Role of GARP Vesicle Tethering Complex in Golgi Physiology
Abstract
1. History and Discovery
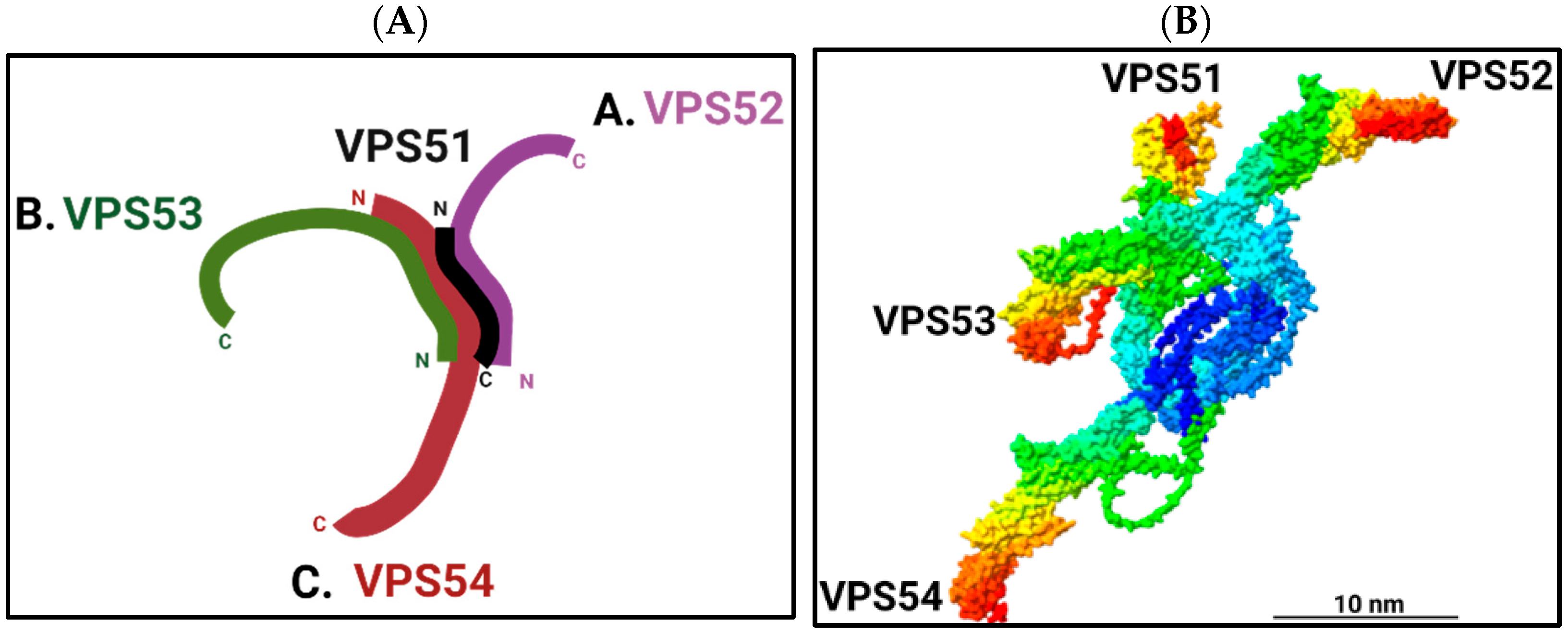
2. Composition and Structure of the GARP Complex
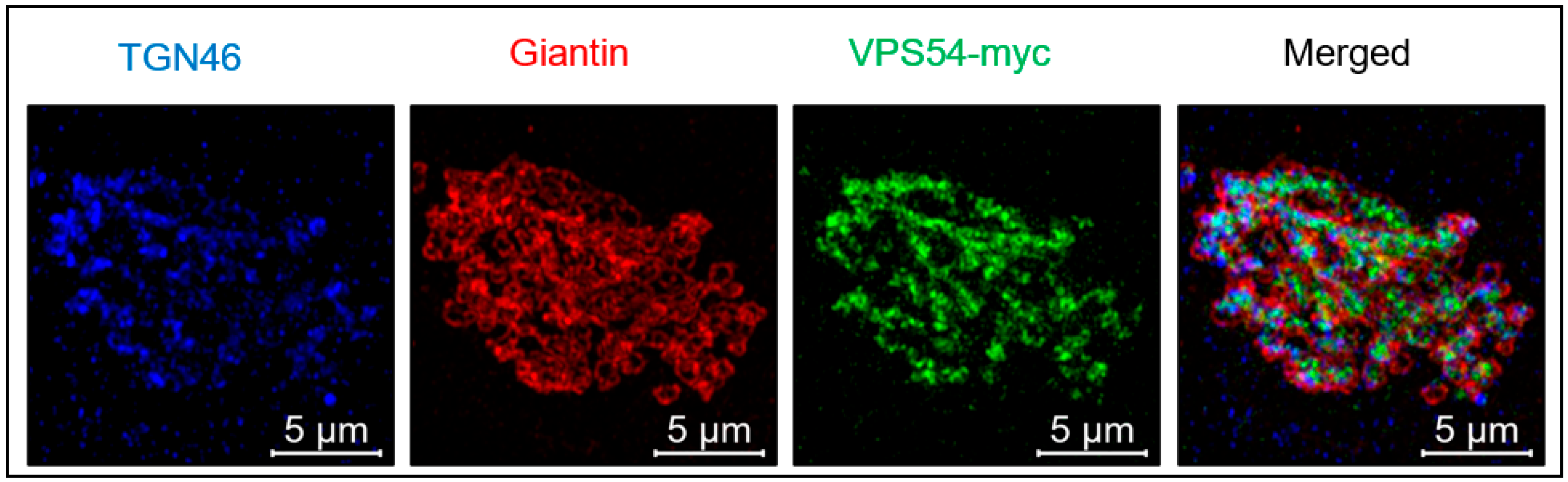
3. Localization of the GARP Complex to the Golgi Membrane
4. The GARP Complex Protein Partners
4.1. SNAREs
4.2. Small GTPases
4.3. Coiled-Coil Tethers
4.4. Other Partners
4.4.1. EIPR1/TSSC1
4.4.2. Vps1p
4.4.3. RNF41
4.4.4. LRRK2
5. Functions of the GARP Complex
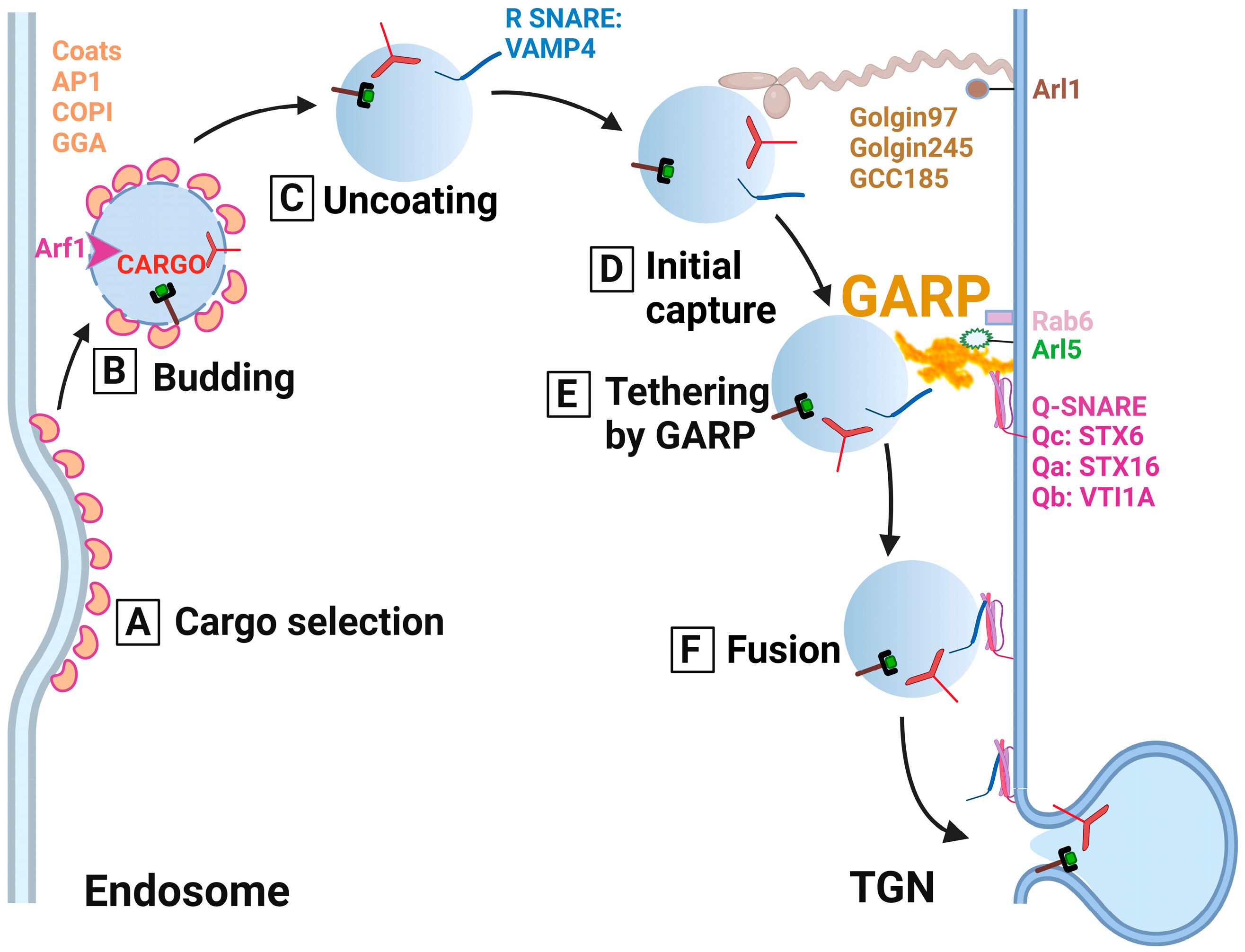
5.1. GARP as a Molecular Tether for the Endosomal-Derived Vesicles
5.2. GARP as a Regulator of SNARE Complexes
5.3. Role of the GARP Complex in the Maintenance of Golgi Glycosylation Machinery
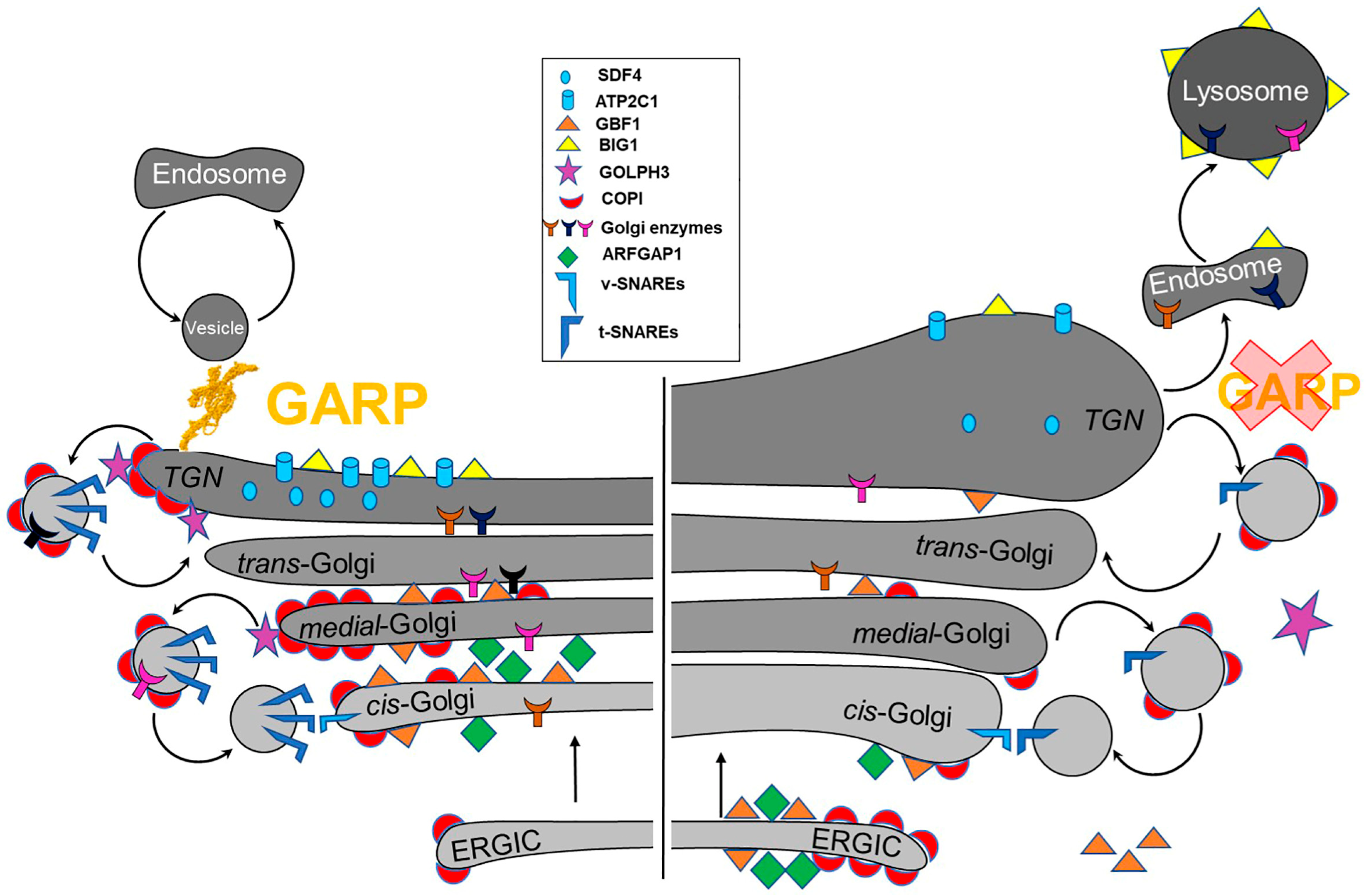
5.4. Role of the GARP Complex in Normal Golgi Physiology
5.5. GARP and Lipid Homeostasis
5.6. Role of GARP Complex in the Secretory Pathway
5.7. Hijacking of GARP by Intracellular Pathogens
6. GARP Complex Mutations and Pathogenesis
6.1. GARP Complex Mutations
6.1.1. Vps54 Null Mutant
6.1.2. Wobbler Mouse
6.2. Pathogenesis of GARP Mutations
7. Future Perspectives in GARP Studies
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Conibear, E.; Stevens, T.H. Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol. Biol. Cell 2000, 11, 305–323. [Google Scholar] [CrossRef] [PubMed]
- Conboy, M.J.; Cyert, M.S. Luv1p/Rki1p/Tcs3p/Vps54p, a yeast protein that localizes to the late Golgi and early endosome, is required for normal vacuolar morphology. Mol. Biol. Cell 2000, 11, 2429–2443. [Google Scholar] [CrossRef] [PubMed]
- Siniossoglou, S.; Pelham, H.R.B. Vps51p links the VFT complex to the SNARE Tlg1p. J. Biol. Chem. 2002, 277, 48318–48324. [Google Scholar] [CrossRef] [PubMed]
- Conibear, E.; Cleck, J.N.; Stevens, T.H. Vps51p mediates the association of the GARP (Vps52/53/54) complex with the late Golgi t-SNARE Tlg1p. Mol. Biol. Cell 2003, 14, 1610–1623. [Google Scholar] [CrossRef] [PubMed]
- Reggiori, F.; Wang, C.-W.; Stromhaug, P.E.; Shintani, T.; Klionsky, D.J. Vps51 is part of the yeast Vps fifty-three tethering complex essential for retrograde traffic from the early endosome and Cvt vesicle completion. J. Biol. Chem. 2003, 278, 5009–5020. [Google Scholar] [CrossRef]
- Pérez-Victoria, F.J.; Schindler, C.; Magadán, J.G.; Mardones, G.A.; Delevoye, C.; Romao, M.; Bonifacino, J.S. Ang2/fat-free is a conserved subunit of the Golgi-associated retrograde protein complex. Mol. Biol. Cell 2010, 21, 3386–3395. [Google Scholar] [CrossRef]
- Oka, T.; Krieger, M. Multi-component protein complexes and Golgi membrane trafficking. J. Biochem. 2005, 137, 109–114. [Google Scholar] [CrossRef]
- Brocker, C.; Engelbrecht-Vandre, S.; Ungermann, C. Multisubunit tethering complexes and their role in membrane fusion. Curr. Biol. 2010, 20, R943–R952. [Google Scholar] [CrossRef]
- Koumandou, V.L.; Dacks, J.B.; Coulson, R.; Field, M.C. Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol. Biol. 2007, 7, 29. [Google Scholar] [CrossRef]
- Bonifacino, J.S.; Hierro, A. Transport according to GARP: Receiving retrograde cargo at the trans-Golgi network. Trends Cell Biol. 2011, 21, 159–167. [Google Scholar] [CrossRef]
