Abstract
As a member of the TRIM (tripartite motif) protein family, TRIM56 can function as an E3 ubiquitin ligase. In addition, TRIM56 has been shown to possess deubiquitinase activity and the ability to bind RNA. This adds to the complexity of the regulatory mechanism of TRIM56. TRIM56 was initially found to be able to regulate the innate immune response. In recent years, its role in direct antiviral and tumor development has also attracted the interest of researchers, but there is no systematic review on TRIM56. Here, we first summarize the structural features and expression of TRIM56. Then, we review the functions of TRIM56 in TLR and cGAS-STING pathways of innate immune response, the mechanisms and structural specificity of TRIM56 against different types of viruses, and the dual roles of TRIM56 in tumorigenesis. Finally, we discuss the future research directions regarding TRIM56.
1. Introduction
The tripartite-motif (TRIM) family of proteins, also known as really interesting new gene (RING)-B-box-Coiled-Coil (RBCC) region proteins, is composed of an N-terminal RING structural domain, one or two B-box patterns, and an α-helical coiled-coil domain, followed by a highly variable carboxyl structural domain from the N-terminus to the C-terminus [1,2,3]. TRIMs are a large family of proteins, and approximately 80 members of the TRIM family have been identified in humans [4,5]. Based on the highly variable C-terminal structural domains, TRIMs with RING structural domains can be classified into subfamilies I to XI (C-I to C-XI). The variable C-terminal regions include the PRY structural domain, the SPRY structural domain, the COS structural domain, the fibronectin type III repeat region (FNIII), the acid-rich region (ACID), the Meprin and TRAF-homologous structural domain (MATH), the ADP-ribosylation factor family structural domain (ARF), the filamine-type IG structural domain (FIL), the NHL structural domain, the PHD structural domain, bromodomain (BROMO), and the transmembrane region (TM) [3,6,7].
TRIM family members are involved in a wide range of cellular activities and biological processes, including DNA damage repair [8], RNA binding [9], autophagy [1,10], apoptosis [11], cell cycle [12], viral infection [13,14], immune activation [7,15], inflammatory processes [16], stem cell differentiation [17], and neurogenesis [18,19]. The aberrant expression of TRIM family members leads to the development of various diseases, including tumors and neurological disorders [19,20,21].
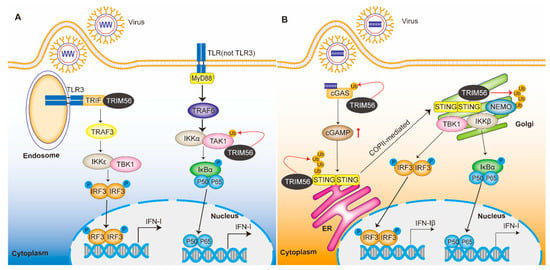
TRIM56 is a member of the TRIM family. TRIM56 was originally reported to regulate the intracellular double-stranded DNA innate immune response [22]. In recent years, an increasing number of studies have shown that TRIM56 is involved in the host response to viral infection. On one hand, TRIM56 acts by regulating host innate immune signaling. TRIM56 is able to cause the transcriptional induction of pro-inflammatory cytokines and type I interferon (IFN) by regulating the toll-like receptor (TLR) signaling pathway and the cyclic GMP-AMP synthase (cGAS)-stimulator interferon gene (STING) signaling pathway to limit viral transmission [23,24,25,26,27]. On the other hand, as a direct antiviral restriction factor, TRIM56 has been shown to have a direct antiviral effect on positive single-stranded RNA viruses of the Flaviviridae, Coronaviridae, and Retroviridae families. In addition, it is also effective against negative single-stranded RNA viruses (influenza A and B) and two DNA viruses [28].
The expression level of TRIM56 is not consistent among different tumor types, and its expression changes are closely related to tumor development and prognosis. This suggests that TRIM56 may play different pro- or anti-cancer functions in different tumor types. In recent years, many studies have revealed the function of TRIM56 in tumor development. TRIM56 is an oncogene in glioma, breast cancer, and Kaposi’s sarcoma [29,30,31,32], but it is a tumor suppressor in ovarian cancer, multiple myeloma, lung adenocarcinoma, hepatocellular carcinoma, and leukemia [33,34,35,36,37].
Here, we first describe the structural features and expression characteristics of TRIM56. Next, we focus on reviewing the role of TRIM56 in innate immunity and antiviral processes. We also summarize the role of TRIM56 in tumors. Finally, we discuss the future directions of TRIM56 research. Reviewing the antiviral and tumor regulatory functions and specific mechanisms of TRIM56 is beneficial to provide new ideas for developing novel antiviral drugs and enriching therapeutic strategies against tumors.
2. Structure and Expression of TRIM56
TRIM56, also known as Ring finger protein 109 (RNF109), is an 81 kDa protein of 755 amino acids encoded by the TRIM56 gene on human chromosome 7. The protein contains three structural domains, a RING domain, a B-box domain, and a coiled-coil domain (Figure 1). Because it lacks a C-terminal structural domain, TRIM56 belongs to the C-V subfamily.
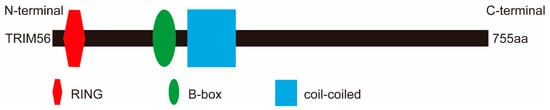
Figure 1.
Schematic representation of the domains of TRIM56. TRIM56 has three structural domains, an N-terminal RING domain (red), a B-box domain (green), and a coiled-coil domain (blue). The human TRIM56 transcript is 755 aa long.
The RING structural domain is a unique linear sequence of cysteine and histidine residues in a zinc finger structural domain that forms the catalytic center of the ubiquitinating enzyme. Ubiquitination is a very important post-translational modification process that plays roles in innate immune and tumorigenic development [38,39,40]. Ubiquitin is a 76-residue polypeptide. The key enzymes required for the ubiquitination process are ubiquitin-activating enzyme E1, ubiquitin-binding enzyme E2, and ubiquitin-ligase E3 [41]. Among them, E3 ubiquitin ligase can catalyze the covalent binding of ubiquitin molecules to substrates. E3 ubiquitin ligases can be classified into several groups according to their specific structural domains: the RING family, the family of homologous to E6AP C-terminus (HECT), RBR E3s, and those of unclassified type [42]. As we mention above, most members of the TRIM family have a RING structural domain, and most TRIM members have been identified as functional E3 ubiquitin ligases [43]. Notably, a few TRIM proteins do not contain any RING domain, such as TRIM14 and TRIM66 [6].
The B-box structural domain consists of small peptide sequences that contain finger-like protrusion. Although the B-box structural domain also contains a “zinc finger” structure, it generally does not exert E3 ubiquitin ligase activity. There are two distinct isoforms of the B-box, B-box1 and B-box2. Most TRIM proteins contain one B-box2 structural domain or two B-box structural domains, while a few TRIMs, such as TRIM69, do not have either structural domain [44]. The B-box structural domain is thought to be involved in the recognition of target proteins by TRIM proteins [4]. The coiled-coil structure domain of TRIMs can serve as a scaffold for mediating the homomeric and heteromeric assembly of TRIMs and other proteins. Additionally, it also exhibits enzymatic or nucleic acid binding activity [21,45].
