatellite Instability (MSI) Phenotype in Endometrial Carcinomas. Gynecol. Oncol. 2015, 137, 306–310. [Google Scholar] [CrossRef]
- Carethers, J.M.; Stoffel, E.M. Lynch Syndrome and Lynch Syndrome Mimics: The Growing Complex Landscape of Hereditary Colon Cancer. World J. Gastroenterol. 2015, 21, 9253–9261. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive Genomic Characterization Defines Human Glioblastoma Genes and Core Pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Yip, S.; Miao, J.; Cahill, D.P.; Iafrate, A.J.; Aldape, K.; Nutt, C.L.; Louis, D.N. MSH6 Mutations Arise in Glioblastomas during Temozolomide Therapy and Mediate Temozolomide Resistance. Clin. Cancer Res. 2009, 15, 4622–4629. [Google Scholar] [CrossRef] [PubMed]
- Felsberg, J.; Thon, N.; Eigenbrod, S.; Hentschel, B.; Sabel, M.C.; Westphal, M.; Schackert, G.; Kreth, F.W.; Pietsch, T.; Löffler, M.; et al. Promoter Methylation and Expression of MGMT and the DNA Mismatch Repair Genes MLH1, MSH2, MSH6 and PMS2 in Paired Primary and Recurrent Glioblastomas. Int. J. Cancer 2011, 129, 659–670. [Google Scholar] [CrossRef]
- Hunter, C.; Smith, R.; Cahill, D.P.; Stephens, P.; Stevens, C.; Teague, J.; Greenman, C.; Edkins, S.; Bignell, G.; Davies, H.; et al. A Hypermutation Phenotype and Somatic MSH6 Mutations in Recurrent Human Malignant Gliomas after Alkylator Chemotherapy. Cancer Res. 2006, 66, 3987–3991. [Google Scholar] [CrossRef] [PubMed]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.-H.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.-M.; Gallia, G.L.; et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science 2008, 321, 1807–1812. [Google Scholar] [CrossRef]
- Huang, L.E. Friend or Foe-IDH1 Mutations in Glioma 10 Years On. Carcinogenesis 2019, 40, 1299–1307. [Google Scholar] [CrossRef]
- Alnahhas, I.; LaHaye, S.; Giglio, P.; Mardis, E.; Puduvalli, V. An Evaluation of MGMT Promoter Methylation within the Methylation Subclasses of Glioblastoma. Neuro-Oncol. Adv. 2020, 2, vdaa117. [Google Scholar] [CrossRef]
- Pinson, H.; Hallaert, G.; Van der Meulen, J.; Dedeurwaerdere, F.; Vanhauwaert, D.; Van den Broecke, C.; Van Dorpe, J.; Van Roost, D.; Kalala, J.P.; Boterberg, T. Weak MGMT Gene Promoter Methylation Confers a Clinically Significant Survival Benefit in Patients with Newly Diagnosed Glioblastoma: A Retrospective Cohort Study. J. Neurooncol. 2020, 146, 55–62. [Google Scholar] [CrossRef]
- Sanai, N.; Polley, M.-Y.; McDermott, M.W.; Parsa, A.T.; Berger, M.S. An Extent of Resection Threshold for Newly Diagnosed Glioblastomas. J. Neurosurg. 2011, 115, 3–8. [Google Scholar] [CrossRef]
- Magrowski, Ł.; Nowicka, E.; Masri, O.; Tukiendorf, A.; Tarnawski, R.; Miszczyk, M. The Survival Impact of Significant Delays between Surgery and Radiochemotherapy in Glioblastoma Patients: A Retrospective Analysis from a Large Tertiary Center. J. Clin. Neurosci. 2021, 90, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Hallaert, G.; Pinson, H.; Vanhauwaert, D.; Van den Broecke, C.; Van Roost, D.; Boterberg, T.; Kalala, J.P. Partial Resection Offers an Overall Survival Benefit over Biopsy in MGMT-Unmethylated IDH-Wildtype Glioblastoma Patients. Surg. Oncol. 2020, 35, 515–519. [Google Scholar] [CrossRef]