- Chou, H.-T.; Dukovski, D.; Chambers, M.G.; Reinisch, K.M.; Walz, T. CATCHR, HOPS and CORVET tethering complexes share a similar architecture. Nat. Struct. Mol. Biol. 2016, 23, 761–763. [Google Scholar] [CrossRef]
- Schindler, C.; Chen, Y.; Pu, J.; Guo, X.; Bonifacino, J.S. EARP is a multisubunit tethering complex involved in endocytic recycling. Nat. Cell Biol. 2015, 17, 639–650. [Google Scholar] [CrossRef]
- Topalidou, I.; Cattin-Ortolá, J.; Pappas, A.L.; Cooper, K.; Merrihew, G.E.; MacCoss, M.J.; Ailion, M. The EARP complex and its interactor EIPR-1 are required for cargo sorting to dense-core vesicles. PLoS Genet. 2016, 12, e1006074. [Google Scholar] [CrossRef]
- Vasan, N.; Hutagalung, A.; Novick, P.; Reinisch, K.M. Structure of a C-terminal fragment of its Vps53 subunit suggests similarity of Golgi-associated retrograde protein (GARP) complex to a family of tethering complexes. Proc. Natl. Acad. Sci. USA 2010, 107, 14176–14181. [Google Scholar] [CrossRef]
- Pei, J.; Ma, C.; Rizo, J.; Grishin, N.V. Remote homology between Munc13 MUN domain and vesicle tethering complexes. J. Mol. Biol. 2009, 391, 509–517. [Google Scholar] [CrossRef]
- Augustin, I.; Rosenmund, C.; Südhof, T.C.; Brose, N. Munc13-1 is essential for fusion competence of glutamatergic synaptic vesicles. Nature 1999, 400, 457–461. [Google Scholar] [CrossRef]
- Basu, J.; Shen, N.; Dulubova, I.; Lu, J.; Guan, R.; Guryev, O.; Grishin, N.V.; Rosenmund, C.; Rizo, J. A minimal domain responsible for Munc13 activity. Nat. Struct. Mol. Biol. 2005, 12, 1017–1018. [Google Scholar] [CrossRef]
- Pérez-Victoria, F.J.; Abascal-Palacios, G.; Tascón, I.; Kajava, A.; Magadán, J.G.; Pioro, E.P.; Bonifacino, J.S.; Hierro, A. Structural basis for the wobbler mouse neurodegenerative disorder caused by mutation in the Vps54 subunit of the GARP complex. Proc. Natl. Acad. Sci. USA 2010, 107, 12860–12865. [Google Scholar] [CrossRef]
- Whyte, J.R.; Munro, S. The Sec34/35 Golgi transport complex is related to the exocyst, defining a family of complexes involved in multiple steps of membrane traffic. Dev. Cell 2001, 1, 527–537. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Victoria, F.J.; Mardones, G.A.; Bonifacino, J.S. Requirement of the human GARP complex for mannose 6-phosphate-receptor-dependent sorting of cathepsin D to lysosomes. Mol. Biol. Cell 2008, 19, 2350–2362. [Google Scholar] [CrossRef] [PubMed]
- Liewen, H.; Meinhold-Heerlein, I.; Oliveira, V.; Schwarzenbacher, R.; Luo, G.; Wadle, A.; Jung, M.; Pfreundschuh, M.; Stenner-Liewen, F. Characterization of the human GARP (Golgi associated retrograde protein) complex. Exp. Cell Res. 2005, 306, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Siniossoglou, S.; Pelham, H.R. An effector of Ypt6p binds the SNARE Tlg1p and mediates selective fusion of vesicles with late Golgi membranes. EMBO J. 2001, 20, 5991–5998. [Google Scholar] [CrossRef]
- Ishida, M.; Bonifacino, J.S. ARFRP1 functions upstream of ARL1 and ARL5 to coordinate recruitment of distinct tethering factors to the trans-Golgi network. J. Cell Biol. 2019, 218, 3681–3696. [Google Scholar] [CrossRef]