TRIM56 can act as an E3 ubiquitin ligase that catalyzes the ubiquitination of Vimentin, DVL2 (Dishevelled-2), ERα, SAP18 (Sin3A associated protein 18), IκBα, STING, cGAS, and TGF-β-activated kinase 1 (TAK1) [23,31,32,33,36]. Interestingly, TRIM56 also has deubiquitinating enzyme activity and the ability to bind RNA [29,30,46]. The relationship between the exertion of these functions and the structure needs to be further investigated.
TRIM56 is widely expressed in various tissues of adult mammals [47]. Similar to many other TRIM proteins, the expression of TRIM56 is regulated by type I IFN. The expression level of TRIM56 was significantly upregulated in cells after type I IFN treatment [22,48]. There are differences in the subcellular distribution of TRIM proteins [49]. Some TRIM proteins are widely distributed in the cytoplasm and nucleus, such as TRIM30 and TRIM32. Some are only present in the nucleus, such as TRIM19, and some are only present in the cytoplasm, such as TRIM29. In resting cells, the TRIM56 protein is only present in the cytoplasm and thus interacts with cytoplasmic proteins [47].
4. The Function of TRIM56 in Tumors
By regulating various signaling pathways and proteins in an E3 ligase-dependent or -independent manner, TRIM56 plays different roles in different tumors. It inhibits ovarian cancer, multiple myeloma, lung adenocarcinoma, hepatocellular carcinoma, and leukemia; however, it promotes the development of glioma, breast cancer, and Kaposi’s sarcoma (Figure 3 and Table 2).
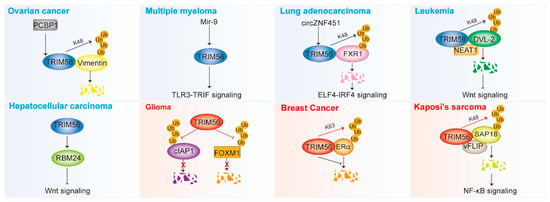
Figure 3.
TRIM56 is involved in the development of cancer (see also Table 2). TRIM56 plays a dual role in tumors. The red color in the figure represents the cancer-promoting function and the blue color represents the cancer-suppressing function. TRIM56 promotes the development of glioma, breast cancer, and Kaposi’s sarcoma, but is an oncogenic repressor in ovarian cancer, multiple myeloma, lung adenocarcinoma, hepatocellular carcinoma, and leukemia. TRIM56 affects multiple signaling pathways, including the TLR3-TRIF pathway, the ELF4-IRF4 pathway, the Wnt pathway, and the NF-κB pathway. TRIM56 mediates the ubiquitin degradation of key proteins, such as Vimentin, FXR1, DVL2, and SAP18. TRIM56 can stabilize some key proteins via K63 ubiquitination (ERα) and deubiquitination (cIAP1 and FOXM1). Ub, ubiquitin.

Table 2.
Expression and clinical significance of TRIM56 in various cancers.
4.1. Tumor Suppression
4.1.1. Ovarian Cancer
Ovarian cancer is a gynecologic oncologic disease and one of the major female lethal cancers [82]. Epithelial-to-mesenchymal transition (EMT) leads to tumor metastasis, which accelerates tumor progression [83]. Vimentin is an important protein that regulates EMT and cancer progression in ovarian cancer [84]. TRIM56 is able to ubiquitinate and downregulate Vimentin. The TRIM56 inhibition of ovarian cancer migration and invasion in vitro occurs via an inhibitory effect on Vimentin [33]. TRIM56 expression is post-transcriptionally regulated at the translational level by RNA-binding protein poly r(c)-binding protein 1 (PCBP1) [85]. PCBP1 promotes ovarian cancer migration and invasion in vitro by inhibiting TRIM56 translation, reducing its protein levels, thereby inducing Vimentin expression [33,85].
4.1.2. Multiple Myeloma
Multiple myeloma (MM) is a group of plasma cell malignancies characterized by the extensive clonal proliferation of tumor plasma cells in the bone marrow [86]. MM accounts for approximately 10% of hematologic neoplastic diseases [87]. The bone marrow microenvironment and cytokines such as interleukin (IL)-6 and TNF (tumor necrosis factor)-α play an important role in the growth and survival of MM cells and are associated with the clinical presentation and prognosis of MM [88]. The expression of TRIM56 is significantly decreased in MM cells. TRIM56 inhibits cell proliferation and produces inflammatory cytokines by activating the TLR3/TRIF signaling pathway [34]. Huang et al. found that cell lines from early MM patients showed upregulated miR-9 expression, which promoted MM cell proliferation and reduced apoptosis. TRIM56 is a target protein of miR-9 that reverses miR-9-mediated proliferation and anti-apoptotic effects. Thus, miR-9 promotes MM development and progression with the regulation of the TRIM56/NF-κB pathway [89].
4.1.3. Lung Cancer
Lung cancer is the most common and lethal malignancy, with lung adenocarcinoma accounting for up to 40% of cases [90]. The reduced expression of TRIM56 in lung adenocarcinoma is associated with poor prognosis. The overexpression of TRIM56 inhibits the invasion and migration of lung adenocarcinoma cells [35]. In the treatment of advanced lung cancer, immunotherapy has achieved some success, but the problem of immunotherapy resistance cannot be ignored [91,92]. Exosomal circZNF451 was upregulated in patients with progressive disease compared with lung adenocarcinoma patients in partial remission after PD1 blockade therapy and was associated with a poor clinical prognosis. Exosomal circZNF451 was able to target RNA-binding protein FXR1 in macrophages and promote the ubiquitination of FXR1 via the E3 ubiquitin ligase TRIM56, which in turn activated the ELF4-IRF4 pathway, leading to M2 polarization and suppressive immune microenvironment in macrophages. Exosomal circZNF451 inhibits anti-PD1 therapy in lung adenocarcinoma by polarizing macrophages in complex with TRIM56 and FXR1 [93]. Thus, TRIM56 may serve as a potential therapeutic target and a novel predictive marker for PD1 inhibitor resistance in lung cancer.
4.1.4. Leukemia
DVL2 is a key regulator of Wnt signaling, which stabilizes β-catenin by catabolizing the APC/Axin/CK1α/GSK3β degradation complex [94]. DVL2 expression levels are closely correlated with Wnt activity and tumor progression [31,95]. The TRIM56-mediated degradation of DVL2 inactivates Wnt signaling and thus inhibits tumor development. Nuclear paraspeckle assembly transcript 1 (NEAT1) localizes to the nucleus and is able to inhibit AML stem cell self-renewal and leukemogenesis by activating Wnt signaling [36]. Alternative splicing (AS) often alters the function of proteins, which in turn affects tumor development [96]. However, heterodimer NEAT1 is localized in the cytoplasm and is able to interact with TRIM56 and DVL2 by enhancing TRIM56-mediated DVL2 degradation, thereby inactivating Wnt signaling [36]. Targeting DVL2 using TRIM56- or DVL2-interacting NEAT1 truncators may be a potential strategy for the treatment of AML.
4.1.5. Hepatocellular Carcinoma
Yang et al. found that downregulated TRIM56 in hepatocellular carcinoma (HCC) patient samples was strongly associated with pathological stage and prognosis [37]. TRIM56 negatively regulated key genes in Wnt signaling, β-catenin, c-Myc, RBM24, MMP-9, and cyclin D1, as well as Wnt. Among them, RBM24 was shown to be a downstream target gene of TRIM56. The overexpression of TRIM56 inhibited cell proliferation, whereas the knockdown of TRIM56 had the opposite effect. TRIM56 inhibited HCC proliferation by inactivating Wnt signaling and targeting RBM24 [37].