- Abhinav, K.; Aquilina, K.; Gbejuade, H.; La, M.; Hopkins, K.; Iyer, V. A Pilot Study of Glioblastoma Multiforme in Elderly Patients: Treatments, O-6-Methylguanine-DNA Methyltransferase (MGMT) Methylation Status and Survival. Clin. Neurol. Neurosurg. 2013, 115, 1375–1378. [Google Scholar] [CrossRef]
- Cao, V.T.; Jung, T.Y.; Jung, S.; Jin, S.G.; Moon, K.S.; Kim, I.Y.; Kang, S.S.; Park, C.S.; Lee, K.H.; Chae, H.J. The Correlation and Prognostic Significance of MGMT Promoter Methylation and MGMT Protein in Glioblastomas. Neurosurgery 2009, 65, 866–875. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, F.J.; Thibodeau, S.N.; Jenkins, R.B.; Schowalter, K.V.; Caron, B.L.; O’neill, B.P.; James, C.D.; David James, C.; Passe, S.; Slezak, J.; et al. MGMT Immunohistochemical Expression and Promoter Methylation in Human Glioblastoma. Appl. Immunohistochem. Mol. Morphol. 2008, 16, 59–65. [Google Scholar] [CrossRef]
- Sonoda, Y.; Yokosawa, M.; Saito, R.; Kanamori, M.; Yamashita, Y.; Kumabe, T.; Watanabe, M.; Tominaga, T. O(6)-Methylguanine DNA Methyltransferase Determined by Promoter Hypermethylation and Immunohistochemical Expression Is Correlated with Progression-Free Survival in Patients with Glioblastoma. Int. J. Clin. Oncol. 2010, 15, 352–358. [Google Scholar] [CrossRef] [PubMed]
- Lalezari, S.; Chou, A.P.; Tran, A.; Solis, O.E.; Khanlou, N.; Chen, W.; Li, S.; Carrillo, J.A.; Chowdhury, R.; Selfridge, J.; et al. Combined Analysis of O6-Methylguanine-DNA Methyltransferase Protein Expression and Promoter Methylation Provides Optimized Prognostication of Glioblastoma Outcome. Neuro Oncol. 2013, 15, 370–381. [Google Scholar] [CrossRef]
- Gao, F.; Cui, Y.; Jiang, H.; Sui, D.; Wang, Y.; Jiang, Z.; Zhao, J.; Lin, S. Circulating Tumor Cell Is a Common Property of Brain Glioma and Promotes the Monitoring System. Oncotarget 2016, 7, 71330–71340. [Google Scholar] [CrossRef]
- Barciszewska, A.-M.; Gurda, D.; Głodowicz, P.; Nowak, S.; Naskręt-Barciszewska, M.Z. A New Epigenetic Mechanism of Temozolomide Action in Glioma Cells. PLoS ONE 2015, 10, e0136669. [Google Scholar] [CrossRef]
- Zinn, P.O.; Colen, R.R.; Kasper, E.M.; Burkhardt, J.-K. Extent of Resection and Radiotherapy in GBM: A 1973 to 2007 Surveillance, Epidemiology and End Results Analysis of 21,783 Patients. Int. J. Oncol. 2013, 42, 929–934. [Google Scholar] [CrossRef]
- Brown, T.J.; Brennan, M.C.; Li, M.; Church, E.W.; Brandmeir, N.J.; Rakszawski, K.L.; Patel, A.S.; Rizk, E.B.; Suki, D.; Sawaya, R.; et al. Association of the Extent of Resection With Survival in Glioblastoma: A Systematic Review and Meta-Analysis. JAMA Oncol. 2016, 2, 1460–1469. [Google Scholar] [CrossRef] [PubMed]
- Awad, A.-W.; Karsy, M.; Sanai, N.; Spetzler, R.; Zhang, Y.; Xu, Y.; Mahan, M.A. Impact of Removed Tumor Volume and Location on Patient Outcome in Glioblastoma. J. Neurooncol. 2017, 135, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Bruno, F.; Pellerino, A.; Pronello, E.; Palmiero, R.; Bertero, L.; Mantovani, C.; Bianconi, A.; Melcarne, A.; Garbossa, D.; Rudà, R. Elderly Gliobastoma Patients: The Impact of Surgery and Adjuvant Treatments on Survival: A Single Institution Experience. Brain Sci. 2022, 12, 632. [Google Scholar] [CrossRef] [PubMed]