- Rosa-Ferreira, C.; Christis, C.; Torres, I.L.; Munro, S. The small G protein Arl5 contributes to endosome-to-Golgi traffic by aiding the recruitment of the GARP complex to the Golgi. Biol. Open 2015, 4, 474–481. [Google Scholar] [CrossRef]
- Fridmann-Sirkis, Y.; Kent, H.M.; Lewis, M.J.; Evans, P.R.; Pelham, H.R.B. Structural analysis of the interaction between the SNARE Tlg1 and Vps51. Traffic 2006, 7, 182–190. [Google Scholar] [CrossRef]
- Pérez-Victoria, F.J.; Bonifacino, J.S. Dual roles of the mammalian GARP complex in tethering and SNARE complex assembly at the trans-golgi network. Mol. Cell. Biol. 2009, 29, 5251–5263. [Google Scholar] [CrossRef]
- D’Souza, Z.; Taher, F.S.; Lupashin, V.V. Golgi inCOGnito: From vesicle tethering to human disease. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2020, 1864, 129694. [Google Scholar] [CrossRef]
- Shestakova, A.; Suvorova, E.; Pavliv, O.; Khaidakova, G.; Lupashin, V. Interaction of the conserved oligomeric Golgi complex with t-SNARE Syntaxin5a/Sed5 enhances intra-Golgi SNARE complex stability. J. Cell Biol. 2007, 179, 1179–1192. [Google Scholar] [CrossRef]
- Willett, R.; Kudlyk, T.; Pokrovskaya, I.; Schönherr, R.; Ungar, D.; Duden, R.; Lupashin, V. COG complexes form spatial landmarks for distinct SNARE complexes. Nat. Commun. 2013, 4, 1–13. [Google Scholar] [CrossRef]
- Laufman, O.; Kedan, A.; Hong, W.; Lev, S. Direct interaction between the COG complex and the SM protein, Sly1, is required for Golgi SNARE pairing. EMBO J. 2009, 28, 2006–2017. [Google Scholar] [CrossRef]
- Wang, S.; Ma, C. Neuronal SNARE complex assembly guided by Munc18-1 and Munc13-1. FEBS Open Bio. 2022, 12, 1939–1957. [Google Scholar] [CrossRef]
- Ma, C.; Su, L.; Seven, A.B.; Xu, Y.; Rizo, J. Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science 2013, 339, 421–425. [Google Scholar] [CrossRef]
- Chen, Y.T.; Wang, I.H.; Wang, Y.H.; Chiu, W.Y.; Hu, J.H.; Chen, W.H.; Lee, F.J.S. Action of Arl1 GTPase and golgin Imh1 in Ypt6-independent retrograde transport from endosomes to the trans-Golgi network. Mol. Biol. Cell 2019, 30, 1008–1019. [Google Scholar] [CrossRef]
- Panic, B.; Whyte, J.R.; Munro, S. The ARF-like GTPases Arl1p and Arl3p act in a pathway that interacts with vesicle-tethering factors at the Golgi apparatus. Curr. Biol. 2003, 13, 405–410. [Google Scholar] [CrossRef]
- Ibuchi, K.; Fukaya, M.; Shinohara, T.; Hara, Y.; Shiroshima, T.; Sugawara, T.; Sakagami, H. The Vps52 subunit of the GARP and EARP complexes is a novel Arf6-interacting protein that negatively regulates neurite outgrowth of hippocampal neurons. Brain Res. 2020, 1745, 146905. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Chiu, W.-Y.; Chen, Y.-T.; Cai, P.-J.; Wu, Y.-C.; Wu, J.-L.; Chen, B.-H.; Liu, Y.-W.; Yu, C.-J.; Lee, F.-J.S. Golgin Imh1 and GARP complex cooperate to restore the impaired SNARE recycling transport induced by ER stress. Cell Rep. 2022, 38, 110488. [Google Scholar] [CrossRef]
- Gershlick, D.C.; Schindler, C.; Chen, Y.; Bonifacino, J.S. TSSC1 is novel component of the endosomal retrieval machinery. Mol. Biol. Cell 2016, 27, 2867–2878. [Google Scholar] [CrossRef]