Hepatocellular carcinoma has been associated with viral infections of type B and C [97]. TRIM56 can inhibit the replication of HBV [77]. In addition, TRIM56 was able to promote the induction of TLR3-mediated chemokines after HCV infection [24].
4.2. Tumor Promotion
4.2.1. Glioma
TRIM56 expression is significantly increased in glioblastoma tissues and cell lines. High TRIM56 expression is associated with a poor prognosis in glioma patients [29,30]. TRIM56 can downregulate the ubiquitination level of cIAP1, thereby reducing the degradation of cIAP1 [30]. cIAP1 belongs to the inhibitors of apoptosis (IAP) family, which regulates the cell cycle and tumor development [98]. Several studies have shown that cIAP1 is highly expressed in various human cancers and plays a key oncogenic role [98,99]. In glioma, TRIM56 does not function as an E3 ligase but as a deubiquitinating enzyme to stabilize the expression of apoptosis inhibitor cIAP1, thereby promoting glioma progression [30]. Recurrent glioblastoma is characterized by resistance to radiotherapy or chemotherapy. TRIM56 increases FOXM1 protein levels and enhances FOXM1 by means of deubiquitination. TRIM56 inhibits the radiosensitivity of human glioblastoma by regulating FOXM1-mediated DNA repair. Targeting TRIM56 may be an effective approach to reverse radioresistance in glioblastoma recurrence [29]. Interestingly, TRIM56 in gliomas function as deubiquitinating enzymes rather than E3 ligases.
4.2.2. Breast Cancer
Breast cancer is the most common cancer in women worldwide [100,101]. The knockdown of TRIM56 enhances the proliferation and metastasis of breast cancer cells. The expression of TRIM56 is positively correlated with ERα and PR in breast cancer samples and is associated with poor prognosis in patients treated with endocrine therapy. Approximately 60–70% of breast cancer patients are Erα-positive [102]. Estrogen-selective modulators, such as tamoxifen, are emerging as effective agents for controlling ERα breast cancer progression [103]. However, tamoxifen resistance develops during long-term treatment and cancer progression [104]. TRIM56 catalyzes the formation of K63-linked polyubiquitin chains of ERα, thereby prolonging the stability of the ERα protein [31]. Breast cancer proliferation requires transduction via the ERα signaling pathway. Therefore, TRIM56-targeted therapy may address treatment resistance, thereby inhibiting cancer cell proliferation.
4.2.3. Kaposi’s Sarcoma
Kaposi’s Sarcoma (KS) is a common AIDS-associated cancer caused by KS-associated herpesvirus (KSHV) infection [105]. KSHV encodes viral FLICE inhibitory protein (vFLIP), a viral oncogenic protein. vFLIP promotes cell migration, invasion, and angiogenesis by downregulating the SAP18-HDAC1 complex. Specifically, vFLIP degrades SAP18 via the ubiquitin–proteasome pathway by recruiting E3 ubiquitin ligase TRIM56, which ultimately activates the NF-κB signaling pathway [32]. Interestingly, KSHV is closely associated with the development of KS, primary exudative lymphoma (PEL), and other diseases [106]. Moreover, the deletion of the TRIM56 gene has been found in PEL patients [107]. The relationship between TRIM56 and KSHV, and KSHV-related tumors needs to be further investigated.
4.3. Regulation of TRIM56 Expression in Tumors
TRIM56 is aberrantly expressed in a variety of tumors. TRIM56 was lowly expressed in multiple myeloma [88], ovarian cancer [85], lung adenocarcinoma [35], and hepatocellular carcinoma [37]. TRIM56 was highly expressed in glioma [29,30]. Furthermore, by analyzing the data in the TCGA database, we found that the expression levels of TRIM56 were significantly low in lung squamous cell carcinoma, uterine corpus endometrial carcinoma, and uterine carcinosarcoma, and significantly high in pancreatic adenocarcinoma, glioblastoma, lower-grade glioma, and thymoma [108] (Figure 4). In addition to the above tumors, TRIM56 was highly expressed in living patients with muscle-invasive bladder cancer (MIBC) [109]. The function and regulatory mechanisms of TRIM56 in the above tumors remain to be investigated. In addition, the upstream regulatory mechanisms of TRIM56 are not well understood. In ovarian cancer, PCBP1 inhibits TRIM56 translation [85]. In multiple myeloma, mir-9 downregulates TRIM56 expression [89]. In lung adenocarcinoma, mir-542 and mir-627 have the potential to inhibit TRIM56 expression [35].
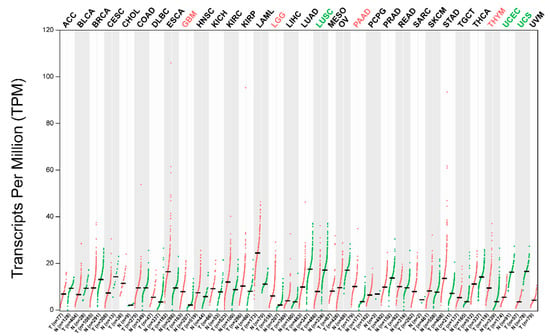
Figure 4.
TRIM56 is aberrantly expressed in several types of tumors. By analyzing the data in the TCGA database, we found that the expression levels of TRIM56 were significantly low in lung squamous cell carcinoma (LUSC), uterine corpus endometrial carcinoma (UCEC), and uterine carcinosarcoma (UCS), and significantly high in pancreatic adenocarcinoma (PAAD), glioblastoma (GBM), lower-grade glioma (LGG), and thymoma (THYM). Data were analyzed using GEPIA 2.0.
5. Concluding Remarks and Future Perspectives
This review summarizes the role of TRIM56 in antiviral processes and the development of tumorigenesis. Elucidating the altered expression of TRIM56 and its potential mechanisms in the pathophysiology of cancer and other diseases may provide insights for the development of new and more effective therapeutic strategies. However, there are still no reports on the clinical applications of TRIM56 in small-molecule therapy. A further understanding of the crystal structure of TRIM56 and its ligand-binding complexes could refine the structure-based design for the development of specific small molecules targeting TRIM56, ultimately leading to therapeutic applications.
As an E3 ubiquitin ligase, TRIM56 catalyzes the ubiquitination modification of substrates [23,26,31,32,33,36,61,77]. The fate of the substrate protein depends on the lysine used to form the ubiquitin molecule of the heteropeptide bond. Different ubiquitination chain lengths (monoubiquitination and polyubiquitination) and a wide variety of ubiquitination chain types (linked by Met1, Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63) play an extremely important role in protein activity, protein–protein interactions, and protein subcellular localization [39,110]. TRIM56 is able to catalyze the formation of K48, K63-linked, or M1-linked ubiquitination [111]. In addition, biological roles of TRIM56 independent of E3 ligases have been identified, including RNA binding and deubiquitinating enzyme activity [29,30,46].