- Almenawer, S.A.; Badhiwala, J.H.; Alhazzani, W.; Greenspoon, J.; Farrokhyar, F.; Yarascavitch, B.; Algird, A.; Kachur, E.; Cenic, A.; Sharieff, W.; et al. Biopsy versus Partial versus Gross Total Resection in Older Patients with High-Grade Glioma: A Systematic Review and Meta-Analysis. Neuro Oncol. 2015, 17, 868–881. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Reifenberger, G.; Hentschel, B.; Felsberg, J.; Schackert, G.; Simon, M.; Schnell, O.; Westphal, M.; Wick, W.; Pietsch, T.; Loeffler, M.; et al. Predictive Impact of MGMT Promoter Methylation in Glioblastoma of the Elderly. Int. J. Cancer 2012, 131, 1342–1350. [Google Scholar] [CrossRef]
- Preusser, M.; Berghoff, A.S.; Manzl, C.; Filipits, M.; Weinhäusel, A.; Pulverer, W.; Dieckmann, K.; Widhalm, G.; Wöhrer, A.; Knosp, E.; et al. Clinical Neuropathology Practice News 1-2014: Pyrosequencing Meets Clinical and Analytical Performance Criteria for Routine Testing of MGMT Promoter Methylation Status in Glioblastoma. Clin. Neuropathol. 2014, 33, 6–14. [Google Scholar] [CrossRef]
| Characteristic | All | <70 Years | ≥70 Years |
|---|---|---|---|
| Patients, n (%) | 42 | 23 (54.8) | 19 (45.2) |
| Sex | |||
| Female, n (%) | 16 (38.1) | 12 (52.2) | 4 (21.1) |
| Male, n (%) | 26 (61.9) | 11 (47.8) | 15 (78.9) |
| Age at surgery, years, median (IQR) | 67.0 (58.0–74.8) | 58.0 (54.0–63.0) | 75.0 (72.5–78.0) |
| IDH1 status | |||
| IDH1 wildtype, n (%) | 34 (81.0) | 1 (4.3) | 2 (10.5) |
| IDH1 mutated, n (%) | 3 (7.1) | 18 (78.3) | 16 (84.2) |
| IDH1 NOS, n (%) | 5 (11.9) | 4 (17.4) | 1 (5.3) |
| KPS | |||
| Preoperative KPS, mean (±SD) | 86.3 (±15.45) | 89.1 (±9.71) | 83.2 (±20.0) |
| Postoperative KPS, mean (±SD) | 77.9 (±27.63) | 82.2(±26.45) | 72.6 (±28.8) |
| Extent of surgery § | |||
| Subtotal resection, n (%) | 13 (31.7) | 7 (69.6) | 12 (66.7) |
| Gross total resection, n (%) | 28 (68.3) | 16 (30.4) | 6 (33.3) |
| First-line therapy | |||
| Stupp protocol, n (%) | 32 (76.2) | 21 (95.5) | 11 (57.9) |
| Radiotherapy, n (%) | 8 (19.0) | 2 (4.5) | 6 (31.6) |
| No further treatment, n (%) | 2 (4.8) | 0 (0.0) | 2 (10.5) |
| All | <70 Years | ≥70 Years | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parameter | n | OS | n | PFS | n | OS | n | PFS | n | OS | n | PFS |
| Total cohort | 42 | 11.5 | 34 | 116.5 | 23 | 15.0 | 20 | 125 | 19 | 4.0 | 14 | 214 |
| Sex | ||||||||||||
| Female | 16 | 14.5 | 13 | 119 | 12 | 17.5 | 10 | 114 | 4 | 1.5 | 3 | 294 |
| Male | 26 | 8.0 | 21 | 190 | 11 | 13.0 | 10 | 130 | 15 | 5.0 | 11 | 214 |
| Age at surgery | ||||||||||||
| <70 years | 23 | 15.0 | 20 | 125 | ||||||||
| ≥70 years | 19 | 4.0 | 14 | 214 | ||||||||
| MGMT methylation status | ||||||||||||
| MGMTmet+ | 21 | 14.0 | 16 | 134 | 11 | 31.0 | 9 | 125 | 10 | 3.0 | 7 | 294 |
| MGMTmet− | 21 | 11.0 | 18 | 190 | 12 | 13.0 | 11 | 125 | 9 | 7.0 | 7 | 214 |
| MGMT protein expression | ||||||||||||
| MGMTlow | 31 | 13.0 | 26 | 190 | 18 | 15.5 | 16 | 125 | 13 | 4.0 | 10 | 214 |
| MGMThigh | 11 | 7.0 | 8 | 134 | 5 | 15.0 | 4 | 118 | 6 | 5.0 | 4 | - * |
| MGMT combined | ||||||||||||
| MGMTmet+/MGMTlow | 16 | 19.5 | 12 | 281 | 8 | 28.0 | 7 | 125 | 8 | 2.5 | 5 | 294 |
| MGMTmet+/MGMThigh | 5 | 12.0 | 4 | 118 | 4 | 17.5 | 2 | 118 | 2 | 7.5 | 2 | 102 |
| MGMTmet−/MGMTlow | 15 | 13.0 | 14 | 149 | 10 | 13.0 | 9 | 125 | 5 | 11.0 | 5 | 214 |
