Molecular Pathways Implicated in Radioresistance of Glioblastoma Multiforme: What Is the Role of Extracellular Vesicles?
Abstract
1. Epidemiology of Glioblastoma Multiforme
1.1. Modern Treatment
1.1.1. Chemotherapy
1.1.2. Radiotherapy
1.2. Radioresistance
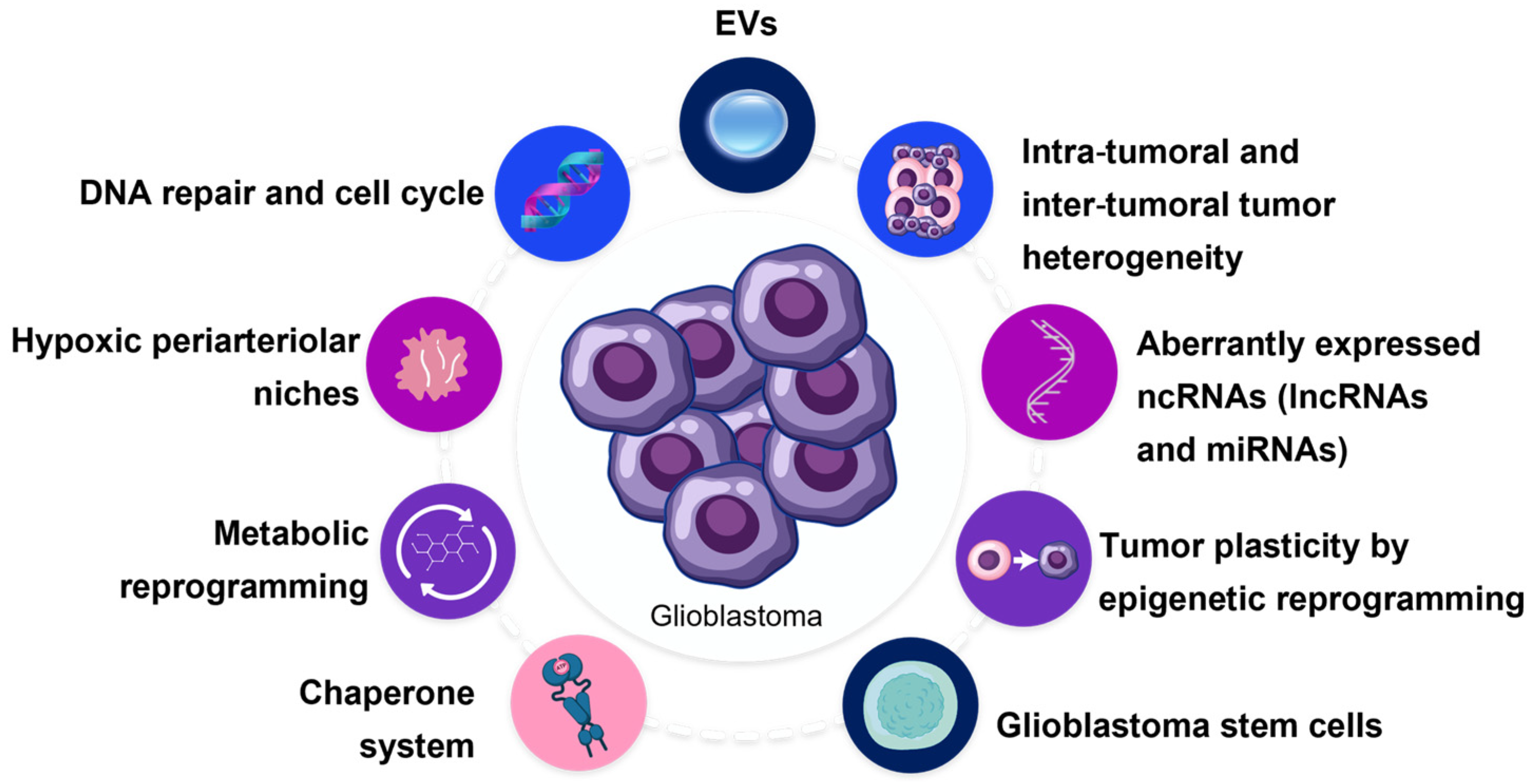
2. Adaptation Mechanisms in Glioblastoma’s Resistance to Radiotherapy
2.1. Glioblastoma Stem Cells
2.2. Tumor Plasticity and Heterogeneity
2.3. Tumor Microenvironment and Hypoxia
2.4. Metabolic Reprogramming and Gene Regulation
GBM Radioresistance and the Chaperone System
2.5. Non-Coding RNAs
2.5.1. miRNAs
2.5.2. lncRNAs
2.6. DNA Repair and Cell Cycle
3. Role of Extracellular Vesicles in Resistance to Radiation Therapy
3.1. Extracellular Vesicles
3.2. Neuron-Derived vs. GBM-Derived EVs
3.2.1. Role of CNS-Derived EVs in Physiological Processes
3.2.2. Role of EVs in Cancer
3.3. Bidirectional Communication between GBM and the Surrounding Tumour Microenvironment
3.4. Role of EVs in Tolerance to Radiation Therapy
4. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ATPase Family AAA Domain Containing 3A | ATAD3A |
| Blood-brain barrier | BBB |
| Cancer stem cells | CSCs |
| Central nervous system | CNS |
| Chaperone system | CS |
| Double-strand DNA breaks | DSBs |
| Endoplasmic reticulum | ER |
| Endosomal sorting complex required for transport | ESCRT |
| Endothelial cells | ECs |
| Epidermal growth factor receptor | EGFR |
| Extracellular vesicles | EVs |
| Glioblastoma multiforme | GBM |
| Glioblastoma stem cells | GSCs |
| Glioma-initiating cells | GICs |
| Histone deacetylase | HDAC |
| Homologous recombination repair | HRR |
| Hypoxia-inducible factor | HIF |
| Hypoxia-inducible factor-1α | AHIF |
| MicroRNAs | miRNAs |
| Multivesicular bodies | MVBs |
| Nicotinamide adenine dinucleotide phosphate | NADPH |
| Non-homologous end-joining | NHEJ |
| O6-methylguanine-DNA methyltransferase gene | MGMT |
| Octamer-binding transcription factor 4 | OCT-4 |
| Poly (ADP-ribose) polymerase | PARP |
| Proliferating cell nuclear antigen | PCNA |
| Proliferating cell nuclear antigen-associated factor | PAF |
| Reactive oxygen species | ROS |
| Replication stress | RS |
| TP53-induced glycolysis and apoptosis regulator | TIGAR |
| Transforming growth factor | TGF |
| Tumor microenvironment | TME |
| Valproic acid | VA |
| Vascular endothelial growth factor | VEGF |
| World Health Organization | WHO |
References
- Hanif, F.; Muzaffar, K.; Perveen, K.; Malhi, S.; Simjee, S. Glioblastoma Multiforme: A Review of Its Epidemiology and Pathogenesis through Clinical Presentation and Treatment. Asian Pac. J. Cancer Prev. 2017, 18, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, A.M.; Taylor, J.W.; Wiencke, J.K.; Wrensch, M.R. Genetic and Molecular Epidemiology of Adult Diffuse Glioma. Nat. Rev. Neurol. 2019, 15, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Cheo, S.; Lim, G.; Lim, K. Glioblastoma Multiforme Outcomes of 107 Patients Treated in Two Singapore Institutions. Singap. Med. J. 2017, 58, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, M.; Shubham, S.; Mandal, K.; Trivedi, V.; Chauhan, R.; Naseera, S. Survival and Prognostic Factors for Glioblastoma Multiforme: Retrospective Single-Institutional Study. Indian J. Cancer 2017, 54, 362. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Patil, N.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2013–2017. Neuro-Oncology 2020, 22, iv1–iv96. [Google Scholar] [CrossRef]
- Li, K.; Lu, D.; Guo, Y.; Wang, C.; Liu, X.; Liu, Y.; Liu, D. Trends and Patterns of Incidence of Diffuse Glioma in Adults in the United States, 1973–2014. Cancer Med. 2018, 7, 5281–5290. [Google Scholar] [CrossRef]
- Darefsky, A.S.; King, J.T.; Dubrow, R. Adult Glioblastoma Multiforme Survival in the Temozolomide Era: A Population-Based Analysis of Surveillance, Epidemiology, and End Results Registries. Cancer 2012, 118, 2163–2172. [Google Scholar] [CrossRef]
- Morgan, E.R.; Norman, A.; Laing, K.; Seal, M.D. Treatment and Outcomes for Glioblastoma in Elderly Compared with Non-Elderly Patients: A Population-Based Study. Curr. Oncol. 2017, 24, 92–98. [Google Scholar] [CrossRef]
- Thomas, R.P.; Recht, L.; Nagpal, S. Advances in the Management of Glioblastoma: The Role of Temozolomide and MGMT Testing. Clin. Pharmacol. 2013, 5, 1–9. [Google Scholar] [CrossRef]
- Choi, S.; Yu, Y.; Grimmer, M.R.; Wahl, M.; Chang, S.M.; Costello, J.F. Temozolomide-Associated Hypermutation in Gliomas. Neuro-Oncology 2018, 20, 1300–1309. [Google Scholar] [CrossRef]
- Momota, H.; Narita, Y.; Miyakita, Y.; Shibui, S. Secondary Hematological Malignancies Associated with Temozolomide in Patients with Glioma. Neuro-Oncology 2013, 15, 1445–1450. [Google Scholar] [CrossRef]
- Thomas, A.; Tanaka, M.; Trepel, J.; Reinhold, W.C.; Rajapakse, V.N.; Pommier, Y. Temozolomide in the Era of Precision Medicine. Cancer Res. 2017, 77, 823–826. [Google Scholar] [CrossRef]
- Campana, D.; Walter, T.; Pusceddu, S.; Gelsomino, F.; Graillot, E.; Prinzi, N.; Spallanzani, A.; Fiorentino, M.; Barritault, M.; Dall’Olio, F.; et al. Correlation between MGMT Promoter Methylation and Response to Temozolomide-Based Therapy in Neuroendocrine Neoplasms: An Observational Retrospective Multicenter Study. Endocrine 2018, 60, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, M.R.; Wang, M.; Aldape, K.D.; Stupp, R.; Hegi, M.E.; Jaeckle, K.A.; Armstrong, T.S.; Wefel, J.S.; Won, M.; Blumenthal, D.T.; et al. Dose-Dense Temozolomide for Newly Diagnosed Glioblastoma: A Randomized Phase III Clinical Trial. J. Clin. Oncol. 2013, 31, 4085–4091. [Google Scholar] [CrossRef] [PubMed]
- Affronti, M.L.; Heery, C.R.; Herndon, J.E., II; Rich, J.N.; Reardon, D.A.; Desjardins, A.; Vredenburgh, J.J.; Friedman, A.H.; Bigner, D.D.; Friedman, H.S. Overall Survival of Newly Diagnosed Glioblastoma Patients Receiving Carmustine Wafers Followed by Radiation and Concurrent Temozolomide plus Rotational Multiagent Chemotherapy. Cancer 2009, 115, 3501–3511. [Google Scholar] [CrossRef] [PubMed]
- Maklad, A.; Sharma, A.; Azimi, I. Calcium Signaling in Brain Cancers: Roles and Therapeutic Targeting. Cancers 2019, 11, 145. [Google Scholar] [CrossRef] [PubMed]
- Grossman, R.; Burger, P.; Soudry, E.; Tyler, B.; Chaichana, K.L.; Weingart, J.; Olivi, A.; Gallia, G.L.; Sidransky, D.; Quiñones-Hinojosa, A.; et al. MGMT Inactivation and Clinical Response in Newly Diagnosed GBM Patients Treated with Gliadel. J. Clin. Neurosci. 2015, 22, 1938–1942. [Google Scholar] [CrossRef] [PubMed]
- Sabel, M.; Giese, A. Safety Profile of Carmustine Wafers in Malignant Glioma: A Review of Controlled Trials and a Decade of Clinical Experience. Curr. Med. Res. Opin. 2008, 24, 3239–3257. [Google Scholar] [CrossRef] [PubMed]
- Bregy, A.; Shah, A.H.; Diaz, M.V.; Pierce, H.E.; Ames, P.L.; Diaz, D.; Komotar, R.J. The Role of Gliadel Wafers in the Treatment of High-Grade Gliomas. Expert Rev. Anticancer Ther. 2013, 13, 1453–1461. [Google Scholar] [CrossRef]
- McGirt, M.J.; Than, K.D.; Weingart, J.D.; Chaichana, K.L.; Attenello, F.J.; Olivi, A.; Laterra, J.; Kleinberg, L.R.; Grossman, S.A.; Brem, H.; et al. Gliadel (BCNU) Wafer plus Concomitant Temozolomide Therapy after Primary Resection of Glioblastoma Multiforme. J. Neurosurg. 2009, 110, 583–588. [Google Scholar] [CrossRef]
- Fleischmann, D.F.; Jenn, J.; Corradini, S.; Ruf, V.; Herms, J.; Forbrig, R.; Unterrainer, M.; Thon, N.; Kreth, F.W.; Belka, C.; et al. Bevacizumab Reduces Toxicity of Reirradiation in Recurrent High-Grade Glioma. Radiother. Oncol. 2019, 138, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Wick, W.; Gorlia, T.; Bendszus, M.; Taphoorn, M.; Sahm, F.; Harting, I.; Brandes, A.A.; Taal, W.; Domont, J.; Idbaih, A.; et al. Lomustine and Bevacizumab in Progressive Glioblastoma. N. Engl. J. Med. 2017, 377, 1954–1963. [Google Scholar] [CrossRef]
- Clarke, J.; Neil, E.; Terziev, R.; Gutin, P.; Barani, I.; Kaley, T.; Lassman, A.B.; Chan, T.A.; Yamada, J.; DeAngelis, L.; et al. Multicenter, Phase 1, Dose Escalation Study of Hypofractionated Stereotactic Radiation Therapy With Bevacizumab for Recurrent Glioblastoma and Anaplastic Astrocytoma. Int. J. Radiat. Oncol. Biol. Phys. 2017, 99, 797–804. [Google Scholar] [CrossRef] [PubMed]
- Kreisl, T.N.; Kim, L.; Moore, K.; Duic, P.; Royce, C.; Stroud, I.; Garren, N.; Mackey, M.; Butman, J.A.; Camphausen, K.; et al. Phase II Trial of Single-Agent Bevacizumab Followed by Bevacizumab plus Irinotecan at Tumor Progression in Recurrent Glioblastoma. J. Clin. Oncol. 2009, 27, 740–745. [Google Scholar] [CrossRef] [PubMed]
- Lovo, E.E.; Moreira, A.; Barahona, K.C.; Ramirez, J.; Campos, F.; Tobar, C.; Caceros, V.; Sallabanda, M.; Sallabanda, K. Stereotactic Radiosurgery for Recurrent Glioblastoma Multiforme: A Retrospective Multi-Institutional Experience. Cureus 2021, 13, e18480. [Google Scholar] [CrossRef]
- Kitange, G.J.; Mladek, A.C.; Carlson, B.L.; Schroeder, M.A.; Pokorny, J.L.; Cen, L.; Decker, P.A.; Wu, W.; Lomberk, G.A.; Gupta, S.K.; et al. Inhibition of Histone Deacetylation Potentiates the Evolution of Acquired Temozolomide Resistance Linked to MGMT Upregulation in Glioblastoma Xenografts. Clin. Cancer Res. 2012, 18, 4070–4079. [Google Scholar] [CrossRef]