- Saimani, U.; Smothers, J.; McDermott, H.; Makaraci, P.; Kim, K. Yeast dynamin associates with the GARP tethering complex for endosome-to-Golgi traffic. Eur. J. Cell Biol. 2017, 96, 612–621. [Google Scholar] [CrossRef]
- Lukehart, J.; Highfill, C.; Kim, K. Vps1, a recycling factor for the traffic from early endosome to the late Golgi. Biochem. Cell Biol. 2013, 91, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Masschaele, D.; De Ceuninck, L.; Wauman, J.; Defever, D.; Stenner, F.; Lievens, S.; Peelman, F.; Tavernier, J. RNF41 interacts with the VPS52 subunit of the GARP and EARP complexes. PLoS ONE 2017, 12, e0178132. [Google Scholar] [CrossRef] [PubMed]
- Zach, S.; Felk, S.; Gillardon, F. Signal transduction protein array analysis links LRRK2 to Ste20 kinases and PKC zeta that modulate neuronal plasticity. PLoS ONE 2010, 5, e13191. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Suaga, P.; Luzón-Toro, B.; Churamani, D.; Zhang, L.; Bloor-Young, D.; Patel, S.; Woodman, P.G.; Churchill, G.C.; Hilfiker, S. Leucine-rich repeat kinase 2 regulates autophagy through a calcium-dependent pathway involving NAADP. Hum. Mol. Genet. 2012, 21, 511–525. [Google Scholar] [CrossRef] [PubMed]
- Steger, M.; Tonelli, F.; Ito, G.; Davies, P.; Trost, M.; Vetter, M.; Wachter, S.; Lorentzen, E.; Duddy, G.; Wilson, S.; et al. Phosphoproteomics reveals that Parkinson’s disease kinase LRRK2 regulates a subset of Rab GTPases. eLife 2016, 5, e12813. [Google Scholar] [CrossRef]
- Beilina, A.; Bonet-Ponce, L.; Kumaran, R.; Kordich, J.J.; Ishida, M.; Mamais, A.; Kaganovich, A.; Saez-Atienzar, S.; Gershlick, D.C.; Roosen, D.A.; et al. The Parkinson’s Disease Protein LRRK2 Interacts with the GARP Complex to Promote Retrograde Transport to the trans-Golgi Network. Cell Rep. 2020, 31, 107614. [Google Scholar] [CrossRef]
- Pahari, S.; Cormark, R.D.; Blackshaw, M.T.; Liu, C.; Erickson, J.L.; Schultz, E.A. Arabidopsis UNHINGED encodes a VPS51 homolog and reveals a role for the GARP complex in leaf shape and vein patterning. Development 2014, 141, 1894–1905. [Google Scholar] [CrossRef]
- Luo, L.; Hannemann, M.; Koenig, S.; Hegermann, J.; Ailion, M.; Cho, M.-K.; Sasidharan, N.; Zweckstetter, M.; Rensing, S.A.; Eimer, S. The Caenorhabditis elegans GARP complex contains the conserved Vps51 subunit and is required to maintain lysosomal morphology. Mol. Biol. Cell 2011, 22, 2564–2578. [Google Scholar] [CrossRef]
- Abascal-Palacios, G.; Schindler, C.; Rojas, A.L.; Bonifacino, J.S.; Hierro, A. Structural Basis for the Interaction of the Golgi-Associated Retrograde Protein Complex with the t-SNARE Syntaxin 6. Structure 2013, 21, 1698–1706. [Google Scholar] [CrossRef]
- Lobstein, E.; Guyon, A.; Férault, M.; Twell, D.; Pelletier, G.; Bonhomme, S. The Putative Arabidopsis Homolog of Yeast Vps52p Is Required for Pollen Tube Elongation, Localizes to Golgi, and Might Be Involved in Vesicle Trafficking. Plant Physiol. 2004, 135, 1480–1490. [Google Scholar] [CrossRef]
- Fiedler, T.; Karpova, T.; Fleig, U.; Young, M.; Cooper, J.; Hegemann, J. The vesicular transport protein Cgp1p/Vps54p/Tcs3p/Luv1p is required for the integrity of the actin cytoskeleton. Mol. Genet. Genom. 2002, 268, 190–205. [Google Scholar] [CrossRef]