The innate immune response is the first line of host defense and is characterized by the production of IFN-I and ISGs to limit viral infection and transmission [112,113,114]. Many studies have shown that TRIM56 plays a key role in the precise coordination of key signaling molecules and their associated pathways. Here, we discuss the current mechanisms regarding the involvement of TRIM56 in the regulation of TLRs, the cGAS-STING pathway, and downstream ISGs [23,26,61]. Whether TRIM56 regulates the RLRs pathway remains to be further investigated. In addition, TRIM56 is able to regulate innate antiviral signaling in a ubiquitination-independent manner, and the specific mechanisms of regulation remain to be explored.
The C-terminal region of TRIM56 mediates protein–protein or protein–RNA interactions between TRIM56 and cellular viral proteins/RNAs and can inhibit viral RNA replication. In addition, the E3 ligase activity of TRIM56 may regulate post-translational modifications of viral proteins and/or host factors to inhibit the replication of positive-stranded RNA viruses. Although TRIM56 is widely expressed in many tissues, the highest expression levels of the protein were detected in the lung and stomach [47]. The presence of pathogenic microorganisms in the respiratory and gastrointestinal tracts, which are continuously exposed to the external environment, may account for the differences in tissue distribution [115].
TRIM56 has been reported to exert oncogenic or tumorigenic potential in solid tumors and hematological cancers [116,117]. The importance of exploring the function of TRIM56 in various malignancies comes not only from the understanding of the key mechanisms of tumor development but also from the important translational potential. In recent years, TRIM family proteins have made some progress in targeted cancer therapy, such as TRIM8-targeted approaches for chemotherapy-resistant colorectal cancer and TRIM24-targeted regimens for glioblastoma [118,119]. TRIM56 can affect tumor cell proliferation, apoptosis, and metastasis by regulating downstream molecules [29,30,31,32,33,34,35,36,37]. However, the effect of TRIM56 on tumor immunity is still unknown. TRIM56 can modulate the innate immune response and promote the production of type I IFNs and ISGs [23,24,25,26,27,65]. Notably, the innate immune response plays an important role in cancer immune escape [120,121]. Therefore, exploring the effect of TRIM56 on tumor immune response is a future research direction.
Author Contributions
L.F. prepared and wrote for this review; X.Z., Q.J. and X.C. commented on the previous versions of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (82273034), Natural Science Foundation of Shandong Province (ZR2022MC035), and Shandong Youth Science and Technology Innovation Team (2022KJ298).
Informed Consent Statement
Not applicable.
Data Availability Statement
All data relevant to this review are included in the text, references, table, and figures.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| TRIM | Tripartite-motif |
| RING | Really interesting new gene |
| RBCC | RING-B-box-Coiled-Coil |
| FNIII | Fibronectin type III repeat region |
| ACID | Acid-rich region |
| MATH | Meprin and TRAF-homologous structural domain |
| ARF | ADP-ribosylation factor family structural domain |
| FIL | Filamine-type IG structural domain |
| BROMO | Bromodomain |
| TM | Transmembrane region |
| IFN | Interferon |
| TLRs | Toll-like receptors |
| cGAS | Cyclic GMP-AMP synthase |
| STING | Stimulator interferon genes HECT: homologous to E6AP C-terminus |
| DVL2 | Dishevelled-2 |
| SAP18 | Sin3A associated protein 18 |
| TAK1 | TGF-β-activated kinase 1 |
| PAMPs | Pathogen-associated molecular patterns |
| PRRs | Pattern recognition receptors |
| RIG-I | Retinoic-acid inducible gene-I |
| RLRs | RIG-I-like receptors |
| ISGs | Interferon-stimulated genes |
| IFN-I | Type I IFN |
| MyD88 | Myeloid differentiation factor 88 |
| IRAK1/4 | IL-1 receptor-associated kinase 1/4 |
| TRAF6 | TNF receptor-associated factor 6 |
| TAB2 | TAK1/MAP3K7 binding protein 2 |
| IκKα/β/γ | IκB kinase α/β/γ |
| TRIF | Toll-IL-1 receptor (TIR) domain-containing adaptor inducing IFN-β |
| cGAMP | Cyclic GMP-AMP |
| ER | Endoplasmic reticulum |
| HSV-1 | Herpes simplex virus-1 |
| UBXN3B | Ubiquitin regulatory X domain-containing proteins 3B |
| VSV | Vesicular stomatitis virus |
| NEMO | NF-κB essential modifier |
| HCV | Hepatitis C virus |
| N(pro) | N-terminal protease |
| BVDV | Bovine viral diarrhea virus |
| IRF3 | Interferon regulatory factor 3 |
| YFV | Yellow fever virus |
| DENV2 | Dengue virus serotype 2 |
| HCoV | Human coronavirus |
| ZIKV | Zika virus |
| PEDV | Porcine epidemic diarrhea virus |
| HIV | Human immunodeficiency virus |
| COVID-19 | Coronavirus Disease 2019 |
| EMCV | Encephalomyocarditis virus |
| HBV | Hepatitis B virus |
| EMT | Epithelial-to-mesenchymal transition |
| PCBP1 | Poly r(c)-binding protein 1 |
| MM | Multiple myeloma |
| IL | Interleukin |
| TNF | Tumor necrosis factor |
| NEAT1 | Nuclear paraspeckle assembly transcript 1 |
| HCC | Hepatocellular carcinoma |
| IAPs | Inhibitors of apoptosis |
| KS | Kaposi’s sarcoma |
| KSHV | KS-associated herpesvirus |
| vFLIP | Viral FLICE inhibitory protein |
| PEL | Primary exudative lymphoma |
| MIBC | Muscle-invasive bladder cancer |
References
- Hatakeyama, S. TRIM Family Proteins: Roles in Autophagy, Immunity, and Carcinogenesis. Trends Biochem. Sci. 2017, 42, 297–311. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.; Sun, X.; Li, P.; Liu, X.; Zhang, X.; Chen, Q.; Xin, H. TRIM family contribute to tumorigenesis, cancer development, and drug resistance. Exp. Hematol. Oncol. 2022, 11, 75. [Google Scholar] [CrossRef] [PubMed]
- Esposito, D.; Koliopoulos, M.G.; Rittinger, K. Structural determinants of TRIM protein function. Biochem. Soc. Trans. 2017, 45, 183–191. [Google Scholar] [CrossRef]