- Gonçalves, R.M.; Agnes, J.P.; Delgobo, M.; de Souza, P.O.; Thomé, M.P.; Heimfarth, L.; Lenz, G.; Moreira, J.C.F.; Zanotto-Filho, A. Late Autophagy Inhibitor Chloroquine Improves Efficacy of the Histone Deacetylase Inhibitor SAHA and Temozolomide in Gliomas. Biochem. Pharmacol. 2019, 163, 440–450. [Google Scholar] [CrossRef]
- Cornago, M.; Garcia-Alberich, C.; Blasco-Angulo, N.; Vall-llaura, N.; Nager, M.; Herreros, J.; Comella, J.X.; Sanchis, D.; Llovera, M. Histone Deacetylase Inhibitors Promote Glioma Cell Death by G2 Checkpoint Abrogation Leading to Mitotic Catastrophe. Cell Death Dis. 2014, 5, e1435. [Google Scholar] [CrossRef]
- Lesueur, P.; Lequesne, J.; Grellard, J.-M.; Dugué, A.; Coquan, E.; Brachet, P.-E.; Geffrelot, J.; Kao, W.; Emery, E.; Berro, D.H.; et al. Phase I/IIa Study of Concomitant Radiotherapy with Olaparib and Temozolomide in Unresectable or Partially Resectable Glioblastoma: OLA-TMZ-RTE-01 Trial Protocol. BMC Cancer 2019, 19, 198. [Google Scholar] [CrossRef]
- Herrlinger, U.; Tzaridis, T.; Mack, F.; Steinbach, J.P.; Schlegel, U.; Sabel, M.; Hau, P.; Kortmann, R.-D.; Krex, D.; Grauer, O.; et al. Lomustine-Temozolomide Combination Therapy versus Standard Temozolomide Therapy in Patients with Newly Diagnosed Glioblastoma with Methylated MGMT Promoter (CeTeG/NOA-09): A Randomised, Open-Label, Phase 3 Trial. Lancet 2019, 393, 678–688. [Google Scholar] [CrossRef]
- Weller, M.; Le Rhun, E. How Did Lomustine Become Standard of Care in Recurrent Glioblastoma? Cancer Treat. Rev. 2020, 87, 102029. [Google Scholar] [CrossRef]
- Taslimi, S.; Ye, V.C.; Wen, P.Y.; Zadeh, G. Lessons Learned from Contemporary Glioblastoma Randomized Clinical Trials through Systematic Review and Network Meta-Analysis: Part 2 Recurrent Glioblastoma. Neuro-Oncol. Adv. 2021, 3, vdab029. [Google Scholar] [CrossRef]
- Berendsen, S.; Broekman, M.; Seute, T.; Snijders, T.; van Es, C.; de Vos, F.; Regli, L.; Robe, P. Valproic Acid for the Treatment of Malignant Gliomas: Review of the Preclinical Rationale and Published Clinical Results. Expert Opin. Investig. Drugs 2012, 21, 1391–1415. [Google Scholar] [CrossRef] [PubMed]
- Krauze, A.V.; Myrehaug, S.D.; Chang, M.G.; Holdford, D.J.; Smith, S.; Shih, J.; Tofilon, P.J.; Fine, H.A.; Camphausen, K. A Phase 2 Study of Concurrent Radiation Therapy, Temozolomide, and the Histone Deacetylase Inhibitor Valproic Acid for Patients with Glioblastoma. Int. J. Radiat. Oncol. Biol. Phys. 2015, 92, 986–992. [Google Scholar] [CrossRef] [PubMed]
- Lu, V.M.; Texakalidis, P.; McDonald, K.L.; Mekary, R.A.; Smith, T.R. The Survival Effect of Valproic Acid in Glioblastoma and Its Current Trend: A Systematic Review and Meta-Analysis. Clin. Neurol. Neurosurg. 2018, 174, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, R.; Braga, C.; Santos, G.; Bronze, M.R.; Perry, M.J.; Moreira, R.; Brites, D.; Falcão, A.S. Targeting Gliomas: Can a New Alkylating Hybrid Compound Make a Difference? ACS Chem. Neurosci. 2017, 8, 50–59. [Google Scholar] [CrossRef]
- Braga, C.; Vaz, A.R.; Oliveira, M.C.; Matilde Marques, M.; Moreira, R.; Brites, D.; Perry, M.J. Targeting Gliomas with Triazene-Based Hybrids: Structure-Activity Relationship, Mechanistic Study and Stability. Eur. J. Med. Chem. 2019, 172, 16–25. [Google Scholar] [CrossRef]
- Hingorani, M.; Colley, W.P.; Dixit, S.; Beavis, A.M. Hypofractionated Radiotherapy for Glioblastoma: Strategy for Poor-Risk Patients or Hope for the Future? Br. J. Radiol. 2012, 85, 1211–1328. [Google Scholar] [CrossRef]
- Barani, I.J.; Larson, D.A. Radiation Therapy of Glioblastoma. In Current Understanding and Treatment of Gliomas; Cancer Treatment and Research; Springer: Cham, Switzerland, 2015; pp. 49–73. [Google Scholar]
- Waters, J.D.; Rose, B.; Gonda, D.D.; Scanderbeg, D.J.; Russell, M.; Alksne, J.F.; Murphy, K.; Carter, B.S.; Lawson, J.; Chen, C.C. Immediate Post-Operative Brachytherapy Prior to Irradiation and Temozolomide for Newly Diagnosed Glioblastoma. J. Neuro-Oncol. 2013, 113, 467–477. [Google Scholar] [CrossRef]
- Kickingereder, P.; Hamisch, C.; Suchorska, B.; Galldiks, N.; Visser-Vandewalle, V.; Goldbrunner, R.; Kocher, M.; Treuer, H.; Voges, J.; Ruge, M.I. Low-Dose Rate Stereotactic Iodine-125 Brachytherapy for the Treatment of Inoperable Primary and Recurrent Glioblastoma: Single-Center Experience with 201 Cases. J. Neuro-Oncol. 2014, 120, 615–623. [Google Scholar] [CrossRef]
- Chatzikonstantinou, G.; Ulrich, P.; Archavlis, E.; Zamboglou, N.; Strouthos, I.; Zoga, E.; Baltas, D.; Tselis, N. Interstitial High-Dose-Rate Brachytherapy in the Primary Treatment of Inoperable Glioblastoma Multiforme. J. Contemp. Brachytherapy 2019, 11, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, C.; Romagna, A.; Thon, N.; Niyazi, M.; Watson, J.; Belka, C.; Tonn, J.-C.; Kreth, F.-W.; Nachbichler, S.B. Outcome and Toxicity Profile of Salvage Low-Dose-Rate Iodine-125 Stereotactic Brachytherapy in Recurrent High-Grade Gliomas. Acta Neurochir. 2015, 157, 1757–1764. [Google Scholar] [CrossRef] [PubMed]
- Chatzikonstantinou, G.; Zamboglou, N.; Archavlis, E.; Strouthos, I.; Zoga, E.; Milickovic, N.; Hilaris, B.; Baltas, D.; Rödel, C.; Tselis, N. CT-Guided Interstitial HDR-Brachytherapy for Recurrent Glioblastoma Multiforme: A 20-Year Single-Institute Experience. Strahlenther Onkol. 2018, 194, 1171–1179. [Google Scholar] [CrossRef] [PubMed]
- Morris, S.-A.L.; Zhu, P.; Rao, M.; Martir, M.; Zhu, J.J.; Hsu, S.; Ballester, L.Y.; Day, A.L.; Tandon, N.; Kim, D.H.; et al. Gamma Knife Stereotactic Radiosurgery in Combination with Bevacizumab for Recurrent Glioblastoma. World Neurosurg. 2019, 127, e523–e533. [Google Scholar] [CrossRef] [PubMed]
- Cuneo, K.C.; Vredenburgh, J.J.; Sampson, J.H.; Reardon, D.A.; Desjardins, A.; Peters, K.B.; Friedman, H.S.; Willett, C.G.; Kirkpatrick, J.P. Safety and Efficacy of Stereotactic Radiosurgery and Adjuvant Bevacizumab in Patients with Recurrent Malignant Gliomas. Int. J. Radiat. Oncol. Biol. Phys. 2012, 82, 2018–2024. [Google Scholar] [CrossRef]
- Ozdemir-Kaynak, E.; Qutub, A.A.; Yesil-Celiktas, O. Advances in Glioblastoma Multiforme Treatment: New Models for Nanoparticle Therapy. Front. Physiol. 2018, 9, 170. [Google Scholar] [CrossRef]
- Siebzehnrubl, F.A.; Reynolds, B.A.; Vescovi, A.; Steindler, D.A.; Deleyrolle, L.P. The Origins of Glioma: E Pluribus Unum? Glia 2011, 59, 1135–1147. [Google Scholar] [CrossRef]
- Uribe, D.; Torres, Á.; Rocha, J.D.; Niechi, I.; Oyarzún, C.; Sobrevia, L.; San Martín, R.; Quezada, C. Multidrug Resistance in Glioblastoma Stem-like Cells: Role of the Hypoxic Microenvironment and Adenosine Signaling. Mol. Asp. Med. 2017, 55, 140–151. [Google Scholar] [CrossRef]
- Todorova, P.K.; Mukherjee, B.; Burma, S. MET Signaling Promotes DNA Repair and Radiation Resistance in Glioblastoma Stem-like Cells. Ann. Transl. Med. 2017, 5, 61. [Google Scholar] [CrossRef]
- Gherardi, E.; Birchmeier, W.; Birchmeier, C.; Woude, G. Vande Targeting MET in Cancer: Rationale and Progress. Nat. Rev. Cancer 2012, 12, 89–103. [Google Scholar] [CrossRef]
- De Bacco, F.; D’Ambrosio, A.; Casanova, E.; Orzan, F.; Neggia, R.; Albano, R.; Verginelli, F.; Cominelli, M.; Poliani, P.L.; Luraghi, P.; et al. MET Inhibition Overcomes Radiation Resistance of Glioblastoma Stem-like Cells. EMBO Mol. Med. 2016, 8, 550–568. [Google Scholar] [CrossRef] [PubMed]
- Kelley, K.; Knisely, J.; Symons, M.; Ruggieri, R. Radioresistance of Brain Tumors. Cancers 2016, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Wei, F.; Wu, Y.; He, Y.; Shi, L.; Xiong, F.; Gong, Z.; Guo, C.; Li, X.; Deng, H.; et al. Role of Metabolism in Cancer Cell Radioresistance and Radiosensitization Methods. J. Exp. Clin. Cancer Res. 2018, 37, 87. [Google Scholar] [CrossRef] [PubMed]
- Dell’Albani, P. Stem Cell Markers in Gliomas. Neurochem. Res. 2008, 33, 2407–2415. [Google Scholar] [CrossRef]
- Vlashi, E.; Pajonk, F. Cancer Stem Cells, Cancer Cell Plasticity and Radiation Therapy. Semin. Cancer Biol. 2015, 31, 28–35. [Google Scholar] [CrossRef]
- Schulz, A.; Meyer, F.; Dubrovska, A.; Borgmann, K. Cancer Stem Cells and Radioresistance: DNA Repair and Beyond. Cancers 2019, 11, 862. [Google Scholar] [CrossRef]
- Lathia, J.D.; Mack, S.C.; Mulkearns-Hubert, E.E.; Valentim, C.L.L.; Rich, J.N. Cancer Stem Cells in Glioblastoma. Genes Dev. 2015, 29, 1203–1217. [Google Scholar] [CrossRef]
- Plaks, V.; Kong, N.; Werb, Z. The Cancer Stem Cell Niche: How Essential Is the Niche in Regulating Stemness of Tumor Cells? Cell Stem Cell 2015, 16, 225–238. [Google Scholar] [CrossRef]
- Galli, R.; Binda, E.; Orfanelli, U.; Cipelletti, B.; Gritti, A.; De Vitis, S.; Fiocco, R.; Foroni, C.; Dimeco, F.; Vescovi, A. Isolation and Characterization of Tumorigenic, Stem-like Neural Precursors from Human Glioblastoma. Cancer Res. 2004, 64, 7011–7021. [Google Scholar] [CrossRef]
- Singh, S.K.; Hawkins, C.; Clarke, I.D.; Squire, J.A.; Bayani, J.; Hide, T.; Henkelman, R.M.; Cusimano, M.D.; Dirks, P.B. Identification of Human Brain Tumour Initiating Cells. Nature 2004, 432, 396–401. [Google Scholar] [CrossRef]
- Ignatova, T.N.; Kukekov, V.G.; Laywell, E.D.; Suslov, O.N.; Vrionis, F.D.; Steindler, D.A. Human Cortical Glial Tumors Contain Neural Stem-like Cells Expressing Astroglial and Neuronal Markers in Vitro. Glia 2002, 39, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Hemmati, H.D.; Nakano, I.; Lazareff, J.A.; Masterman-Smith, M.; Geschwind, D.H.; Bronner-Fraser, M.; Kornblum, H.I. Cancerous Stem Cells Can Arise from Pediatric Brain Tumors. Proc. Natl. Acad. Sci. USA 2003, 100, 15178–15183. [Google Scholar] [CrossRef] [PubMed]