- Patel, P.H.; Wilkinson, E.C.; Starke, E.L.; McGimsey, M.R.; Blankenship, J.T.; Barbee, S.A. Vps54 regulates Drosophila neuromuscular junction development and interacts genetically with Rab7 to control composition of the postsynaptic density. Biol. Open 2020, 9, bio053421. [Google Scholar] [CrossRef]
- Wilkinson, E.C.; Starke, E.L.; Barbee, S.A. Vps54 Regulates Lifespan and Locomotor Behavior in Adult Drosophila melanogaster. Front. Genet. 2021, 12, 762012. [Google Scholar] [CrossRef]
- Fabrizio, J.; Hime, G.; Lemmon, S.; Bazinet, C. Genetic dissection of sperm individualization in Drosophila melanogaster. Development 1998, 125, 1833–1843. [Google Scholar] [CrossRef]
- Schmitt-John, T.; Drepper, C.; Mußmann, A.; Hahn, P.; Kuhlmann, M.; Thiel, C.; Hafner, M.; Lengeling, A.; Heimann, P.; Jones, J.M.; et al. Mutation of Vps54 causes motor neuron disease and defective spermiogenesis in the wobbler mouse. Nat. Genet. 2005, 37, 1213–1215. [Google Scholar] [CrossRef]
- Khakurel, A.; Kudlyk, T.; Bonifacino, J.S.; Lupashin, V.V. The Golgi-associated retrograde protein (GARP) complex plays an essential role in the maintenance of the Golgi glycosylation machinery. Mol. Biol. Cell 2021, 32, 1594–1610. [Google Scholar] [CrossRef]
- Peer, M.; Yuan, H.; Zhang, Y.; Korbula, K.; Novick, P.; Dong, G. Double NPY motifs at the N-terminus of the yeast t-SNARE Sso2 synergistically bind Sec3 to promote membrane fusion. eLife 2022, 11, e82041. [Google Scholar] [CrossRef]
- Travis, S.M.; Damico, K.; Yu, I.-M.; McMahon, C.; Hamid, S.; Ramirez-Arellano, G.; Jeffrey, P.D.; Hughson, F.M. Structural basis for the binding of SNAREs to the multisubunit tethering complex Dsl1. J. Biol. Chem. 2020, 295, 10125–10135. [Google Scholar] [CrossRef]
- Torng, T.; Wickner, W. Phosphatidylinositol and phosphatidylinositol-3-phosphate activate HOPS to catalyze SNARE assembly, allowing small headgroup lipids to support the terminal steps of membrane fusion. Mol. Biol. Cell 2021, 32, ar19. [Google Scholar] [CrossRef]
- Hong, W.; Lev, S. Tethering the assembly of SNARE complexes. Trends Cell Biol. 2014, 24, 35–43. [Google Scholar] [CrossRef]
- Khakurel, A.; Kudlyk, T.; Pokrovskaya, I.; D’Souza, Z.; Lupashin, V.V. GARP dysfunction results in COPI displacement, depletion of Golgi v-SNAREs and calcium homeostasis proteins. Front. Cell Dev. Biol. 2022, 10, 1066504. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.D.; Lupashin, V.V. Role of the conserved oligomeric Golgi (COG) complex in protein glycosylation. Carbohydr. Res. 2008, 343, 2024–2031. [Google Scholar] [CrossRef] [PubMed]
- Brasil, S.; Pascoal, C.; Francisco, R.; Marques-da-Silva, D.; Andreotti, G.; Videira, P.A.; Morava, E.; Jaeken, J.; Dos Reis Ferreira, V. CDG therapies: From bench to bedside. Int. J. Mol. Sci. 2018, 19, 1304. [Google Scholar] [CrossRef] [PubMed]
- Stanley, P. Golgi glycosylation. Cold Spring Harb. Perspect. Biol. 2011, 3, a005199. [Google Scholar] [CrossRef]
- Gershlick, D.; Ishida, M.; Jones, J.R.; Bellomo, A.; Bonifacino, J.S.; Everman, D.B. A neurodevelopmental disorder caused by mutations in the VPS51 subunit of the GARP and EARP complexes. Hum. Mol. Genet. 2019, 28, 1548–1560. [Google Scholar] [CrossRef]