- Van Gent, M.; Sparrer, K.M.J.; Gack, M.U. TRIM Proteins and Their Roles in Antiviral Host Defenses. Annu. Rev. Virol. 2018, 5, 385–405. [Google Scholar] [CrossRef] [PubMed]
- Vunjak, M.; Versteeg, G.A. TRIM proteins. Curr. Biol. 2019, 29, R42–R44. [Google Scholar] [CrossRef]
- Meroni, G. Genomics and evolution of the TRIM gene family. Adv. Exp. Med. Biol. 2012, 770, 1–9. [Google Scholar] [CrossRef]
- Kimura, T.; Jain, A.; Choi, S.W.; Mandell, M.A.; Johansen, T.; Deretic, V. TRIM-directed selective autophagy regulates immune activation. Autophagy 2017, 13, 989–990. [Google Scholar] [CrossRef]
- Meroni, G. TRIM E3 Ubiquitin Ligases in Rare Genetic Disorders. Adv. Exp. Med. Biol. 2020, 1233, 311–325. [Google Scholar] [CrossRef]
- Connacher, R.P.; Goldstrohm, A.C. Molecular and biological functions of TRIM-NHL RNA-binding proteins. Wiley Interdiscip. Rev. RNA 2021, 12, e1620. [Google Scholar] [CrossRef]
- Mandell, M.A.; Saha, B.; Thompson, T.A. The Tripartite Nexus: Autophagy, Cancer, and Tripartite Motif-Containing Protein Family Members. Front. Pharmacol. 2020, 11, 308. [Google Scholar] [CrossRef]
- Kumar, S.; Chauhan, S.; Jain, A.; Ponpuak, M.; Choi, S.W.; Mudd, M.; Peters, R.; Mandell, M.A.; Johansen, T.; Deretic, V. Galectins and TRIMs directly interact and orchestrate autophagic response to endomembrane damage. Autophagy 2017, 13, 1086–1087. [Google Scholar] [CrossRef] [PubMed]
- Venuto, S.; Merla, G. E3 Ubiquitin Ligase TRIM Proteins, Cell Cycle and Mitosis. Cells 2019, 8, 510. [Google Scholar] [CrossRef]
- Koepke, L.; Gack, M.U.; Sparrer, K.M. The antiviral activities of TRIM proteins. Curr. Opin. Microbiol. 2021, 59, 50–57. [Google Scholar] [CrossRef]
- Fletcher, A.J.; Towers, G.J. Inhibition of retroviral replication by members of the TRIM protein family. Curr. Top. Microbiol. Immunol. 2013, 371, 29–66. [Google Scholar] [CrossRef]
- Khan, R.; Khan, A.; Ali, A.; Idrees, M. The interplay between viruses and TRIM family proteins. Rev. Med. Virol. 2019, 29, e2028. [Google Scholar] [CrossRef] [PubMed]
- Tomar, D.; Singh, R. TRIM family proteins: Emerging class of RING E3 ligases as regulator of NF-kappaB pathway. Biol. Cell 2015, 107, 22–40. [Google Scholar] [CrossRef]
- Zhu, Y.; Afolabi, L.O.; Wan, X.; Shim, J.S.; Chen, L. TRIM family proteins: Roles in proteostasis and neurodegenerative diseases. Open Biol. 2022, 12, 220098. [Google Scholar] [CrossRef]
- Jaworska, A.M.; Wlodarczyk, N.A.; Mackiewicz, A.; Czerwinska, P. The role of TRIM family proteins in the regulation of cancer stem cell self-renewal. Stem Cells 2020, 38, 165–173. [Google Scholar] [CrossRef]
- Kedia, S.; Aghanoori, M.R.; Burns, K.M.L.; Subha, M.; Williams, L.; Wen, P.; Kopp, D.; Erickson, S.L.; Harvey, E.M.; Chen, X.; et al. Ubiquitination and deubiquitination of 4E-T regulate neural progenitor cell maintenance and neurogenesis by controlling P-body formation. Cell Rep. 2022, 40, 111070. [Google Scholar] [CrossRef]
- Cambiaghi, V.; Giuliani, V.; Lombardi, S.; Marinelli, C.; Toffalorio, F.; Pelicci, P.G. TRIM proteins in cancer. Adv. Exp. Med. Biol. 2012, 770, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Giannopoulou, A.I.; Xanthopoulos, C.; Piperi, C.; Kostareli, E. Emerging Roles of TRIM Family Proteins in Gliomas Pathogenesis. Cancers 2022, 14, 4536. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, T.; Zou, J.; Saitoh, T.; Kumar, H.; Abe, T.; Matsuura, Y.; Kawai, T.; Akira, S. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity 2010, 33, 765–776. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, Y.; Ding, C.; Zhu, X.; Song, X.; Ren, Y.; Wang, Q.; Zhang, Y.; Sun, X. TRIM56 positively regulates TNFalpha-induced NF-kappaB signaling by enhancing the ubiquitination of TAK1. Int. J. Biol. Macromol. 2022, 219, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Li, N.L.; Wang, J.; Liu, B.; Lester, S.; Li, K. TRIM56 is an essential component of the TLR3 antiviral signaling pathway. J. Biol. Chem. 2012, 287, 36404–36413. [Google Scholar] [CrossRef]
- Seo, G.J.; Kim, C.; Shin, W.J.; Sklan, E.H.; Eoh, H.; Jung, J.U. TRIM56-mediated monoubiquitination of cGAS for cytosolic DNA sensing. Nat. Commun. 2018, 9, 613. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wang, L.; Ketkar, H.; Ma, J.; Yang, G.; Cui, S.; Geng, T.; Mordue, D.G.; Fujimoto, T.; Cheng, G.; et al. UBXN3B positively regulates STING-mediated antiviral immune responses. Nat. Commun. 2018, 9, 2329. [Google Scholar] [CrossRef]
- Fang, R.; Wang, C.; Jiang, Q.; Lv, M.; Gao, P.; Yu, X.; Mu, P.; Zhang, R.; Bi, S.; Feng, J.M.; et al. NEMO-IKKbeta Are Essential for IRF3 and NF-kappaB Activation in the cGAS-STING Pathway. J. Immunol. 2017, 199, 3222–3233. [Google Scholar] [CrossRef]
- Heidary, F.; Gharebaghi, R. Systematic review of the antiviral properties of TRIM56: A potential therapeutic intervention for COVID-19. Expert Rev. Clin. Immunol. 2020, 16, 973–984. [Google Scholar] [CrossRef]
- Dong, Y.; Xiong, Y.; Zhou, D.; Yao, M.; Wang, X.; Bi, W.; Zhang, J. TRIM56 Reduces Radiosensitization of Human Glioblastoma by Regulating FOXM1-Mediated DNA Repair. Mol. Neurobiol. 2022, 59, 5312–5325. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, Y.; Xue, Z.; Hu, Y.; Zhou, W.; Xue, Z.; Liu, X.; Liu, G.; Li, W.; Liu, X.; et al. TRIM56 promotes malignant progression of glioblastoma by stabilizing cIAP1 protein. J. Exp. Clin. Cancer Res. 2022, 41, 336. [Google Scholar] [CrossRef]
- Xue, M.; Zhang, K.; Mu, K.; Xu, J.; Yang, H.; Liu, Y.; Wang, B.; Wang, Z.; Li, Z.; Kong, Q.; et al. Regulation of estrogen signaling and breast cancer proliferation by an ubiquitin ligase TRIM56. Oncogenesis 2019, 8, 30. [Google Scholar] [CrossRef]