- Piccirillo, S.G.M.; Combi, R.; Cajola, L.; Patrizi, A.; Redaelli, S.; Bentivegna, A.; Baronchelli, S.; Maira, G.; Pollo, B.; Mangiola, A.; et al. Distinct Pools of Cancer Stem-like Cells Coexist within Human Glioblastomas and Display Different Tumorigenicity and Independent Genomic Evolution. Oncogene 2009, 28, 1807–1811. [Google Scholar] [CrossRef] [PubMed]
- Lathia, J.D.; Gallagher, J.; Heddleston, J.M.; Wang, J.; Eyler, C.E.; Macswords, J.; Wu, Q.; Vasanji, A.; McLendon, R.E.; Hjelmeland, A.B.; et al. Integrin Alpha 6 Regulates Glioblastoma Stem Cells. Cell Stem Cell 2010, 6, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Stopschinski, B.E.; Beier, C.P.; Beier, D. Glioblastoma Cancer Stem Cells—From Concept to Clinical Application. Cancer Lett. 2013, 338, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Zeppernick, F.; Ahmadi, R.; Campos, B.; Dictus, C.; Helmke, B.M.; Becker, N.; Lichter, P.; Unterberg, A.; Radlwimmer, B.; Herold-Mende, C.C. Stem Cell Marker CD133 Affects Clinical Outcome in Glioma Patients. Clin. Cancer Res. 2008, 14, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Aoyagi, M.; Wakimoto, H.; Ando, N.; Nariai, T.; Yamamoto, M.; Ohno, K. Accumulation of CD133-Positive Glioma Cells after High-Dose Irradiation by Gamma Knife Surgery plus External Beam Radiation. J. Neurosurg. 2010, 113, 310–318. [Google Scholar] [CrossRef]
- Bao, S.; Wu, Q.; McLendon, R.E.; Hao, Y.; Shi, Q.; Hjelmeland, A.B.; Dewhirst, M.W.; Bigner, D.D.; Rich, J.N. Glioma Stem Cells Promote Radioresistance by Preferential Activation of the DNA Damage Response. Nature 2006, 444, 756–760. [Google Scholar] [CrossRef]
- Wang, W.; Long, L.; Wang, L.; Tan, C.; Fei, X.; Chen, L.; Huang, Q.; Liang, Z. Knockdown of Cathepsin L Promotes Radiosensitivity of Glioma Stem Cells Both in Vivo and in Vitro. Cancer Lett. 2016, 371, 274–284. [Google Scholar] [CrossRef]
- Dean, M.; Fojo, T.; Bates, S. Tumour Stem Cells and Drug Resistance. Nat. Rev. Cancer 2005, 5, 275–284. [Google Scholar] [CrossRef]
- Ong, D.S.T.; Hu, B.; Ho, Y.W.; Sauvé, C.-E.G.; Bristow, C.A.; Wang, Q.; Multani, A.S.; Chen, P.; Nezi, L.; Jiang, S.; et al. PAF Promotes Stemness and Radioresistance of Glioma Stem Cells. Proc. Natl. Acad. Sci. USA 2017, 114, E9086–E9095. [Google Scholar] [CrossRef] [PubMed]
- Almendro, V.; Marusyk, A.; Polyak, K. Cellular Heterogeneity and Molecular Evolution in Cancer. Annu. Rev. Pathol. 2013, 8, 277–302. [Google Scholar] [CrossRef] [PubMed]
- De Sousa E Melo, F.; Vermeulen, L.; Fessler, E.; Medema, J.P. Cancer Heterogeneity—A Multifaceted View. EMBO Rep. 2013, 14, 686–695. [Google Scholar] [CrossRef] [PubMed]
- Wick, W.; Kessler, T. New Glioblastoma Heterogeneity Atlas—A Shared Resource. Nat. Rev. Neurol. 2018, 14, 453–454. [Google Scholar] [CrossRef] [PubMed]
- Davidson, N.E.; Armstrong, S.A.; Coussens, L.M.; Cruz-Correa, M.R.; DeBerardinis, R.J.; Doroshow, J.H.; Foti, M.; Hwu, P.; Kensler, T.W.; Morrow, M.; et al. AACR Cancer Progress Report 2016. Clin. Cancer Res. 2016, 22, S1–S137. [Google Scholar] [CrossRef] [PubMed]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Navin, N.; Kendall, J.; Troge, J.; Andrews, P.; Rodgers, L.; McIndoo, J.; Cook, K.; Stepansky, A.; Levy, D.; Esposito, D.; et al. Tumour Evolution Inferred by Single-Cell Sequencing. Nature 2011, 472, 90–94. [Google Scholar] [CrossRef]
- Parker, N.R.; Hudson, A.L.; Khong, P.; Parkinson, J.F.; Dwight, T.; Ikin, R.J.; Zhu, Y.; Cheng, Z.J.; Vafaee, F.; Chen, J.; et al. Intratumoral Heterogeneity Identified at the Epigenetic, Genetic and Transcriptional Level in Glioblastoma. Sci. Rep. 2016, 6, 22477. [Google Scholar] [CrossRef]
- Soeda, A.; Hara, A.; Kunisada, T.; Yoshimura, S.; Iwama, T.; Park, D.M. The Evidence of Glioblastoma Heterogeneity. Sci. Rep. 2015, 5, 7979. [Google Scholar] [CrossRef]
- Xiong, Z.; Yang, Q.; Li, X. Effect of Intra- and Inter-tumoral Heterogeneity on Molecular Characteristics of Primary IDH-wild Type Glioblastoma Revealed by Single-cell Analysis. CNS Neurosci. Ther. 2020, 26, 981–989. [Google Scholar] [CrossRef]
- Bonavia, R.; Inda, M.-M.; Cavenee, W.K.; Furnari, F.B. Heterogeneity Maintenance in Glioblastoma: A Social Network. Cancer Res. 2011, 71, 4055–4060. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Sottoriva, A.; Spiteri, I.; Piccirillo, S.G.M.; Touloumis, A.; Collins, V.P.; Marioni, J.C.; Curtis, C.; Watts, C.; Tavaré, S. Intratumor Heterogeneity in Human Glioblastoma Reflects Cancer Evolutionary Dynamics. Proc. Natl. Acad. Sci. USA 2013, 110, 4009–4014. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-Cell RNA-Seq Highlights Intratumoral Heterogeneity in Primary Glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef]
- Piccirillo, S.G.M.; Colman, S.; Potter, N.E.; van Delft, F.W.; Lillis, S.; Carnicer, M.-J.; Kearney, L.; Watts, C.; Greaves, M. Genetic and Functional Diversity of Propagating Cells in Glioblastoma. Stem Cell Rep. 2015, 4, 7–15. [Google Scholar] [CrossRef]
- Suvà, M.L.; Rheinbay, E.; Gillespie, S.M.; Patel, A.P.; Wakimoto, H.; Rabkin, S.D.; Riggi, N.; Chi, A.S.; Cahill, D.P.; Nahed, B.V.; et al. Reconstructing and Reprogramming the Tumor-Propagating Potential of Glioblastoma Stem-like Cells. Cell 2014, 157, 580–594. [Google Scholar] [CrossRef]
- Friedmann-Morvinski, D.; Bushong, E.A.; Ke, E.; Soda, Y.; Marumoto, T.; Singer, O.; Ellisman, M.H.; Verma, I.M. Dedifferentiation of Neurons and Astrocytes by Oncogenes Can Induce Gliomas in Mice. Science 2012, 338, 1080–1084. [Google Scholar] [CrossRef]
- Soda, Y.; Marumoto, T.; Friedmann-Morvinski, D.; Soda, M.; Liu, F.; Michiue, H.; Pastorino, S.; Yang, M.; Hoffman, R.M.; Kesari, S.; et al. Transdifferentiation of Glioblastoma Cells into Vascular Endothelial Cells. Proc. Natl. Acad. Sci. USA 2011, 108, 4274–4280. [Google Scholar] [CrossRef]
- Schiffer, D.; Annovazzi, L.; Mazzucco, M.; Mellai, M. The Microenvironment in Gliomas: Phenotypic Expressions. Cancers 2015, 7, 2352–2359. [Google Scholar] [CrossRef]
- Schiffer, D.; Annovazzi, L.; Casalone, C.; Corona, C.; Mellai, M. Glioblastoma: Microenvironment and Niche Concept. Cancers 2018, 11, 5. [Google Scholar] [CrossRef]
- Mannino, M.; Chalmers, A.J. Radioresistance of Glioma Stem Cells: Intrinsic Characteristic or Property of the ‘Microenvironment-Stem Cell Unit’? Mol. Oncol. 2011, 5, 374–386. [Google Scholar] [CrossRef] [PubMed]
- Jamal, M.; Rath, B.H.; Tsang, P.S.; Camphausen, K.; Tofilon, P.J. The Brain Microenvironment Preferentially Enhances the Radioresistance of CD133+ Glioblastoma Stem-like Cells. Neoplasia 2012, 14, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Lyssiotis, C.A.; Kimmelman, A.C. Metabolic Interactions in the Tumor Microenvironment. Trends Cell Biol. 2017, 27, 863–875. [Google Scholar] [CrossRef] [PubMed]
- Roper, J.; Yilmaz, Ö.H. Metabolic Teamwork in the Stem Cell Niche. Cell Metab. 2017, 25, 993–994. [Google Scholar] [CrossRef]
- Borovski, T.; De Sousa E Melo, F.; Vermeulen, L.; Medema, J.P. Cancer Stem Cell Niche: The Place to Be. Cancer Res. 2011, 71, 634–639. [Google Scholar] [CrossRef]
- Calabrese, C.; Poppleton, H.; Kocak, M.; Hogg, T.L.; Fuller, C.; Hamner, B.; Oh, E.Y.; Gaber, M.W.; Finklestein, D.; Allen, M.; et al. A Perivascular Niche for Brain Tumor Stem Cells. Cancer Cell 2007, 11, 69–82. [Google Scholar] [CrossRef]
- Hira, V.V.V.; Ploegmakers, K.J.; Grevers, F.; Verbovšek, U.; Silvestre-Roig, C.; Aronica, E.; Tigchelaar, W.; Turnšek, T.L.; Molenaar, R.J.; Van Noorden, C.J.F. CD133+ and Nestin+ Glioma Stem-like Cells Reside around CD31+ Arterioles in Niches That Express SDF-1α, CXCR4, Osteopontin and Cathepsin K. J. Histochem. Cytochem. 2015, 63, 481–493. [Google Scholar] [CrossRef]
- Charles, N.; Holland, E.C. The Perivascular Niche Microenvironment in Brain Tumor Progression. Cell Cycle 2010, 9, 3012–3021. [Google Scholar] [CrossRef]
- Kierszenbaum, A.L.; Tres, L. Histology and Cell Biology: An Introduction in Pathology; Saunders: Philadelphia, PA, USA, 2015; ISBN 0-323-31335-3. [Google Scholar]
- Gray, L.H.; Conger, A.D.; Ebert, M.; Hornsey, S.; Scott, O.C. The Concentration of Oxygen Dissolved in Tissues at the Time of Irradiation as a Factor in Radiotherapy. Br. J. Radiol. 1953, 26, 638–648. [Google Scholar] [CrossRef]
- Hira, V.V.V.; Aderetti, D.A.; van Noorden, C.J.F. Glioma Stem Cell Niches in Human Glioblastoma Are Periarteriolar? J. Histochem. Cytochem. 2018, 66, 349–358. [Google Scholar] [CrossRef]
- Elming, P.; Sørensen, B.; Oei, A.; Franken, N.; Crezee, J.; Overgaard, J.; Horsman, M. Hyperthermia: The Optimal Treatment to Overcome Radiation Resistant Hypoxia. Cancers 2019, 11, 60. [Google Scholar] [CrossRef]
- Baumann, R.; Depping, R.; Delaperriere, M.; Dunst, J. Targeting Hypoxia to Overcome Radiation Resistance in Head & Neck Cancers: Real Challenge or Clinical Fairytale? Expert Rev. Anticancer Ther. 2016, 16, 751–758. [Google Scholar] [CrossRef] [PubMed]
- Epel, B.; Maggio, M.C.; Barth, E.D.; Miller, R.C.; Pelizzari, C.A.; Krzykawska-Serda, M.; Sundramoorthy, S.V.; Aydogan, B.; Weichselbaum, R.R.; Tormyshev, V.M.; et al. Oxygen-Guided Radiation Therapy. Int. J. Radiat. Oncol. Biol. Phys. 2019, 103, 977–984. [Google Scholar] [CrossRef] [PubMed]
- Holley, A.K.; Miao, L.; St. Clair, D.K.; St. Clair, W.H. Redox-Modulated Phenomena and Radiation Therapy: The Central Role of Superoxide Dismutases. Antioxid. Redox Signal. 2014, 20, 1567–1589. [Google Scholar] [CrossRef] [PubMed]