- Khakurel, A.; Kudlyk, T.; Lupashin, V.V. Generation and Analysis of hTERT-RPE1 VPS54 Knock-Out and Rescued Cell Lines, In Golgi: Methods and Protocols 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 349–364. [Google Scholar]
- Dell’Angelica, E.C.; Bonifacino, J.S. Coatopathies: Genetic disorders of protein coats. Annu. Rev. Cell Dev. Biol. 2019, 35, 131–168. [Google Scholar] [CrossRef]
- Fröhlich, F.; Petit, C.; Kory, N.; Christiano, R.; Hannibal-Bach, H.K.; Graham, M.; Liu, X.; Ejsing, C.S.; Farese, R.V., Jr.; Walther, T.C. The GARP complex is required for cellular sphingolipid homeostasis. eLife 2015, 4, e08712. [Google Scholar] [CrossRef]
- Takagi, K.; Iwamoto, K.; Kobayashi, S.; Horiuchi, H.; Fukuda, R.; Ohta, A. Involvement of Golgi-associated retrograde protein complex in the recycling of the putative Dnf aminophospholipid flippases in yeast. Biochem. Biophys. Res. Commun. 2012, 417, 490–494. [Google Scholar] [CrossRef]
- Eising, S.; Thiele, L.; Fröhlich, F. A systematic approach to identify recycling endocytic cargo depending on the GARP complex. eLife 2019, 8, e42837. [Google Scholar] [CrossRef]
- O’Brien, C.E.; Younger, S.H.; Jan, L.Y.; Jan, Y.N. The GARP complex prevents sterol accumulation at the trans-Golgi network during dendrite remodeling. J. Cell Biol. 2022, 222, e202112108. [Google Scholar] [CrossRef]
- Hossain, S.; Robbins, N.; Cowen, E.L. The GARP complex is required for filamentation in Candida albicans. Genetics 2022, 222, iyac152. [Google Scholar] [CrossRef]
- Wei, J.; Zhang, Y.-Y.; Luo, J.; Wang, J.-Q.; Zhou, Y.-X.; Miao, H.-H.; Shi, X.-J.; Qu, Y.-X.; Xu, J.; Li, B.-L.; et al. The GARP Complex Is Involved in Intracellular Cholesterol Transport via Targeting NPC2 to Lysosomes. Cell Rep. 2017, 19, 2823–2835. [Google Scholar] [CrossRef]
- Homma, Y.; Fukuda, M. Knockout analysis of Rab6 effector proteins revealed the role of VPS52 in the secretory pathway. Biochem. Biophys. Res. Commun. 2021, 561, 151–157. [Google Scholar] [CrossRef]
- Hirata, T.; Fujita, M.; Nakamura, S.; Gotoh, K.; Motooka, D.; Murakami, Y.; Maeda, Y.; Kinoshita, T. Post-Golgi anterograde transport requires GARP-dependent endosome-to-TGN retrograde transport. Mol. Biol. Cell 2015, 26, 3071–3084. [Google Scholar] [CrossRef]
- Realegeno, S.; Priyamvada, L.; Kumar, A.; Blackburn, J.; Hartloge, C.; Puschnik, A.; Sambhara, S.; Olson, V.; Carette, J.; Lupashin, V.; et al. Conserved Oligomeric Golgi (COG) Complex Proteins Facilitate Orthopoxvirus Entry, Fusion and Spread. Viruses 2020, 12, 707. [Google Scholar] [CrossRef]
- Realegeno, S.; Puschnik, A.S.; Kumar, A.; Goldsmith, C.; Burgado, J.; Sambhara, S.; Olson, V.A.; Carroll, D.; Damon, I.; Hirata, T.; et al. Monkeypox virus host factor screen using haploid cells identifies essential role of GARP complex in extracellular virus formation. J. Virol. 2017, 91, e00011-17. [Google Scholar] [CrossRef]
- Boillée, S.; Peschanski, M.; Junier, M.-P. The wobbler mouse: A neurodegeneration jigsaw puzzle. Mol. Neurobiol. 2003, 28, 65–106. [Google Scholar] [CrossRef]
- Bird, M.T.; Shuttleworth, E.; Koestner, A.; Reinglass, J. The wobbler mouse mutant: An animal model of hereditary motor system disease. Acta Neuropathol. 1971, 19, 39–50. [Google Scholar] [CrossRef]