- Ding, X.; Xu, J.; Wang, C.; Feng, Q.; Wang, Q.; Yang, Y.; Lu, H.; Wang, F.; Zhu, K.; Li, W.; et al. Suppression of the SAP18/HDAC1 complex by targeting TRIM56 and Nanog is essential for oncogenic viral FLICE-inhibitory protein-induced acetylation of p65/RelA, NF-kappaB activation, and promotion of cell invasion and angiogenesis. Cell Death Differ. 2019, 26, 1970–1986. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, P.; Su, X.J.; Zhang, B. The ubiquitin ligase TRIM56 inhibits ovarian cancer progression by targeting vimentin. J. Cell Physiol. 2018, 233, 2420–2425. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhao, J.; Li, D.; Hao, J.; He, P.; Wang, H.; Zhang, M. TRIM56 Suppresses Multiple Myeloma Progression by Activating TLR3/TRIF Signaling. Yonsei Med. J. 2018, 59, 43–50. [Google Scholar] [CrossRef]
- Lu, K.; Sui, Y.; Fu, L. Identification of TRIM56 as a Potential Biomarker for Lung Adenocarcinoma. Cancer Manag. Res. 2021, 13, 2201–2213. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Wang, Z.; Sun, Y.; Hu, L.; Bu, P. Cytoplasmic NEAT1 Suppresses AML Stem Cell Self-Renewal and Leukemogenesis through Inactivation of Wnt Signaling. Adv. Sci. 2021, 8, e2100914. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Mao, F.F.; Guo, L.; Guo, W.X. TRIM56 suppresses the malignant development of hepatocellular carcinoma via targeting RBM24 and inactivating the Wnt signaling. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 722–730. [Google Scholar] [CrossRef]
- Popovic, D.; Vucic, D.; Dikic, I. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 2014, 20, 1242–1253. [Google Scholar] [CrossRef]
- Fu, L.; Cui, C.P.; Zhang, X.; Zhang, L. The functions and regulation of Smurfs in cancers. Semin. Cancer Biol. 2020, 67, 102–116. [Google Scholar] [CrossRef]
- Cockram, P.E.; Kist, M.; Prakash, S.; Chen, S.H.; Wertz, I.E.; Vucic, D. Ubiquitination in the regulation of inflammatory cell death and cancer. Cell Death Differ. 2021, 28, 591–605. [Google Scholar] [CrossRef]
- Pickart, C.M.; Eddins, M.J. Ubiquitin: Structures, functions, mechanisms. Biochim. Biophys. Acta 2004, 1695, 55–72. [Google Scholar] [CrossRef]
- Berndsen, C.E.; Wolberger, C. New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 2014, 21, 301–307. [Google Scholar] [CrossRef]
- Ikeda, K.; Inoue, S. TRIM proteins as RING finger E3 ubiquitin ligases. Adv. Exp. Med. Biol. 2012, 770, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Nguyen, X.N.; Kumar, A.; da Silva, C.; Picard, L.; Etienne, L.; Cimarelli, A. Trim69 is a microtubule regulator that acts as a pantropic viral inhibitor. Proc. Natl. Acad. Sci. USA 2022, 119, e2211467119. [Google Scholar] [CrossRef] [PubMed]
- Napolitano, L.M.; Meroni, G. TRIM family: Pleiotropy and diversification through homomultimer and heteromultimer formation. IUBMB Life 2012, 64, 64–71. [Google Scholar] [CrossRef]
- Yang, D.; Li, N.L.; Wei, D.; Liu, B.; Guo, F.; Elbahesh, H.; Zhang, Y.; Zhou, Z.; Chen, G.Y.; Li, K. The E3 ligase TRIM56 is a host restriction factor of Zika virus and depends on its RNA-binding activity but not miRNA regulation, for antiviral function. PLoS Negl. Trop. Dis. 2019, 13, e0007537. [Google Scholar] [CrossRef]
- Wang, J.; Liu, B.; Wang, N.; Lee, Y.M.; Liu, C.; Li, K. TRIM56 is a virus- and interferon-inducible E3 ubiquitin ligase that restricts pestivirus infection. J. Virol. 2011, 85, 3733–3745. [Google Scholar] [CrossRef]
- Carthagena, L.; Bergamaschi, A.; Luna, J.M.; David, A.; Uchil, P.D.; Margottin-Goguet, F.; Mothes, W.; Hazan, U.; Transy, C.; Pancino, G.; et al. Human TRIM gene expression in response to interferons. PLoS ONE 2009, 4, e4894. [Google Scholar] [CrossRef]
- Reymond, A.; Meroni, G.; Fantozzi, A.; Merla, G.; Cairo, S.; Luzi, L.; Riganelli, D.; Zanaria, E.; Messali, S.; Cainarca, S.; et al. The tripartite motif family identifies cell compartments. EMBO J. 2001, 20, 2140–2151. [Google Scholar] [CrossRef] [PubMed]
- Brubaker, S.W.; Bonham, K.S.; Zanoni, I.; Kagan, J.C. Innate immune pattern recognition: A cell biological perspective. Annu. Rev. Immunol. 2015, 33, 257–290. [Google Scholar] [CrossRef] [PubMed]
- Brennan, K.; Bowie, A.G. Activation of host pattern recognition receptors by viruses. Curr. Opin. Microbiol. 2010, 13, 503–507. [Google Scholar] [CrossRef]
- Paludan, S.R.; Bowie, A.G.; Horan, K.A.; Fitzgerald, K.A. Recognition of herpesviruses by the innate immune system. Nat. Rev. Immunol. 2011, 11, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [PubMed]
- Carty, M.; Guy, C.; Bowie, A.G. Detection of Viral Infections by Innate Immunity. Biochem. Pharmacol. 2021, 183, 114316. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Kawai, T. Toll-like receptor signaling pathways. Front. Immunol. 2014, 5, 461. [Google Scholar] [CrossRef]
- Fitzgerald, K.A.; Kagan, J.C. Toll-like Receptors and the Control of Immunity. Cell 2020, 180, 1044–1066. [Google Scholar] [CrossRef]
- Kishida, S.; Sanjo, H.; Akira, S.; Matsumoto, K.; Ninomiya-Tsuji, J. TAK1-binding protein 2 facilitates ubiquitination of TRAF6 and assembly of TRAF6 with IKK in the IL-1 signaling pathway. Genes Cells 2005, 10, 447–454. [Google Scholar] [CrossRef]
- Kumar, H.; Kawai, T.; Akira, S. Toll-like receptors and innate immunity. Biochem. Biophys. Res. Commun. 2009, 388, 621–625. [Google Scholar] [CrossRef]
- Xu, X.; Wang, L.; Liu, Y.; Shi, X.; Yan, Y.; Zhang, S.; Zhang, Q. TRIM56 overexpression restricts porcine epidemic diarrhoea virus replication in Marc-145 cells by enhancing TLR3-TRAF3-mediated IFN-beta antiviral response. J. Gen. Virol. 2022, 103, 001748. [Google Scholar] [CrossRef]
- Andreeva, L.; Hiller, B.; Kostrewa, D.; Lassig, C.; de Oliveira Mann, C.C.; Jan Drexler, D.; Maiser, A.; Gaidt, M.; Leonhardt, H.; Hornung, V.; et al. cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders. Nature 2017, 549, 394–398. [Google Scholar] [CrossRef]
- Wu, J.; Sun, L.; Chen, X.; Du, F.; Shi, H.; Chen, C.; Chen, Z.J. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 2013, 339, 826–830. [Google Scholar] [CrossRef]