- Hira, V.V.V.; Wormer, J.R.; Kakar, H.; Breznik, B.; van der Swaan, B.; Hulsbos, R.; Tigchelaar, W.; Tonar, Z.; Khurshed, M.; Molenaar, R.J.; et al. Periarteriolar Glioblastoma Stem Cell Niches Express Bone Marrow Hematopoietic Stem Cell Niche Proteins. J. Histochem. Cytochem. 2018, 66, 155–173. [Google Scholar] [CrossRef]
- Qiang, L.; Wu, T.; Zhang, H.-W.; Lu, N.; Hu, R.; Wang, Y.-J.; Zhao, L.; Chen, F.-H.; Wang, X.-T.; You, Q.-D.; et al. HIF-1α Is Critical for Hypoxia-Mediated Maintenance of Glioblastoma Stem Cells by Activating Notch Signaling Pathway. Cell Death Differ. 2012, 19, 284–294. [Google Scholar] [CrossRef]
- Li, Z.; Bao, S.; Wu, Q.; Wang, H.; Eyler, C.; Sathornsumetee, S.; Shi, Q.; Cao, Y.; Lathia, J.; McLendon, R.E.; et al. Hypoxia-Inducible Factors Regulate Tumorigenic Capacity of Glioma Stem Cells. Cancer Cell 2009, 15, 501–513. [Google Scholar] [CrossRef]
- Marampon, F.; Gravina, G.L.; Zani, B.M.; Popov, V.M.; Fratticci, A.; Cerasani, M.; Di Genova, D.; Mancini, M.; Ciccarelli, C.; Ficorella, C.; et al. Hypoxia Sustains Glioblastoma Radioresistance through ERKs/DNA-PKcs/HIF-1α Functional Interplay. Int. J. Oncol. 2014, 44, 2121–2131. [Google Scholar] [CrossRef]
- Covello, K.L.; Kehler, J.; Yu, H.; Gordan, J.D.; Arsham, A.M.; Hu, C.-J.; Labosky, P.A.; Simon, M.C.; Keith, B. HIF-2alpha Regulates Oct-4: Effects of Hypoxia on Stem Cell Function, Embryonic Development, and Tumor Growth. Genes Dev. 2006, 20, 557–570. [Google Scholar] [CrossRef]
- Li, P.; Zhou, C.; Xu, L.; Xiao, H. Hypoxia Enhances Stemness of Cancer Stem Cells in Glioblastoma: An in Vitro Study. Int. J. Med. Sci. 2013, 10, 399–407. [Google Scholar] [CrossRef]
- Michiels, C.; Tellier, C.; Feron, O. Cycling Hypoxia: A Key Feature of the Tumor Microenvironment. Biochim. Biophys. Acta 2016, 1866, 76–86. [Google Scholar] [CrossRef]
- Hsieh, C.-H.; Lee, C.-H.; Liang, J.-A.; Yu, C.-Y.; Shyu, W.-C. Cycling Hypoxia Increases U87 Glioma Cell Radioresistance via ROS Induced Higher and Long-Term HIF-1 Signal Transduction Activity. Oncol. Rep. 2010, 24, 1629–1636. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Xiao, Y.; Zhou, Z.; Mao, X.; Cai, J.; Xiong, L.; Liao, C.; Huang, F.; Liu, Z.; Ali Sheikh, M.S.; et al. Extensive Metabolic Disorders Are Present in APC(Min) Tumorigenesis Mice. Mol. Cell. Endocrinol. 2016, 427, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Rani, V.; Deep, G.; Singh, R.K.; Palle, K.; Yadav, U.C.S. Oxidative Stress and Metabolic Disorders: Pathogenesis and Therapeutic Strategies. Life Sci. 2016, 148, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, G.J. Metabolic Reprogramming: The Emerging Concept and Associated Therapeutic Strategies. J. Exp. Clin. Cancer Res. 2015, 34, 111. [Google Scholar] [CrossRef] [PubMed]
- Marin-Valencia, I.; Yang, C.; Mashimo, T.; Cho, S.; Baek, H.; Yang, X.-L.; Rajagopalan, K.N.; Maddie, M.; Vemireddy, V.; Zhao, Z.; et al. Analysis of Tumor Metabolism Reveals Mitochondrial Glucose Oxidation in Genetically Diverse Human Glioblastomas in the Mouse Brain in Vivo. Cell Metab. 2012, 15, 827–837. [Google Scholar] [CrossRef] [PubMed]
- Mashimo, T.; Pichumani, K.; Vemireddy, V.; Hatanpaa, K.J.; Singh, D.K.; Sirasanagandla, S.; Nannepaga, S.; Piccirillo, S.G.; Kovacs, Z.; Foong, C.; et al. Acetate Is a Bioenergetic Substrate for Human Glioblastoma and Brain Metastases. Cell 2014, 159, 1603–1614. [Google Scholar] [CrossRef]
- Venneti, S.; Thompson, C.B. Metabolic Reprogramming in Brain Tumors. Annu. Rev. Pathol. 2017, 12, 515–545. [Google Scholar] [CrossRef]
- Zhou, W.; Wahl, D.R. Metabolic Abnormalities in Glioblastoma and Metabolic Strategies to Overcome Treatment Resistance. Cancers 2019, 11, 1231. [Google Scholar] [CrossRef]
- Libby, C.J.; Tran, A.N.; Scott, S.E.; Griguer, C.; Hjelmeland, A.B. The Pro-Tumorigenic Effects of Metabolic Alterations in Glioblastoma Including Brain Tumor Initiating Cells. Biochim. Biophys. Acta Rev. Cancer 2018, 1869, 175–188. [Google Scholar] [CrossRef]
- Oliva, C.R.; Moellering, D.R.; Gillespie, G.Y.; Griguer, C.E. Acquisition of Chemoresistance in Gliomas Is Associated with Increased Mitochondrial Coupling and Decreased ROS Production. PLoS ONE 2011, 6, e24665. [Google Scholar] [CrossRef] [PubMed]
- Oliva, C.R.; Nozell, S.E.; Diers, A.; McClugage, S.G.; Sarkaria, J.N.; Markert, J.M.; Darley-Usmar, V.M.; Bailey, S.M.; Gillespie, G.Y.; Landar, A.; et al. Acquisition of Temozolomide Chemoresistance in Gliomas Leads to Remodeling of Mitochondrial Electron Transport Chain. J. Biol. Chem. 2010, 285, 39759–39767. [Google Scholar] [CrossRef] [PubMed]
- Wolf, A.; Agnihotri, S.; Micallef, J.; Mukherjee, J.; Sabha, N.; Cairns, R.; Hawkins, C.; Guha, A. Hexokinase 2 Is a Key Mediator of Aerobic Glycolysis and Promotes Tumor Growth in Human Glioblastoma Multiforme. J. Exp. Med. 2011, 208, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Vartanian, A.; Agnihotri, S.; Wilson, M.R.; Burrell, K.E.; Tonge, P.D.; Alamsahebpour, A.; Jalali, S.; Taccone, M.S.; Mansouri, S.; Golbourn, B.; et al. Targeting Hexokinase 2 Enhances Response to Radio-Chemotherapy in Glioblastoma. Oncotarget 2016, 7, 69518–69535. [Google Scholar] [CrossRef] [PubMed]
- Ong, A.J.; Saeidi, S.; Chi, N.H.K.; Kim, S.J.; Kim, D.-H.; Kim, S.H.; Park, S.-A.; Cha, Y.-N.; Na, H.-K.; Surh, Y.-J. The Positive Feedback Loop between Nrf2 and Phosphogluconate Dehydrogenase Stimulates Proliferation and Clonogenicity of Human Hepatoma Cells. Free Radic. Res. 2020, 54, 906–917. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Levine, A.J.; Puzio-Kuter, A.M. The Control of the Metabolic Switch in Cancers by Oncogenes and Tumor Suppressor Genes. Science 2010, 330, 1340–1344. [Google Scholar] [CrossRef]
- Ying, W. NAD+/NADH and NADP+/NADPH in Cellular Functions and Cell Death: Regulation and Biological Consequences. Antioxid. Redox Signal. 2008, 10, 179–206. [Google Scholar] [CrossRef]
- Bleeker, F.E.; Atai, N.A.; Lamba, S.; Jonker, A.; Rijkeboer, D.; Bosch, K.S.; Tigchelaar, W.; Troost, D.; Vandertop, W.P.; Bardelli, A.; et al. The Prognostic IDH1(R132) Mutation Is Associated with Reduced NADP+-Dependent IDH Activity in Glioblastoma. Acta Neuropathol. 2010, 119, 487–494. [Google Scholar] [CrossRef]
- Wahl, D.R.; Dresser, J.; Wilder-Romans, K.; Parsels, J.D.; Zhao, S.G.; Davis, M.; Zhao, L.; Kachman, M.; Wernisch, S.; Burant, C.F.; et al. Glioblastoma Therapy Can Be Augmented by Targeting IDH1-Mediated NADPH Biosynthesis. Cancer Res. 2017, 77, 960–970. [Google Scholar] [CrossRef]
- Calvert, A.E.; Chalastanis, A.; Wu, Y.; Hurley, L.A.; Kouri, F.M.; Bi, Y.; Kachman, M.; May, J.L.; Bartom, E.; Hua, Y.; et al. Cancer-Associated IDH1 Promotes Growth and Resistance to Targeted Therapies in the Absence of Mutation. Cell Rep. 2017, 19, 1858–1873. [Google Scholar] [CrossRef] [PubMed]
- Vousden, K.H.; Lu, X. Live or Let Die: The Cell’s Response to P53. Nat. Rev. Cancer 2002, 2, 594–604. [Google Scholar] [CrossRef] [PubMed]
- Bensaad, K.; Tsuruta, A.; Selak, M.A.; Vidal, M.N.C.; Nakano, K.; Bartrons, R.; Gottlieb, E.; Vousden, K.H. TIGAR, a P53-Inducible Regulator of Glycolysis and Apoptosis. Cell 2006, 126, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Peña-Rico, M.A.; Calvo-Vidal, M.N.; Villalonga-Planells, R.; Martínez-Soler, F.; Giménez-Bonafé, P.; Navarro-Sabaté, À.; Tortosa, A.; Bartrons, R.; Manzano, A. TP53 Induced Glycolysis and Apoptosis Regulator (TIGAR) Knockdown Results in Radiosensitization of Glioma Cells. Radiother. Oncol. 2011, 101, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.-P.; Xie, J.-M.; Li, B.; Sun, Y.-H.; Gao, Q.-G.; Ding, Z.-H.; Wu, H.-R.; Qin, Z.-H. TIGAR Regulates DNA Damage and Repair through Pentosephosphate Pathway and Cdk5-ATM Pathway. Sci. Rep. 2015, 5, 9853. [Google Scholar] [CrossRef] [PubMed]
- Gilquin, B.; Cannon, B.R.; Hubstenberger, A.; Moulouel, B.; Falk, E.; Merle, N.; Assard, N.; Kieffer, S.; Rousseau, D.; Wilder, P.T.; et al. The Calcium-Dependent Interaction between S100B and the Mitochondrial AAA ATPase ATAD3A and the Role of This Complex in the Cytoplasmic Processing of ATAD3A. Mol. Cell. Biol. 2010, 30, 2724–2736. [Google Scholar] [CrossRef]
- Fang, H.-Y.; Chang, C.-L.; Hsu, S.-H.; Huang, C.-Y.; Chiang, S.-F.; Chiou, S.-H.; Huang, C.-H.; Hsiao, Y.-T.; Lin, T.-Y.; Chiang, I.-P.; et al. ATPase Family AAA Domain-Containing 3A Is a Novel Anti-Apoptotic Factor in Lung Adenocarcinoma Cells. J. Cell Sci. 2010, 123, 1171–1180. [Google Scholar] [CrossRef]
- You, W.-C.; Chiou, S.-H.; Huang, C.-Y.; Chiang, S.-F.; Yang, C.-L.; Sudhakar, J.N.; Lin, T.-Y.; Chiang, I.-P.; Shen, C.-C.; Cheng, W.-Y.; et al. Mitochondrial Protein ATPase Family, AAA Domain Containing 3A Correlates with Radioresistance in Glioblastoma. Neuro-Oncology 2013, 15, 1342–1352. [Google Scholar] [CrossRef]
- Macario, A.J.L.; Conway de Macario, E. Chapter 12—Chaperone Proteins and Chaperonopathies. In Stress: Physiology, Biochemistry, and Pathology; Fink, G., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 135–152. ISBN 978-0-12-813146-6. [Google Scholar]
- Macario, A.J.L.; Conway de Macario, E. Chaperonins in Cancer: Expression, Function, and Migration in Extracellular Vesicles. Semin. Cancer Biol. 2022, 86, 26–35. [Google Scholar] [CrossRef]