- Kaupmann, K.; Simon-Chazottes, D.; Guénet, J.-L.; Jockusch, H. Wobbler, a mutation affecting motoneuron survival and gonadal functions in the mouse, maps to proximal chromosome 11. Genomics 1992, 13, 39–43. [Google Scholar] [CrossRef]
- Andrews, J.M.; Gardner, M.B.; Wolfgram, F.J.; Ellison, G.W.; Porter, D.D.; Brandkamp, W.W. Studies on a murine form of spontaneous lower motor neuron degeneration—The wobbler (wr) mouse. Am. J. Pathol. 1974, 76, 63. [Google Scholar]
- Fuchs, S.; Resch, K.; Thiel, C.; Ulbrich, M.; Platzer, M.; Jockusch, H.; Schmitt-John, T. Comparative transcription map of the wobbler critical region on mouse chromosome 11 and the homologous region on human chromosome 2p13-14. BMC Genet. 2002, 3, 40. [Google Scholar] [CrossRef]
- Moser, J.M.; Bigini, P.; Schmitt-John, T. The wobbler mouse, an ALS animal model. Mol. Genet. Genom. 2013, 288, 207–229. [Google Scholar] [CrossRef] [PubMed]
- Jockusch, H.; Holland, A.; Staunton, L.; Schmitt-John, T.; Heimann, P.; Dowling, P.; Ohlendieck, K. Pathoproteomics of testicular tissue deficient in the GARP component VPS54: The wobbler mouse model of globozoospermia. Proteomics 2014, 14, 839–852. [Google Scholar] [CrossRef] [PubMed]
- Uwineza, A.; Caberg, J.-H.; Hitayezu, J.; Wenric, S.; Mutesa, L.; Vial, Y.; Drunat, S.; Passemard, S.; Verloes, A.; El Ghouzzi, V.; et al. VPS51 biallelic variants cause microcephaly with brain malformations: A confirmatory report. Eur. J. Med Genet. 2019, 62, 103704. [Google Scholar] [CrossRef]
- Zhang, J.; Lin, Y.; Hu, X.; Wu, Z.; Guo, W. VPS52 induces apoptosis via cathepsin D in gastric cancer. J. Mol. Med. 2017, 95, 1107–1116. [Google Scholar] [CrossRef]
- Rodriguez, P.A.; Escudero-Martinez, C.; Bos, J.I. An Aphid Effector Targets Trafficking Protein VPS52 in a Host-Specific Manner to Promote Virulence. Plant Physiol. 2017, 173, 1892–1903. [Google Scholar] [CrossRef]
- Feinstein, M.; Flusser, H.; Lerman-Sagie, T.; Ben-Zeev, B.; Lev, D.; Agamy, O.; Cohen, I.; Kadir, R.; Sivan, S.; Leshinsky-Silver, E.; et al. VPS53mutations cause progressive cerebello-cerebral atrophy type 2 (PCCA2). J. Med. Genet. 2014, 51, 303–308. [Google Scholar] [CrossRef]
- Hausman-Kedem, M.; Ben-Shachar, S.; Menascu, S.; Geva, K.; Sagie, L.; Fattal-Valevski, A. VPS53 gene is associated with a new phenotype of complicated hereditary spastic paraparesis. Neurogenetics 2019, 20, 187–195. [Google Scholar] [CrossRef]
- Peng, H.; Zheng, J.; Su, Q.; Feng, X.; Peng, M.; Gong, L.; Wu, H.; Pan, X. VPS53 Suppresses Malignant Properties in Colorectal Cancer by Inducing the Autophagy Signaling Pathway. OncoTargets Ther. 2020, 13, 10667. [Google Scholar] [CrossRef]
- Kim, T.E.; Kim, Y.W.; Hwang, S.Y.; Shin, S.M.; Shin, J.W.; Lee, Y.H.; Shin, S.Y.; Han, K.T.; Lee, J.M.; Namkoong, S.E.; et al. Candidate tumor suppressor, HCCS-1, is downregulated in human cancers and induces apoptosis in cervical cancer. Int. J. Cancer 2002, 97, 780–786. [Google Scholar] [CrossRef]