- Ritchie, C.; Carozza, J.A.; Li, L. Biochemistry, Cell Biology, and Pathophysiology of the Innate Immune cGAS-cGAMP-STING Pathway. Annu. Rev. Biochem. 2022, 91, 599–628. [Google Scholar] [CrossRef] [PubMed]
- Hussain, B.; Xie, Y.; Jabeen, U.; Lu, D.; Yang, B.; Wu, C.; Shang, G. Activation of STING Based on Its Structural Features. Front. Immunol. 2022, 13, 808607. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, X.; Cui, Y.; Tang, Y.; Chen, W.; Li, S.; Yu, H.; Pan, Y.; Wang, C. The E3 ubiquitin ligase AMFR and INSIG1 bridge the activation of TBK1 kinase by modifying the adaptor STING. Immunity 2014, 41, 919–933. [Google Scholar] [CrossRef]
- Tao, J.; Zhou, X.; Jiang, Z. cGAS-cGAMP-STING: The three musketeers of cytosolic DNA sensing and signaling. IUBMB Life 2016, 68, 858–870. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W. Interferon-Stimulated Genes: What Do They All Do? Annu. Rev. Virol. 2019, 6, 567–584. [Google Scholar] [CrossRef]
- Kane, M.; Zang, T.M.; Rihn, S.J.; Zhang, F.; Kueck, T.; Alim, M.; Schoggins, J.; Rice, C.M.; Wilson, S.J.; Bieniasz, P.D. Identification of Interferon-Stimulated Genes with Antiretroviral Activity. Cell Host Microbe 2016, 20, 392–405. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Li, N.L.; Wang, J.; Shi, P.Y.; Wang, T.; Miller, M.A.; Li, K. Overlapping and distinct molecular determinants dictating the antiviral activities of TRIM56 against flaviviruses and coronavirus. J. Virol. 2014, 88, 13821–13835. [Google Scholar] [CrossRef]
- Musso, D.; Gubler, D.J. Zika Virus. Clin. Microbiol. Rev. 2016, 29, 487–524. [Google Scholar] [CrossRef]
- Wolf, E.; Tuomilehto, J.; Luonamaa, R. Can the high risk of type I diabetes in Finland be explained by familial aggregation and by HLA haplotype distribution? Study Group on Childhood Diabetes in Finland. Adv. Exp. Med. Biol. 1988, 246, 235–239. [Google Scholar] [CrossRef]
- Lucas, S.; Nelson, A.M. HIV and the spectrum of human disease. J. Pathol. 2015, 235, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Uchil, P.D.; Quinlan, B.D.; Chan, W.T.; Luna, J.M.; Mothes, W. TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS Pathog. 2008, 4, e16. [Google Scholar] [CrossRef] [PubMed]
- Baskol, G.; Ozel, M.; Saracoglu, H.; Ulger, B.; Kalin Unuvar, G.; Onuk, S.; Bayram, A.; Karayol Akin, A.; Muhtaroglu, S.; Sagiroglu, P.; et al. New Avenues to Explore in SARS-CoV-2 Infection: Both TRIM25 and TRIM56 Positively Correlate with VEGF, GAS6, and sAXL in COVID-19 Patients. Viral. Immunol. 2022, 35, 690–699. [Google Scholar] [CrossRef]
- Javanian, M.; Barary, M.; Ghebrehewet, S.; Koppolu, V.; Vasigala, V.; Ebrahimpour, S. A brief review of influenza virus infection. J. Med. Virol. 2021, 93, 4638–4646. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.I.; Holmes, E.C. The evolution of epidemic influenza. Nat. Rev. Genet. 2007, 8, 196–205. [Google Scholar] [CrossRef]
- Liu, B.; Li, N.L.; Shen, Y.; Bao, X.; Fabrizio, T.; Elbahesh, H.; Webby, R.J.; Li, K. The C-Terminal Tail of TRIM56 Dictates Antiviral Restriction of Influenza A and B Viruses by Impeding Viral RNA Synthesis. J. Virol. 2016, 90, 4369–4382. [Google Scholar] [CrossRef]
- Tian, X.; Dong, H.; Lai, X.; Ou, G.; Cao, J.; Shi, J.; Xiang, C.; Wang, L.; Zhang, X.; Zhang, K.; et al. TRIM56 impairs HBV infection and replication by inhibiting HBV core promoter activity. Antiviral Res. 2022, 207, 105406. [Google Scholar] [CrossRef]
- Dougan, G.; Baker, S. Salmonella enterica serovar Typhi and the pathogenesis of typhoid fever. Annu. Rev. Microbiol. 2014, 68, 317–336. [Google Scholar] [CrossRef]
- Diao, J.; Zhang, Y.; Huibregtse, J.M.; Zhou, D.; Chen, J. Crystal structure of SopA, a Salmonella effector protein mimicking a eukaryotic ubiquitin ligase. Nat. Struct. Mol. Biol. 2008, 15, 65–70. [Google Scholar] [CrossRef]
- Kamanova, J.; Sun, H.; Lara-Tejero, M.; Galan, J.E. The Salmonella Effector Protein SopA Modulates Innate Immune Responses by Targeting TRIM E3 Ligase Family Members. PLoS Pathog. 2016, 12, e1005552. [Google Scholar] [CrossRef]
- Fiskin, E.; Bhogaraju, S.; Herhaus, L.; Kalayil, S.; Hahn, M.; Dikic, I. Structural basis for the recognition and degradation of host TRIM proteins by Salmonella effector SopA. Nat. Commun. 2017, 8, 14004. [Google Scholar] [CrossRef] [PubMed]
- Penny, S.M. Ovarian Cancer: An Overview. Radiol. Technol. 2020, 91, 561–575. [Google Scholar] [PubMed]
- Mittal, V. Epithelial Mesenchymal Transition in Tumor Metastasis. Annu. Rev. Pathol. 2018, 13, 395–412. [Google Scholar] [CrossRef] [PubMed]
- Das, V.; Bhattacharya, S.; Chikkaputtaiah, C.; Hazra, S.; Pal, M. The basics of epithelial-mesenchymal transition (EMT): A study from a structure, dynamics, and functional perspective. J. Cell. Physiol. 2019, 234, 14535–14555. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, Z.G.; Zhang, P.; Yu, X.F.; Su, X.J. Poly r(C) Binding Protein 1 Regulates Posttranscriptional Expression of the Ubiquitin Ligase TRIM56 in Ovarian Cancer. IUBMB Life 2019, 71, 177–182. [Google Scholar] [CrossRef]
- Cowan, A.J.; Green, D.J.; Kwok, M.; Lee, S.; Coffey, D.G.; Holmberg, L.A.; Tuazon, S.; Gopal, A.K.; Libby, E.N. Diagnosis and Management of Multiple Myeloma: A Review. JAMA 2022, 327, 464–477. [Google Scholar] [CrossRef]
- Rodriguez-Otero, P.; Paiva, B.; San-Miguel, J.F. Roadmap to cure multiple myeloma. Cancer Treat. Rev. 2021, 100, 102284. [Google Scholar] [CrossRef]
- Manier, S.; Sacco, A.; Leleu, X.; Ghobrial, I.M.; Roccaro, A.M. Bone marrow microenvironment in multiple myeloma progression. J. Biomed. Biotechnol. 2012, 2012, 157496. [Google Scholar] [CrossRef]
- Huang, G.; Liu, X.; Zhao, X.; Zhao, J.; Hao, J.; Ren, J.; Chen, Y. MiR-9 promotes multiple myeloma progression by regulating TRIM56/NF-kappaB pathway. Cell Biol. Int. 2019, 43, 1223–1233. [Google Scholar] [CrossRef]