- Graziano, F.; Caruso Bavisotto, C.; Marino Gammazza, A.; Rappa, F.; Conway de Macario, E.; Macario, A.J.L.; Cappello, F.; Campanella, C.; Maugeri, R.; Iacopino, D. Chaperonology: The Third Eye on Brain Gliomas. Brain Sci. 2018, 8, 110. [Google Scholar] [CrossRef]
- Campanella, C.; Caruso Bavisotto, C.; Marino Gammazza, A.; Nikolic, D.; Rappa, F.; David, S.; Cappello, F.; Bucchieri, F.; Fais, S. Exosomal Heat Shock Proteins as New Players in Tumour Cell-to-Cell Communication. J. Circ. Biomark. 2014, 3, 1–10. [Google Scholar] [CrossRef]
- Marino Gammazza, A.; Campanella, C.; Barone, R.; Caruso Bavisotto, C.; Gorska, M.; Wozniak, M.; Carini, F.; Cappello, F.; D’Anneo, A.; Lauricella, M.; et al. Doxorubicin Anti-Tumor Mechanisms Include Hsp60 Post-Translational Modifications Leading to the Hsp60/P53 Complex Dissociation and Instauration of Replicative Senescence. Cancer Lett. 2017, 385, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.B.; Cai, L.; Lin, S.J.; Leng, Z.G.; Guo, Y.H.; Yang, W.L.; Chu, Y.W.; Yang, S.-H.; Zhao, W.G. Heat Shock Protein 47 Promotes Glioma Angiogenesis. Brain Pathol. 2016, 26, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Belkacemi, L.; Hebb, M.O. Hsp27 Knockdown Produces Synergistic Induction of Apoptosis by Hsp90 and Kinase Inhibitors in Glioblastoma Multiforme. Anticancer Res. 2014, 34, 4915–4927. [Google Scholar] [PubMed]
- Trentin, G.A.; He, Y.; Wu, D.C.; Tang, D.; Rozakis-Adcock, M. Identification of a HTid-1 Mutation Which Sensitizes Gliomas to Apoptosis. FEBS Lett. 2004, 578, 323–330. [Google Scholar] [CrossRef]
- Li, G.; Xu, Y.; Guan, D.; Liu, Z.; Liu, D.X. HSP70 Protein Promotes Survival of C6 and U87 Glioma Cells by Inhibition of ATF5 Degradation. J. Biol. Chem. 2011, 286, 20251–20259. [Google Scholar] [CrossRef]
- Condelli, V.; Crispo, F.; Pietrafesa, M.; Lettini, G.; Matassa, D.S.; Esposito, F.; Landriscina, M.; Maddalena, F. Hsp90 Molecular Chaperones, Metabolic Rewiring, and Epigenetics: Impact on Tumor Progression and Perspective for Anticancer Therapy. Cells 2019, 8, 532. [Google Scholar] [CrossRef]
- Machida, H.; Nakajima, S.; Shikano, N.; Nishio, J.; Okada, S.; Asayama, M.; Shirai, M.; Kubota, N. Heat Shock Protein 90 Inhibitor 17-Allylamino-17-Demethoxygeldanamycin Potentiates the Radiation Response of Tumor Cells Grown as Monolayer Cultures and Spheroids by Inducing Apoptosis. Cancer Sci. 2005, 96, 911–917. [Google Scholar] [CrossRef]
- Yun, H.S.; Baek, J.-H.; Yim, J.-H.; Um, H.-D.; Park, J.K.; Song, J.-Y.; Park, I.-C.; Kim, J.-S.; Lee, S.-J.; Lee, C.-W.; et al. Radiotherapy Diagnostic Biomarkers in Radioresistant Human H460 Lung Cancer Stem-like Cells. Cancer Biol. Ther. 2016, 17, 208–218. [Google Scholar] [CrossRef]
- Brondani Da Rocha, A.; Regner, A.; Grivicich, I.; Pretto Schunemann, D.; Diel, C.; Kovaleski, G.; Brunetto De Farias, C.; Mondadori, E.; Almeida, L.; Braga Filho, A.; et al. Radioresistance Is Associated to Increased Hsp70 Content in Human Glioblastoma Cell Lines. Int. J. Oncol. 2004, 25, 777–785. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Agabalazadeh, A.; Abak, A.; Shoorei, H.; Hassanzadeh Taheri, M.M.; Taheri, M.; Sharifi, G. Role of Long Non-Coding RNAs in Conferring Resistance in Tumors of the Nervous System. Front. Oncol. 2021, 11, 670917. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Zhang, S.; He, J. The Mechanism of Long Non-Coding RNA in Cancer Radioresistance/Radiosensitivity: A Systematic Review. Front. Pharmacol. 2022, 13, 879704. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; He, X. Involvement of Non-Coding RNAs in Chemo-and Radioresistance of Nasopharyngeal Carcinoma. Cancer Manag. Res. 2021, 13, 8781–8794. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Wang, H.; Wang, W.; Wang, Y.; Ouyang, L.; Pan, C.; Xia, L.; Cao, D.; Liao, Q. LncRNAs in DNA Damage Response and Repair in Cancer Cells. Acta Biochim. Biophys. Sin. 2018, 50, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.X.; Rothenberg, M.E. MicroRNA. J. Allergy Clin. Immunol. 2018, 141, 1202–1207. [Google Scholar] [CrossRef] [PubMed]
- Ciafrè, S.A.; Galardi, S.; Mangiola, A.; Ferracin, M.; Liu, C.-G.; Sabatino, G.; Negrini, M.; Maira, G.; Croce, C.M.; Farace, M.G. Extensive Modulation of a Set of MicroRNAs in Primary Glioblastoma. Biochem. Biophys. Res. Commun. 2005, 334, 1351–1358. [Google Scholar] [CrossRef]
- Møller, H.G.; Rasmussen, A.P.; Andersen, H.H.; Johnsen, K.B.; Henriksen, M.; Duroux, M. A Systematic Review of MicroRNA in Glioblastoma Multiforme: Micro-Modulators in the Mesenchymal Mode of Migration and Invasion. Mol. Neurobiol. 2013, 47, 131–144. [Google Scholar] [CrossRef]
- Moskwa, P.; Zinn, P.O.; Choi, Y.E.; Shukla, S.A.; Fendler, W.; Chen, C.C.; Lu, J.; Golub, T.R.; Hjelmeland, A.; Chowdhury, D. A Functional Screen Identifies MiRs That Induce Radioresistance in Glioblastomas. Mol. Cancer Res. 2014, 12, 1767–1778. [Google Scholar] [CrossRef]
- Chao, T.; Xiong, H.; Liu, W.; Chen, Y.; Zhang, J. MiR-21 Mediates the Radiation Resistance of Glioblastoma Cells by Regulating PDCD4 and HMSH2. J. Huazhong Univ. Sci. Technol. Med. Sci. 2013, 33, 525–529. [Google Scholar] [CrossRef]
- Xiao, S.; Yang, Z.; Lv, R.; Zhao, J.; Wu, M.; Liao, Y.; Liu, Q. MiR-135b Contributes to the Radioresistance by Targeting GSK3β in Human Glioblastoma Multiforme Cells. PLoS ONE 2014, 9, e108810. [Google Scholar] [CrossRef]
- Yang, W.; Wei, J.; Guo, T.; Shen, Y.; Liu, F. Knockdown of MiR-210 Decreases Hypoxic Glioma Stem Cells Stemness and Radioresistance. Exp. Cell Res. 2014, 326, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long Non-Coding RNAs: Insights into Functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Lu, X.; Yuan, L. LncRNA: A Link between RNA and Cancer. Biochim. Biophys. Acta 2014, 1839, 1097–1109. [Google Scholar] [CrossRef] [PubMed]
- Podralska, M.; Ciesielska, S.; Kluiver, J.; van den Berg, A.; Dzikiewicz-Krawczyk, A.; Slezak-Prochazka, I. Non-Coding RNAs in Cancer Radiosensitivity: MicroRNAs and LncRNAs as Regulators of Radiation-Induced Signaling Pathways. Cancers 2020, 12, 1662. [Google Scholar] [CrossRef]
- Li, J.; Ji, X.; Wang, H. Targeting Long Noncoding RNA HMMR-AS1 Suppresses and Radiosensitizes Glioblastoma. Neoplasia 2018, 20, 456–466. [Google Scholar] [CrossRef]
- Li, W.; Cui, Y.; Ma, W.; Wang, M.; Cai, Y.; Jiang, Y. LncRNA RBPMS-AS1 Promotes NRGN Transcription to Enhance the Radiosensitivity of Glioblastoma through the MicroRNA-301a-3p/CAMTA1 Axis. Transl. Oncol. 2022, 15, 101282. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Yu, H.; Shen, Y.; Liu, Y.; Yang, Z.; Sun, T. MiR-146b-5p Overexpression Attenuates Stemness and Radioresistance of Glioma Stem Cells by Targeting HuR/LincRNA-P21/β-Catenin Pathway. Oncotarget 2016, 7, 41505–41526. [Google Scholar] [CrossRef]
- Zheng, R.; Yao, Q.; Ren, C.; Liu, Y.; Yang, H.; Xie, G.; Du, S.; Yang, K.; Yuan, Y. Upregulation of Long Noncoding RNA Small Nucleolar RNA Host Gene 18 Promotes Radioresistance of Glioma by Repressing Semaphorin 5A. Int. J. Radiat. Oncol. Biol. Phys. 2016, 96, 877–887. [Google Scholar] [CrossRef]
- Wang, B.; Wang, K.; Jin, T.; Xu, Q.; He, Y.; Cui, B.; Wang, Y. NCK1-AS1 Enhances Glioma Cell Proliferation, Radioresistance and Chemoresistance via MiR-22-3p/IGF1R CeRNA Pathway. Biomed. Pharmacother. 2020, 129, 110395. [Google Scholar] [CrossRef]
- Wang, Y.-P.; Li, H.-Q.; Chen, J.-X.; Kong, F.-G.; Mo, Z.-H.; Wang, J.-Z.; Huang, K.-M.; Li, X.-N.; Yan, Y. Overexpression of XIST Facilitates Cell Proliferation, Invasion and Suppresses Cell Apoptosis by Reducing Radio-Sensitivity of Glioma Cells via MiR-329-3p/CREB1 Axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 3190–3203. [Google Scholar] [CrossRef]
- Tang, T.; Wang, L.-X.; Yang, M.-L.; Zhang, R.-M. LncRNA TPTEP1 Inhibits Stemness and Radioresistance of Glioma through MiR-106a-5p-Mediated P38 MAPK Signaling. Mol. Med. Rep. 2020, 22, 4857–4867. [Google Scholar] [CrossRef]
- Zheng, J.; Wang, B.; Zheng, R.; Zhang, J.; Huang, C.; Zheng, R.; Huang, Z.; Qiu, W.; Liu, M.; Yang, K.; et al. Linc-RA1 Inhibits Autophagy and Promotes Radioresistance by Preventing H2Bub1/USP44 Combination in Glioma Cells. Cell Death Dis. 2020, 11, 758. [Google Scholar] [CrossRef]
- Skvortsova, I.; Debbage, P.; Kumar, V.; Skvortsov, S. Radiation Resistance: Cancer Stem Cells (CSCs) and Their Enigmatic pro-Survival Signaling. Semin. Cancer Biol. 2015, 35, 39–44. [Google Scholar] [CrossRef]
- Hur, W.; Yoon, S. Molecular Pathogenesis of Radiation-Induced Cell Toxicity in Stem Cells. Int. J. Mol. Sci. 2017, 18, 2749. [Google Scholar] [CrossRef] [PubMed]
- Mladenov, E.; Magin, S.; Soni, A.; Iliakis, G. DNA Double-Strand Break Repair as Determinant of Cellular Radiosensitivity to Killing and Target in Radiation Therapy. Front. Oncol. 2013, 3, 113. [Google Scholar] [CrossRef] [PubMed]
- Saleh-Gohari, N. Conservative Homologous Recombination Preferentially Repairs DNA Double-Strand Breaks in the S Phase of the Cell Cycle in Human Cells. Nucleic Acids Res. 2004, 32, 3683–3688. [Google Scholar] [CrossRef] [PubMed]
- van Gent, D.C.; van der Burg, M. Non-Homologous End-Joining, a Sticky Affair. Oncogene 2007, 26, 7731–7740. [Google Scholar] [CrossRef] [PubMed]