- Shi, L.; Zhao, S.-M.; Luo, Y.; Zhang, A.-W.; Wei, L.-H.; Xie, Z.-Y.; Li, Y.-Y.; Ma, W. MiR-375: A prospective regulator in medullary thyroid cancer based on microarray data and bioinformatics analyses. Pathol. Res. Pract. 2017, 213, 1344–1354. [Google Scholar] [CrossRef]
- Rebbeck, T.R. Prostate Cancer Genetics: Variation by Race, Ethnicity, and Geography. Semin. Radiat. Oncol. 2016, 27, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Goh, C.L.; Eeles, R.A. Germline genetic variants associated with prostate cancer and potential relevance to clinical practice. Prostate Cancer Prev. 2014, 202, 9–26. [Google Scholar]
- Zhang, Z.; Liu, F.; Xu, Y.; Huang, H.; Zou, Y.; Yang, B.; Luo, Y.; Zhang, Q.; Xiong, A.; Wang, L.; et al. GZFLW Induces Apoptosis of Ectopic Endometrial Stromal Cells via Promoting VPS53 Protein Stability. Evidence-Based Complement. Altern. Med. 2018, 2018, 1–10. [Google Scholar]
| Gene Name | Organisms | Common Names | Length of the Protein | Protein Partners | References | |||
|---|---|---|---|---|---|---|---|---|
| SNAREs | GTPases | EIPR1/ TSSC1 | Others | |||||
| VPS51 | Yeast | Vps51/Vps67/Whi6/Api3/YKR020W | 164 | Tlg1p [3,4,27] | Vps1 [40] | |||
| Plant | VPS51, UNH | 780 | [47] | |||||
| Worm | vps-51, B0414.8 | 700 | [48] | |||||
| Fly | Vps51 | 740 | Arl5 [26] | |||||
| Fish | vps51, ffr | 827 | ||||||
| Human: | VPS51, ANG2/C11orf2, C11orf3 | 782 | STX6 [18,49] | [39] | [6] | |||
| VPS52 | Yeast | VPS52/Sac2/YDR484W, D8035.27 | 641 | Ypt6 [24] | ||||
| Plant | VPS52, POK, TTD8, At1g71270, F3I17.8 | 707 | [50] | |||||
| Worm | vps-52 | 702 | Rab6 [48] | |||||
| Fly | Vps52 | 662 | Arl5 [26] | |||||
| Fish | Vps52 | 724 | ||||||
| Human | VPS52, SACM2L | 723 | STX6 [46]; STX16; VAMP4; VTI1A [28]; STX10 [23] | Rab6 [23] Arf6 [37] | RNF41 [42] | |||
| VPS53 | Yeast | VPS53, YJL029C, J1258 | 822 | Arl1 [36] | ||||
| Plant | VPS53, HIT1, At1g50500, F11F12.15, F17J6.4 | 828 | ||||||
| Worm | vps-53 | 798 | ||||||
| Fly | Vps53 | 683 | ||||||
| Fish | vps53 | 831 | ||||||
| Human | VPS53, PP13624 | 832 | STX6; STX16; VAMP4; VTI1A [28] | |||||
| VPS54 | Yeast | VPS54, CGP1, LUV1, RKI1, TCS3, YDR027C, PZF889, YD9813.05C | 889 | [51] | ||||
| Plant | VPS54, At4g19490, F24J7.50 | 1034 | ||||||
| Worm | vps-54, T21C9.2 | 1058 | ||||||
| Fly | scat, CG3766 | 940 | Rab5,7,11 [52] | [53] | ||||
| Fish | vps54 | 998 | ||||||
| Human | VPS54, HCC8 | 977 | STX6; STX16; VAMP4; VTI1A [28] | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khakurel, A.; Lupashin, V.V. Role of GARP Vesicle Tethering Complex in Golgi Physiology. Int. J. Mol. Sci. 2023, 24, 6069. https://doi.org/10.3390/ijms24076069
Khakurel A, Lupashin VV. Role of GARP Vesicle Tethering Complex in Golgi Physiology. International Journal of Molecular Sciences. 2023; 24(7):6069. https://doi.org/10.3390/ijms24076069
Chicago/Turabian StyleKhakurel, Amrita, and Vladimir V. Lupashin. 2023. "Role of GARP Vesicle Tethering Complex in Golgi Physiology" International Journal of Molecular Sciences 24, no. 7: 6069. https://doi.org/10.3390/ijms24076069
APA StyleKhakurel, A., & Lupashin, V. V. (2023). Role of GARP Vesicle Tethering Complex in Golgi Physiology. International Journal of Molecular Sciences, 24(7), 6069. https://doi.org/10.3390/ijms24076069