| MGMTmet−/MGMThigh | 6 | 5.0 | 4 | 211 | 1 | 15.0 | 2 | 124 | 4 | 4.0 | 2 | - * |
| Extent of surgery § | ||||||||||||
| Subtotal resection | 13 | 4 | 11 | 102 | 7 | 17.0 | 14 | 172 | 6 | 3 | 5 | 294 |
| Gross total resection | 28 | 13 | 23 | 214 | 16 | 14.5 | 6 | 70 | 12 | 9 | 9 | - * |
| All | <70 Years | ≥70 Years | ||||
|---|---|---|---|---|---|---|
| Parameter | HR (95% CI) | p | HR (95% CI) | p | HR (95% CI) | p |
| Sex (male) | 1.34 (0.70–2.58) | 0.379 | 1.24 (0.52–2.94) | 0.623 | 0.75 (0.24–2.36) | 0.620 |
| Unmethylated MGMT promoter | 2.20 (1.06–4.55) | 0.034 | 4.85 (1.59–14.74) | 0.005 | 0.96 (0.37–2.53) | 0.941 |
| Expressed MGMT protein | 1.48 (0.73–2.99) | 0.278 | 1.19 (0.43–3.31) | 0.743 | 1.40 (0.49–3.97) | 0.531 |
| Subtotal resection | 1.41 (0.72–2.77) | 0.317 | 1.03 (0.41–2.57) | 0.950 | 4.05 (1.11–14.79) | 0.035 |
| KPS pre-surgery (score > 70) | 0.80 (0.31–2.06) | 0.647 | 1.99 (0.26–15.20) | 0.508 | 0.78 (0.25–2.42) | 0.666 |
| All | <70 Years | ≥70 Years | ||||
|---|---|---|---|---|---|---|
| Parameter | HR (95% CI) | p | HR (95% CI) | p | HR (95% CI) | p |
| Sex (male) | 1.36 (0.65–2.87) | 0.415 | 1.00 (0.39–2.51) | 0.993 | 0.70 (0.15–3.23) | 0.652 |
| Unmethylated MGMT promoter | 2.69 (1.19–6.07) | 0.017 | 6.14 (1.78–21.21) | 0.004 | 1.20 (0.39–3.72) | 0.753 |
| Expressed MGMT protein | 1.67 (0.81–3.44) | 0.165 | 1.53 (0.53–4.43) | 0.438 | 2.56 (0.79–8.32) | 0.117 |
| Subtotal resection | 1.92 (0.94–3.93) | 0.074 | 1.60 (0.60–4.29) | 0.350 | 5.74 (1.43–23.05) | 0.012 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brawanski, K.R.; Sprung, S.; Freyschlag, C.F.; Hoeftberger, R.; Ströbel, T.; Haybaeck, J.; Thomé, C.; Manzl, C.; Birkl-Toeglhofer, A.M. Influence of MMR, MGMT Promotor Methylation and Protein Expression on Overall and Progression-Free Survival in Primary Glioblastoma Patients Treated with Temozolomide. Int. J. Mol. Sci. 2023, 24, 6184. https://doi.org/10.3390/ijms24076184
Brawanski KR, Sprung S, Freyschlag CF, Hoeftberger R, Ströbel T, Haybaeck J, Thomé C, Manzl C, Birkl-Toeglhofer AM. Influence of MMR, MGMT Promotor Methylation and Protein Expression on Overall and Progression-Free Survival in Primary Glioblastoma Patients Treated with Temozolomide. International Journal of Molecular Sciences. 2023; 24(7):6184. https://doi.org/10.3390/ijms24076184
Chicago/Turabian StyleBrawanski, Konstantin R., Susanne Sprung, Christian F. Freyschlag, Romana Hoeftberger, Thomas Ströbel, Johannes Haybaeck, Claudius Thomé, Claudia Manzl, and Anna M. Birkl-Toeglhofer. 2023. "Influence of MMR, MGMT Promotor Methylation and Protein Expression on Overall and Progression-Free Survival in Primary Glioblastoma Patients Treated with Temozolomide" International Journal of Molecular Sciences 24, no. 7: 6184. https://doi.org/10.3390/ijms24076184
APA StyleBrawanski, K. R., Sprung, S., Freyschlag, C. F., Hoeftberger, R., Ströbel, T., Haybaeck, J., Thomé, C., Manzl, C., & Birkl-Toeglhofer, A. M. (2023). Influence of MMR, MGMT Promotor Methylation and Protein Expression on Overall and Progression-Free Survival in Primary Glioblastoma Patients Treated with Temozolomide. International Journal of Molecular Sciences, 24(7), 6184. https://doi.org/10.3390/ijms24076184