- Karanam, K.; Kafri, R.; Loewer, A.; Lahav, G. Quantitative Live Cell Imaging Reveals a Gradual Shift between DNA Repair Mechanisms and a Maximal Use of HR in Mid S Phase. Mol. Cell 2012, 47, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Marampon, F.; Megiorni, F.; Camero, S.; Crescioli, C.; McDowell, H.P.; Sferra, R.; Vetuschi, A.; Pompili, S.; Ventura, L.; De Felice, F.; et al. HDAC4 and HDAC6 Sustain DNA Double Strand Break Repair and Stem-like Phenotype by Promoting Radioresistance in Glioblastoma Cells. Cancer Lett. 2017, 397, 1–11. [Google Scholar] [CrossRef]
- Zhang, J.-P.; Yang, Z.-X.; Zhang, F.; Fu, Y.-W.; Dai, X.-Y.; Wen, W.; Zhang, B.; Choi, H.; Chen, W.; Brown, M.; et al. HDAC Inhibitors Improve CRISPR-Mediated HDR Editing Efficiency in IPSCs. Sci. China Life Sci. 2021, 64, 1449–1462. [Google Scholar] [CrossRef]
- Mukherjee, B.; McEllin, B.; Camacho, C.V.; Tomimatsu, N.; Sirasanagandala, S.; Nannepaga, S.; Hatanpaa, K.J.; Mickey, B.; Madden, C.; Maher, E.; et al. EGFRvIII and DNA Double-Strand Break Repair: A Molecular Mechanism for Radioresistance in Glioblastoma. Cancer Res. 2009, 69, 4252–4259. [Google Scholar] [CrossRef]
- Golding, S.E.; Morgan, R.N.; Adams, B.R.; Hawkins, A.J.; Povirk, L.F.; Valerie, K. Pro-Survival AKT and ERK Signaling from EGFR and Mutant EGFRvIII Enhances DNA Double-Strand Break Repair in Human Glioma Cells. Cancer Biol. Ther. 2009, 8, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Facchino, S.; Abdouh, M.; Chatoo, W.; Bernier, G. BMI1 Confers Radioresistance to Normal and Cancerous Neural Stem Cells through Recruitment of the DNA Damage Response Machinery. J. Neurosci. 2010, 30, 10096–10111. [Google Scholar] [CrossRef] [PubMed]
- Zeng, A.-L.; Yan, W.; Liu, Y.-W.; Wang, Z.; Hu, Q.; Nie, E.; Zhou, X.; Li, R.; Wang, X.-F.; Jiang, T.; et al. Tumour Exosomes from Cells Harbouring PTPRZ1—MET Fusion Contribute to a Malignant Phenotype and Temozolomide Chemoresistance in Glioblastoma. Oncogene 2017, 36, 5369–5381. [Google Scholar] [CrossRef] [PubMed]
- Garnier, D.; Meehan, B.; Kislinger, T.; Daniel, P.; Sinha, A.; Abdulkarim, B.; Nakano, I.; Rak, J. Divergent Evolution of Temozolomide Resistance in Glioblastoma Stem Cells Is Reflected in Extracellular Vesicles and Coupled with Radiosensitization. Neuro-Oncology 2018, 20, 236–248. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.C.; da Silva Lima, N.; Sarian, L.O.; Matheu, A.; Ribeiro, M.L.; Derchain, S.F.M. Exosome-Mediated Breast Cancer Chemoresistance via MiR-155 Transfer. Sci. Rep. 2018, 8, 829. [Google Scholar] [CrossRef] [PubMed]
- Pavlyukov, M.S.; Yu, H.; Bastola, S.; Minata, M.; Shender, V.O.; Lee, Y.; Zhang, S.; Wang, J.; Komarova, S.; Wang, J.; et al. Apoptotic Cell-Derived Extracellular Vesicles Promote Malignancy of Glioblastoma via Intercellular Transfer of Splicing Factors. Cancer Cell 2018, 34, 119–135.e10. [Google Scholar] [CrossRef]
- Ramakrishnan, V.; Xu, B.; Akers, J.; Nguyen, T.; Ma, J.; Dhawan, S.; Ning, J.; Mao, Y.; Hua, W.; Kokkoli, E.; et al. Radiation-Induced Extracellular Vesicle (EV) Release of MiR-603 Promotes IGF1-Mediated Stem Cell State in Glioblastomas. eBioMedicine 2020, 55, 102736. [Google Scholar] [CrossRef]
- Dai, X.; Liao, K.; Zhuang, Z.; Chen, B.; Zhou, Z.; Zhou, S.; Lin, G.; Zhang, F.; Lin, Y.; Miao, Y.; et al. AHIF Promotes Glioblastoma Progression and Radioresistance via Exosomes. Int. J. Oncol. 2019, 54, 261–270. [Google Scholar] [CrossRef]
- Yu, T.; Wang, X.; Zhi, T.; Zhang, J.; Wang, Y.; Nie, E.; Zhou, F.; You, Y.; Liu, N. Delivery of MGMT MRNA to Glioma Cells by Reactive Astrocyte-Derived Exosomes Confers a Temozolomide Resistance Phenotype. Cancer Lett. 2018, 433, 210–220. [Google Scholar] [CrossRef]
- Caruso Bavisotto, C.; Marino Gammazza, A.; Rappa, F.; Fucarino, A.; Pitruzzella, A.; David, S.; Campanella, C. Exosomes: Can Doctors Still Ignore Their Existence? Euromediterranean Biomed. J. 2013, 8, 136–139. [Google Scholar] [CrossRef]
- Zaborowski, M.P.; Balaj, L.; Breakefield, X.O.; Lai, C.P. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Bioscience 2015, 65, 783–797. [Google Scholar] [CrossRef] [PubMed]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Bedina Zavec, A.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological Properties of Extracellular Vesicles and Their Physiological Functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed]
- Caby, M.-P.; Lankar, D.; Vincendeau-Scherrer, C.; Raposo, G.; Bonnerot, C. Exosomal-like Vesicles Are Present in Human Blood Plasma. Int. Immunol. 2005, 17, 879–887. [Google Scholar] [CrossRef]
- Pisitkun, T.; Shen, R.-F.; Knepper, M.A. Identification and Proteomic Profiling of Exosomes in Human Urine. Proc. Natl. Acad. Sci. USA 2004, 101, 13368–13373. [Google Scholar] [CrossRef]
- Michael, A.; Bajracharya, S.; Yuen, P.; Zhou, H.; Star, R.; Illei, G.; Alevizos, I. Exosomes from Human Saliva as a Source of MicroRNA Biomarkers: MicroRNA Biomarkers in Salivary Exosomes. Oral Dis. 2010, 16, 34–38. [Google Scholar] [CrossRef]
- Manek, R.; Moghieb, A.; Yang, Z.; Kumar, D.; Kobeissy, F.; Sarkis, G.A.; Raghavan, V.; Wang, K.K.W. Protein Biomarkers and Neuroproteomics Characterization of Microvesicles/Exosomes from Human Cerebrospinal Fluid Following Traumatic Brain Injury. Mol. Neurobiol. 2018, 55, 6128–6129, Correction to: Mol. Neurobiol. 2018, 55, 6129. [Google Scholar] [CrossRef]
- Admyre, C.; Johansson, S.M.; Qazi, K.R.; Filén, J.-J.; Lahesmaa, R.; Norman, M.; Neve, E.P.A.; Scheynius, A.; Gabrielsson, S. Exosomes with Immune Modulatory Features Are Present in Human Breast Milk. J. Immunol. 2007, 179, 1969–1978. [Google Scholar] [CrossRef]
- Théry, C. Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle Formation during Reticulocyte Maturation. Association of Plasma Membrane Activities with Released Vesicles (Exosomes). J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar] [CrossRef]
- Zitvogel, L.; Regnault, A.; Lozier, A.; Wolfers, J.; Flament, C.; Tenza, D.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Eradication of Established Murine Tumors Using a Novel Cell-Free Vaccine: Dendritic Cell Derived Exosomes. Nat. Med. 1998, 4, 594–600. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B Lymphocytes Secrete Antigen-Presenting Vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef] [PubMed]
- Akers, J.C.; Gonda, D.; Kim, R.; Carter, B.S.; Chen, C.C. Biogenesis of Extracellular Vesicles (EV): Exosomes, Microvesicles, Retrovirus-like Vesicles, and Apoptotic Bodies. J. Neuro-Oncol. 2013, 113, 1–11. [Google Scholar] [CrossRef]
- Kowal, J.; Tkach, M.; Théry, C. Biogenesis and Secretion of Exosomes. Curr. Opin. Cell Biol. 2014, 29, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Piper, R.C.; Katzmann, D.J. Biogenesis and Function of Multivesicular Bodies. Annu. Rev. Cell Dev. Biol. 2007, 23, 519–547. [Google Scholar] [CrossRef] [PubMed]
- Stuffers, S.; Brech, A.; Stenmark, H. ESCRT Proteins in Physiology and Disease. Exp. Cell Res. 2009, 315, 1619–1626. [Google Scholar] [CrossRef]
- Luzio, J.P.; Gray, S.R.; Bright, N.A. Endosome-Lysosome Fusion. Biochem. Soc. Trans. 2010, 38, 1413–1416. [Google Scholar] [CrossRef]
- Mathivanan, S.; Simpson, R.J. ExoCarta: A Compendium of Exosomal Proteins and RNA. Proteomics 2009, 9, 4997–5000. [Google Scholar] [CrossRef]
- Scott, S.; Pendlebury, S.A.; Green, C. Lipid Organization in Erythrocyte Membrane Microvesicles. Biochem. J. 1984, 224, 285–290. [Google Scholar] [CrossRef]
- Zwaal, R.F.A.; Schroit, A.J. Pathophysiologic Implications of Membrane Phospholipid Asymmetry in Blood Cells. Blood 1997, 89, 1121–1132. [Google Scholar] [CrossRef] [PubMed]
- Hugel, B.; Martínez, M.C.; Kunzelmann, C.; Freyssinet, J.-M. Membrane Microparticles: Two Sides of the Coin. Physiology 2005, 20, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Lai, Y.; Hua, Z.-C. Apoptosis and Apoptotic Body: Disease Message and Therapeutic Target Potentials. Biosci. Rep. 2019, 39, BSR20180992. [Google Scholar] [CrossRef] [PubMed]
- Vlassov, A.V.; Magdaleno, S.; Setterquist, R.; Conrad, R. Exosomes: Current Knowledge of Their Composition, Biological Functions, and Diagnostic and Therapeutic Potentials. Biochim. Biophys. Acta 2012, 1820, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Fraser, K.; Jo, A.; Giedt, J.; Vinegoni, C.; Yang, K.S.; Peruzzi, P.; Chiocca, E.A.; Breakefield, X.O.; Lee, H.; Weissleder, R. Characterization of Single Microvesicles in Plasma from Glioblastoma Patients. Neuro-Oncology 2019, 21, 606–615. [Google Scholar] [CrossRef]
- Cappello, F.; Logozzi, M.; Campanella, C.; Caruso Bavisotto, C.; Marcilla, A.; Properzi, F.; Fais, S. Exosome Levels in Human Body Fluids: A Tumor Marker by Themselves? Eur. J. Pharm. Sci. 2017, 96, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, K.; Ezrin, A.; Hadjipanayis, C. Small Extracellular Vesicles as Tumor Biomarkers for Glioblastoma. Mol. Asp. Med. 2015, 45, 97–102. [Google Scholar] [CrossRef]
- Zhu, Q.; Ling, X.; Yang, Y.; Zhang, J.; Li, Q.; Niu, X.; Hu, G.; Chen, B.; Li, H.; Wang, Y.; et al. Embryonic Stem Cells-Derived Exosomes Endowed with Targeting Properties as Chemotherapeutics Delivery Vehicles for Glioblastoma Therapy. Adv. Sci. 2019, 6, 1801899. [Google Scholar] [CrossRef]
- Men, Y.; Yelick, J.; Jin, S.; Tian, Y.; Chiang, M.S.R.; Higashimori, H.; Brown, E.; Jarvis, R.; Yang, Y. Exosome Reporter Mice Reveal the Involvement of Exosomes in Mediating Neuron to Astroglia Communication in the CNS. Nat. Commun. 2019, 10, 4136. [Google Scholar] [CrossRef]