- Nicholson, A.G.; Tsao, M.S.; Beasley, M.B.; Borczuk, A.C.; Brambilla, E.; Cooper, W.A.; Dacic, S.; Jain, D.; Kerr, K.M.; Lantuejoul, S.; et al. The 2021 WHO Classification of Lung Tumors: Impact of Advances Since 2015. J. Thorac. Oncol. 2022, 17, 362–387. [Google Scholar] [CrossRef]
- Rotow, J.; Bivona, T.G. Understanding and targeting resistance mechanisms in NSCLC. Nat. Rev. Cancer 2017, 17, 637–658. [Google Scholar] [CrossRef]
- Hughes, P.E.; Caenepeel, S.; Wu, L.C. Targeted Therapy and Checkpoint Immunotherapy Combinations for the Treatment of Cancer. Trends Immunol. 2016, 37, 462–476. [Google Scholar] [CrossRef]
- Gao, J.; Ao, Y.Q.; Zhang, L.X.; Deng, J.; Wang, S.; Wang, H.K.; Jiang, J.H.; Ding, J.Y. Exosomal circZNF451 restrains anti-PD1 treatment in lung adenocarcinoma via polarizing macrophages by complexing with TRIM56 and FXR1. J. Exp. Clin. Cancer Res. 2022, 41, 295. [Google Scholar] [CrossRef] [PubMed]
- Nusse, R.; Clevers, H. Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017, 169, 985–999. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Castro-Piedras, I.; Simmons, G.E., Jr.; Pruitt, K. Dishevelled: A masterful conductor of complex Wnt signals. Cell Signal. 2018, 47, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Che, Y.; Bai, M.; Lu, K.; Fu, L. Splicing factor SRSF3 promotes the progression of cervical cancer through regulating DDX5. Mol. Carcinog. 2023, 62, 210–223. [Google Scholar] [CrossRef]
- Han, C.L.; Tian, B.W.; Yang, C.C.; Yang, Y.F.; Ma, Y.L.; Ding, Z.N.; Yan, L.J.; Liu, H.; Dong, Z.R.; Chen, Z.Q.; et al. The association of fatty liver and risk of hepatocellular carcinoma in HBV or HCV infected individuals: A systematic review and meta-analysis. Expert Rev. Gastroenterol. Hepatol. 2023, 17, 189–198. [Google Scholar] [CrossRef]
- Zadoroznyj, A.; Dubrez, L. Cytoplasmic and Nuclear Functions of cIAP1. Biomolecules 2022, 12, 322. [Google Scholar] [CrossRef]
- Estornes, Y.; Bertrand, M.J. IAPs, regulators of innate immunity and inflammation. Semin. Cell Dev. Biol. 2015, 39, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Harbeck, N.; Gnant, M. Breast cancer. Lancet 2017, 389, 1134–1150. [Google Scholar] [CrossRef]
- Bai, M.; Che, Y.; Lu, K.; Fu, L. Analysis of deubiquitinase OTUD5 as a biomarker and therapeutic target for cervical cancer by bioinformatic analysis. PeerJ 2020, 8, e9146. [Google Scholar] [CrossRef] [PubMed]
- Rouzier, R.; Perou, C.M.; Symmans, W.F.; Ibrahim, N.; Cristofanilli, M.; Anderson, K.; Hess, K.R.; Stec, J.; Ayers, M.; Wagner, P.; et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin. Cancer Res. 2005, 11, 5678–5685. [Google Scholar] [CrossRef] [PubMed]
- Onitilo, A.A.; Engel, J.M.; Greenlee, R.T.; Mukesh, B.N. Breast cancer subtypes based on ER/PR and Her2 expression: Comparison of clinicopathologic features and survival. Clin. Med. Res. 2009, 7, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Srivastava, A.; Pateriya, A.; Tomar, M.S.; Mishra, A.K.; Shrivastava, A. Metabolic reprograming confers tamoxifen resistance in breast cancer. Chem. Biol. Interact. 2021, 347, 109602. [Google Scholar] [CrossRef]
- Antman, K.; Chang, Y. Kaposi’s sarcoma. N. Engl. J. Med. 2000, 342, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
- Barrett, L.; Dai, L.; Wang, S.; Qin, Z. Kaposi’s sarcoma-associated herpesvirus and extracellular vesicles. J. Med. Virol. 2021, 93, 3294–3299. [Google Scholar] [CrossRef]
- Roy, D.; Sin, S.H.; Damania, B.; Dittmer, D.P. Tumor suppressor genes FHIT and WWOX are deleted in primary effusion lymphoma (PEL) cell lines. Blood 2011, 118, e32–e39. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Zhang, P.B.; Huang, Z.L.; Xu, Y.H.; Huang, J.; Huang, X.Y.; Huang, X.Y. Systematic analysis of gene expression profiles reveals prognostic stratification and underlying mechanisms for muscle-invasive bladder cancer. Cancer Cell Int. 2019, 19, 337. [Google Scholar] [CrossRef]
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef]
- Akutsu, M.; Dikic, I.; Bremm, A. Ubiquitin chain diversity at a glance. J. Cell Sci. 2016, 129, 875–880. [Google Scholar] [CrossRef] [PubMed]
- Ivashkiv, L.B.; Donlin, L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014, 14, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Lv, M.; Zhou, X.; Cui, Y. Roles of peripheral immune cells in the recovery of neurological function after ischemic stroke. Front. Cell Neurosci. 2022, 16, 1013905. [Google Scholar] [CrossRef] [PubMed]
- Corrales, L.; Matson, V.; Flood, B.; Spranger, S.; Gajewski, T.F. Innate immune signaling and regulation in cancer immunotherapy. Cell Res. 2017, 27, 96–108. [Google Scholar] [CrossRef]
- Nisole, S.; Stoye, J.P.; Saib, A. TRIM family proteins: Retroviral restriction and antiviral defence. Nat. Rev. Microbiol. 2005, 3, 799–808. [Google Scholar] [CrossRef]
- Crawford, L.J.; Johnston, C.K.; Irvine, A.E. TRIM proteins in blood cancers. J. Cell Commun. Signal. 2018, 12, 21–29. [Google Scholar] [CrossRef] [PubMed]
- McAvera, R.M.; Crawford, L.J. TIF1 Proteins in Genome Stability and Cancer. Cancers 2020, 12, 2094. [Google Scholar] [CrossRef]
- Marzano, F.; Caratozzolo, M.F.; Pesole, G.; Sbisa, E.; Tullo, A. TRIM Proteins in Colorectal Cancer: TRIM8 as a Promising Therapeutic Target in Chemo Resistance. Biomedicines 2021, 9, 241. [Google Scholar] [CrossRef]
- Gechijian, L.N.; Buckley, D.L.; Lawlor, M.A.; Reyes, J.M.; Paulk, J.; Ott, C.J.; Winter, G.E.; Erb, M.A.; Scott, T.G.; Xu, M.; et al. Functional TRIM24 degrader via conjugation of ineffectual bromodomain and VHL ligands. Nat. Chem. Biol. 2018, 14, 405–412. [Google Scholar] [CrossRef]
- Gajewski, T.F.; Schreiber, H.; Fu, Y.X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013, 14, 1014–1022. [Google Scholar] [CrossRef]
- Vesely, M.D.; Kershaw, M.H.; Schreiber, R.D.; Smyth, M.J. Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol. 2011, 29, 235–271. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).