- Caruso Bavisotto, C.; Graziano, F.; Rappa, F.; Marino Gammazza, A.; Logozzi, M.; Fais, S.; Maugeri, R.; Bucchieri, F.; Conway de Macario, E.; Macario, A.J.L.; et al. Exosomal Chaperones and MiRNAs in Gliomagenesis: State-of-Art and Theranostics Perspectives. Int. J. Mol. Sci. 2018, 19, 2626. [Google Scholar] [CrossRef]
- Pascual, M.; Ibáñez, F.; Guerri, C. Exosomes as Mediators of Neuron-Glia Communication in Neuroinflammation. Neural Regen. Res. 2020, 15, 796–801. [Google Scholar] [CrossRef] [PubMed]
- Korkut, C.; Ataman, B.; Ramachandran, P.; Ashley, J.; Barria, R.; Gherbesi, N.; Budnik, V. Trans-Synaptic Transmission of Vesicular Wnt Signals through Evi/Wntless. Cell 2009, 139, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Korkut, C.; Li, Y.; Koles, K.; Brewer, C.; Ashley, J.; Yoshihara, M.; Budnik, V. Regulation of Postsynaptic Retrograde Signaling by Presynaptic Exosome Release. Neuron 2013, 77, 1039–1046. [Google Scholar] [CrossRef]
- Frühbeis, C.; Fröhlich, D.; Kuo, W.P.; Amphornrat, J.; Thilemann, S.; Saab, A.S.; Kirchhoff, F.; Möbius, W.; Goebbels, S.; Nave, K.-A.; et al. Neurotransmitter-Triggered Transfer of Exosomes Mediates Oligodendrocyte-Neuron Communication. PLoS Biol. 2013, 11, e1001604. [Google Scholar] [CrossRef] [PubMed]
- Glebov, K.; Löchner, M.; Jabs, R.; Lau, T.; Merkel, O.; Schloss, P.; Steinhäuser, C.; Walter, J. Serotonin Stimulates Secretion of Exosomes from Microglia Cells. Glia 2015, 63, 626–634. [Google Scholar] [CrossRef]
- Lachenal, G.; Pernet-Gallay, K.; Chivet, M.; Hemming, F.J.; Belly, A.; Bodon, G.; Blot, B.; Haase, G.; Goldberg, Y.; Sadoul, R. Release of Exosomes from Differentiated Neurons and Its Regulation by Synaptic Glutamatergic Activity. Mol. Cell. Neurosci. 2011, 46, 409–418. [Google Scholar] [CrossRef]
- Qian, M.; Wang, S.; Guo, X.; Wang, J.; Zhang, Z.; Qiu, W.; Gao, X.; Chen, Z.; Xu, J.; Zhao, R.; et al. Hypoxic Glioma-Derived Exosomes Deliver MicroRNA-1246 to Induce M2 Macrophage Polarization by Targeting TERF2IP via the STAT3 and NF-ΚB Pathways. Oncogene 2020, 39, 428–442. [Google Scholar] [CrossRef]
- Melo, S.A.; Sugimoto, H.; O’Connell, J.T.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A.; et al. Cancer Exosomes Perform Cell-Independent MicroRNA Biogenesis and Promote Tumorigenesis. Cancer Cell 2014, 26, 707–721. [Google Scholar] [CrossRef]
- Graner, M.W.; Alzate, O.; Dechkovskaia, A.M.; Keene, J.D.; Sampson, J.H.; Mitchell, D.A.; Bigner, D.D. Proteomic and Immunologic Analyses of Brain Tumor Exosomes. FASEB J. 2009, 23, 1541–1557. [Google Scholar] [CrossRef]
- Azambuja, J.H.; Ludwig, N.; Yerneni, S.S.; Braganhol, E.; Whiteside, T.L. Arginase-1+ Exosomes from Reprogrammed Macrophages Promote Glioblastoma Progression. Int. J. Mol. Sci. 2020, 21, 3990. [Google Scholar] [CrossRef]
- Shao, H.; Chung, J.; Balaj, L.; Charest, A.; Bigner, D.D.; Carter, B.S.; Hochberg, F.H.; Breakefield, X.O.; Weissleder, R.; Lee, H. Protein Typing of Circulating Microvesicles Allows Real-Time Monitoring of Glioblastoma Therapy. Nat. Med. 2012, 18, 1835–1840. [Google Scholar] [CrossRef]
- David, S.; Vitale, A.M.; Fucarino, A.; Scalia, F.; Vergilio, G.; Conway de Macario, E.; Macario, A.J.L.; Caruso Bavisotto, C.; Pitruzzella, A. The Challenging Riddle about the Janus-Type Role of Hsp60 and Related Extracellular Vesicles and MiRNAs in Carcinogenesis and the Promises of Its Solution. Appl. Sci. 2021, 11, 1175. [Google Scholar] [CrossRef]
- Vitale, A.M.; Santonocito, R.; Vergilio, G.; Marino Gammazza, A.; Campanella, C.; Conway de Macario, E.; Bucchieri, F.; Macario, A.J.L.; Caruso Bavisotto, C. Brain Tumor-Derived Extracellular Vesicles as Carriers of Disease Markers: Molecular Chaperones and MicroRNAs. Appl. Sci. 2020, 10, 6961. [Google Scholar] [CrossRef]
- Graziano, F.; Iacopino, D.G.; Cammarata, G.; Scalia, G.; Campanella, C.; Giannone, A.G.; Porcasi, R.; Florena, A.M.; Conway de Macario, E.; Macario, A.J.L.; et al. The Triad Hsp60-MiRNAs-Extracellular Vesicles in Brain Tumors: Assessing Its Components for Understanding Tumorigenesis and Monitoring Patients. Appl. Sci. 2021, 11, 2867. [Google Scholar] [CrossRef]
- Manterola, L.; Guruceaga, E.; Gállego Pérez-Larraya, J.; González-Huarriz, M.; Jauregui, P.; Tejada, S.; Diez-Valle, R.; Segura, V.; Samprón, N.; Barrena, C.; et al. A Small Noncoding RNA Signature Found in Exosomes of GBM Patient Serum as a Diagnostic Tool. Neuro-Oncology 2014, 16, 520–527. [Google Scholar] [CrossRef]
- Shi, R.; Wang, P.-Y.; Li, X.-Y.; Chen, J.-X.; Li, Y.; Zhang, X.-Z.; Zhang, C.-G.; Jiang, T.; Li, W.-B.; Ding, W.; et al. Exosomal Levels of MiRNA-21 from Cerebrospinal Fluids Associated with Poor Prognosis and Tumor Recurrence of Glioma Patients. Oncotarget 2015, 6, 26971–26981. [Google Scholar] [CrossRef]
- Walker, S.; Busatto, S.; Pham, A.; Tian, M.; Suh, A.; Carson, K.; Quintero, A.; Lafrence, M.; Malik, H.; Santana, M.X.; et al. Extracellular Vesicle-Based Drug Delivery Systems for Cancer Treatment. Theranostics 2019, 9, 8001–8017. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, R.; Maresh, G.; Zhang, X.; Salomon, C.; Hooper, J.; Margolin, D.; Li, L. The Emerging Roles of Extracellular Vesicles as Communication Vehicles within the Tumor Microenvironment and Beyond. Front. Endocrinol. 2017, 8, 194. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, A.R.; Hill, R.; Pilkington, G.J.; Madureira, P.A. The Role of Hypoxia in Glioblastoma Invasion. Cells 2017, 6, 45. [Google Scholar] [CrossRef] [PubMed]
- Nareshkumar, R.N.; Sulochana, K.N.; Coral, K. Inhibition of Angiogenesis in Endothelial Cells by Human Lysyl Oxidase Propeptide. Sci. Rep. 2018, 8, 10426. [Google Scholar] [CrossRef] [PubMed]
- Fidoamore, A.; Cristiano, L.; Antonosante, A.; D’Angelo, M.; Di Giacomo, E.; Astarita, C.; Giordano, A.; Ippoliti, R.; Benedetti, E.; Cimini, A. Glioblastoma Stem Cells Microenvironment: The Paracrine Roles of the Niche in Drug and Radioresistance. Stem Cells Int. 2016, 2016, 6809105. [Google Scholar] [CrossRef] [PubMed]
- Heffernan, J.M.; Sirianni, R.W. Modeling Microenvironmental Regulation of Glioblastoma Stem Cells: A Biomaterials Perspective. Front. Mater. 2018, 5, 7. [Google Scholar] [CrossRef]
- Kucharzewska, P.; Christianson, H.C.; Welch, J.E.; Svensson, K.J.; Fredlund, E.; Ringnér, M.; Mörgelin, M.; Bourseau-Guilmain, E.; Bengzon, J.; Belting, M. Exosomes Reflect the Hypoxic Status of Glioma Cells and Mediate Hypoxia-Dependent Activation of Vascular Cells during Tumor Development. Proc. Natl. Acad. Sci. USA 2013, 110, 7312–7317. [Google Scholar] [CrossRef] [PubMed]
- Brandao, M.; Simon, T.; Critchley, G.; Giamas, G. Astrocytes, the Rising Stars of the Glioblastoma Microenvironment. Glia 2019, 67, 779–790. [Google Scholar] [CrossRef] [PubMed]
- Hallal, S.; Mallawaaratchy, D.M.; Wei, H.; Ebrahimkhani, S.; Stringer, B.W.; Day, B.W.; Boyd, A.W.; Guillemin, G.J.; Buckland, M.E.; Kaufman, K.L. Extracellular Vesicles Released by Glioblastoma Cells Stimulate Normal Astrocytes to Acquire a Tumor-Supportive Phenotype via P53 and MYC Signaling Pathways. Mol. Neurobiol. 2019, 56, 4566–4581. [Google Scholar] [CrossRef] [PubMed]
- Watkins, S.; Robel, S.; Kimbrough, I.F.; Robert, S.M.; Ellis-Davies, G.; Sontheimer, H. Disruption of Astrocyte-Vascular Coupling and the Blood-Brain Barrier by Invading Glioma Cells. Nat. Commun. 2014, 5, 4196. [Google Scholar] [CrossRef]
- Zhao, C.; Wang, H.; Xiong, C.; Liu, Y. Hypoxic Glioblastoma Release Exosomal VEGF-A Induce the Permeability of Blood-Brain Barrier. Biochem. Biophys. Res. Commun. 2018, 502, 324–331. [Google Scholar] [CrossRef]
- Ricklefs, F.L.; Alayo, Q.; Krenzlin, H.; Mahmoud, A.B.; Speranza, M.C.; Nakashima, H.; Hayes, J.L.; Lee, K.; Balaj, L.; Passaro, C.; et al. Immune Evasion Mediated by PD-L1 on Glioblastoma-Derived Extracellular Vesicles. Sci. Adv. 2018, 4, eaar2766. [Google Scholar] [CrossRef]
- Namee, N.M.; O’Driscoll, L. Extracellular Vesicles and Anti-Cancer Drug Resistance. Biochim. Biophys. Acta Rev. Cancer 2018, 1870, 123–136. [Google Scholar] [CrossRef]
- Jeelani, S.; Reddy, R.C.J.; Maheswaran, T.; Asokan, G.S.; Dany, A.; Anand, B. Theranostics: A Treasured Tailor for Tomorrow. J. Pharm. Bioallied Sci. 2014, 6, S6–S8. [Google Scholar] [CrossRef]
- Maheswaran, T.; Ganapathy, N.; DineshShankar, J.; Mohanapriya, S.; Ilayaraja, V.; Yoithapprabhunath, T.R.; Yamunadevi, A.M. Theranostics an Emerging Paradigm—A Review. IOSR J. Dent. Med. Sci. 2018, 17, 19–24. [Google Scholar]
- Kang, S.J.; Jeong, H.Y.; Kim, M.W.; Jeong, I.H.; Choi, M.J.; You, Y.M.; Im, C.S.; Song, I.H.; Lee, T.S.; Park, Y.S. Anti-EGFR Lipid Micellar Nanoparticles Co-Encapsulating Quantum Dots and Paclitaxel for Tumor-Targeted Theranosis. Nanoscale 2018, 10, 19338–19350. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, S.; Jain, S.; Kumar, D.; Soni, S.L.; Sharma, M. A Review on Theranostics: An Approach to Targeted Diagnosis and Therapy. Asian J. Pharm. Res. Dev. 2019, 7, 63–69. [Google Scholar] [CrossRef]
- Jeyamogan, S.; Khan, N.A.; Siddiqui, R. Application and Importance of Theranostics in the Diagnosis and Treatment of Cancer. Arch. Med. Res. 2021, 52, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.Y.; Liu, G.; Lee, S.; Chen, X. Theranostic Nanoplatforms for Simultaneous Cancer Imaging and Therapy: Current Approaches and Future Perspectives. Nanoscale 2012, 4, 330–342. [Google Scholar] [CrossRef] [PubMed]
- Vilos, C.; Velasquez, L.A. Therapeutic Strategies Based on Polymeric Microparticles. J. Biomed. Biotechnol. 2012, 2012, 672760. [Google Scholar] [CrossRef] [PubMed]
- Bhojani, M.S.; Van Dort, M.; Rehemtulla, A.; Ross, B.D. Targeted Imaging and Therapy of Brain Cancer Using Theranostic Nanoparticles. Mol. Pharm. 2010, 7, 1921–1929. [Google Scholar] [CrossRef]
- Pan, B.; Cui, D.; Sheng, Y.; Ozkan, C.; Gao, F.; He, R.; Li, Q.; Xu, P.; Huang, T. Dendrimer-Modified Magnetic Nanoparticles Enhance Efficiency of Gene Delivery System. Cancer Res. 2007, 67, 8156–8163. [Google Scholar] [CrossRef]
- Picone, P.; Porcelli, G.; Bavisotto, C.; Nuzzo, D.; Galizzi, G.; Biagio, P.L.S.; Bulone, D.; Di Carlo, M. Synaptosomes: New Vesicles for Neuronal Mitochondrial Transplantation. J. Nanobiotechnol. 2021, 19, 6. [Google Scholar] [CrossRef]
- Wang, R.; Wang, X.; Zhang, Y.; Zhao, H.; Cui, J.; Li, J.; Di, L. Emerging Prospects of Extracellular Vesicles for Brain Disease Theranostics. J. Control. Release 2022, 341, 844–868. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The Biology, Function, and Biomedical Applications of Exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Müller Bark, J.; Kulasinghe, A.; Chua, B.; Day, B.W.; Punyadeera, C. Circulating Biomarkers in Patients with Glioblastoma. Br. J. Cancer 2020, 122, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Westphal, M.; Lamszus, K. Circulating Biomarkers for Gliomas. Nat. Rev. Neurol. 2015, 11, 556–566. [Google Scholar] [CrossRef] [PubMed]
- Joshi, B.S.; de Beer, M.A.; Giepmans, B.N.G.; Zuhorn, I.S. Endocytosis of Extracellular Vesicles and Release of Their Cargo from Endosomes. ACS Nano 2020, 14, 4444–4455. [Google Scholar] [CrossRef]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J.A. Delivery of SiRNA to the Mouse Brain by Systemic Injection of Targeted Exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef]
- Basu, B.; Ghosh, M.K. Extracellular Vesicles in Glioma: From Diagnosis to Therapy. Bioessays 2019, 41, e1800245. [Google Scholar] [CrossRef]
- Ma, C.; Nguyen, H.P.T.; Jones, J.J.; Stylli, S.S.; Whitehead, C.A.; Paradiso, L.; Luwor, R.B.; Areeb, Z.; Hanssen, E.; Cho, E.; et al. Extracellular Vesicles Secreted by Glioma Stem Cells Are Involved in Radiation Resistance and Glioma Progression. Int. J. Mol. Sci. 2022, 23, 2770. [Google Scholar] [CrossRef]
- Burgio, S.; Noori, L.; Marino Gammazza, A.; Campanella, C.; Logozzi, M.; Fais, S.; Bucchieri, F.; Cappello, F.; Caruso Bavisotto, C. Extracellular Vesicles-Based Drug Delivery Systems: A New Challenge and the Exemplum of Malignant Pleural Mesothelioma. Int. J. Mol. Sci. 2020, 21, 5432. [Google Scholar] [CrossRef]
| Drug | Target | Biological Effects | Advantages | Limitations/Concerns | References |
|---|---|---|---|---|---|
| Temozolomide | Alkylation or methylation of guanine N7 or O6 and adenine N3 | Induction of guanine binding to thymine instead of cytosine, leading to extensive DNA damage and, eventually, apoptosis | Rapid and complete absorption. Weak plasma protein binding. Blood-brain barrier permeability. Use in patients with kidney and liver malfunction | Myelosuppression and lymphopenia. Downregulation of the O6-methylguanine-DNA. methyltransferase gene (MGMT). Lymphoblastic leukemia | [9,10,11,12,13,14] |
| Carmustine | DNA and RNA alkylating agent | Binds to and modifies glutathione reductase, which leads to cell death in tumor cells | Recurrent GBM | Pulmonary fibrosis, bone marrow suppression, optical toxicity | [15,16] |
| Carmustine in biodegradable polymer | Placement of wafers directly into the resection area allows more effective local treatment, resulting in improved outcomes and reduced toxicity | Cerebral oedema, intracranial hypertension, infections, seizures, and thromboembolic events | [17,18,19,20] | ||
| Bevacizumab | Vascular endothelial growth factor (VEGF) | Inhibition of VEGF | Inhibition of Vascular Endothelial Growth Factor A leads to a decrease in the growth of new blood vessels, reducing the vascularization of GBM. Increases recurrence-free period in recurrent GBM | Pulmonary embolism, arterial hypertension, and hematologic toxic effects | [21,22,23,24,25] |
| Vorinostat | Inhibitor of histone deacetylases 1, 2, 3, and 6 | Inhibition of tumor growth | Inhibition of growth of tumor cells resistant to alkylating drugs. A combination of Vorinostat and Temozolomide inhibits glioblastoma growth in experimental mice | Stimulation of autophagy and inhibition of tumor cells apoptosis | [26,27,28] |
| Olaparib | Poly (ADP-ribose) polymerase (PARP) inhibitor | Enhance drug delivery to tumor | Higher survival rates and no damage to healthy tissues in combination with Temozolomide, and radiotherapy | Poor brain penetration | [29] |
| Lomustine | Alkylating agent | Formation of O6-chloroethyl-guanine, can be reverted by O6-methyl-guanine DNA methyltransferase (MGMT) | Recurrent GBM | Restricted to patients with MGMT promoter-methylated tumors. Thrombocytopenia | [30,31,32] |
| Valproic acid | Short-chain fatty acid | Inhibition of histone deacetylase | Chemical and metabolic stability. Increasing tumor cell sensitivity to ionizing radiation | Thrombocytopenia, fatigue, and hypertension | [33,34,35,36,37] |
| Factor or Pathway | Properties/Mechanism | Reference |
|---|---|---|
| EVs | Transfer the genomic and proteomic cargo (mRNA, miRNA, lncRNA, spliceosomes, and proteins). | [187,188,189,190,191,192] |
| Transfer the transcripts of DNA repair enzymes. | [193] | |
| Glioblastoma stem cells | Ability to initiate carcinogenesis, sustain tumor proliferation, differentiate into all cellular subpopulations of the primary tumor, and unlimited self-renewal. | [55] |
| Expression of a particular marker CD133 (prominin-1). | [55] | |
| Cathepsin L co-expression. | [70] | |
| PAF Overexpression. | [72] | |
| Intra-tumoral and inter-tumoral tumor heterogeneity | Heterogeneity at the transcriptional, methylation, and mutational levels. | [79] |
| Tumor plasticity | Epigenetic reprogramming. | [87] |
| Hypoxic periarteriolar niches | Reduction of ROS formation and up-regulation of ROS scavenging. | [57,106] |
| Increased expression of the VEGF and HIF-1α. | [98,107] | |
| Activation of Hedgehog pathway, Notch, wingless, and INT-1 (WNT). | [108,109] | |
| Mediates the functional regulation of DNA-PKcs and ERKs. | [110] | |
| Activation of the OCT-4. | [111,112] | |
| Cyclic periods of hypoxia. | [113,114] | |
| Metabolic reprogramming | Surplus production of lactate, acetate, and increase of glucose oxidation to generate macromolecular precursors and energy. | [118,119] |
| Overexpression of the heat shock proteins Hsp27, Hsp40, Hsp47, Hsp70, and Hsp90. | [146,147,148,149,150] | |
| Warburg effect. | [127,128] | |
| Activation of a pathway that is the source of excess NADPH with extra promotion by the IDH1 gene. | [129,132] | |
| Activation of TIGAR. | [135,136] | |
| Activation of ATAD3A. | [140] | |
| Aberrantly expressed ncRNAs (lncRNAs and miRNAs) | Control of cell cycle, apoptosis, DNA damage checkpoints, and other critical signaling paths. | [141,142,154,155,156,157] |
| DNA repair and cell cycle | Cells’ enlarged DNA repair potential | [175] |
| Use of HRR, NHEJ, and alternative NHEJ as main pathways for DSBs processing. | [178] |
| EVs Subtype | Size (nm) | Biogenesis Mechanism | Molecular Composition | Reference |
|---|---|---|---|---|
| Exosomes Small/medium EVs | 40–120 | Endocytic origin | ALIX, TSG101, GTPase, annexins, flotillin, and tetraspanin proteins (CD9, CD63, CD81). | [204,205,206,207,208,209,210,211,212] |
| Microvesicles Medium/large EVs | 100–1000 | Outward budding and fission of the plasma membrane | mRNA, non-coding RNAs. Selectins, integrins. CD40. ARF6. | [203,213,214,215] |
| Apoptotic bodies large EVs | >1000 | Cell fragmentation during apoptotic cell death | VAMP3. Cytoplasmic and membrane proteins, amino-phospholipids, phosphatidylserine, and ethanolamine. | [216] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Burko, P.; D’Amico, G.; Miltykh, I.; Scalia, F.; Conway de Macario, E.; Macario, A.J.L.; Giglia, G.; Cappello, F.; Caruso Bavisotto, C. Molecular Pathways Implicated in Radioresistance of Glioblastoma Multiforme: What Is the Role of Extracellular Vesicles? Int. J. Mol. Sci. 2023, 24, 4883. https://doi.org/10.3390/ijms24054883
Burko P, D’Amico G, Miltykh I, Scalia F, Conway de Macario E, Macario AJL, Giglia G, Cappello F, Caruso Bavisotto C. Molecular Pathways Implicated in Radioresistance of Glioblastoma Multiforme: What Is the Role of Extracellular Vesicles? International Journal of Molecular Sciences. 2023; 24(5):4883. https://doi.org/10.3390/ijms24054883
Chicago/Turabian StyleBurko, Pavel, Giuseppa D’Amico, Ilia Miltykh, Federica Scalia, Everly Conway de Macario, Alberto J. L. Macario, Giuseppe Giglia, Francesco Cappello, and Celeste Caruso Bavisotto. 2023. "Molecular Pathways Implicated in Radioresistance of Glioblastoma Multiforme: What Is the Role of Extracellular Vesicles?" International Journal of Molecular Sciences 24, no. 5: 4883. https://doi.org/10.3390/ijms24054883
APA StyleBurko, P., D’Amico, G., Miltykh, I., Scalia, F., Conway de Macario, E., Macario, A. J. L., Giglia, G., Cappello, F., & Caruso Bavisotto, C. (2023). Molecular Pathways Implicated in Radioresistance of Glioblastoma Multiforme: What Is the Role of Extracellular Vesicles? International Journal of Molecular Sciences, 24(5), 4883. https://doi.org/10.3390/ijms24054883











