PARP-1 Expression Influences Cancer Stem Cell Phenotype in Colorectal Cancer Depending on p53
Abstract
1. Introduction
2. Results
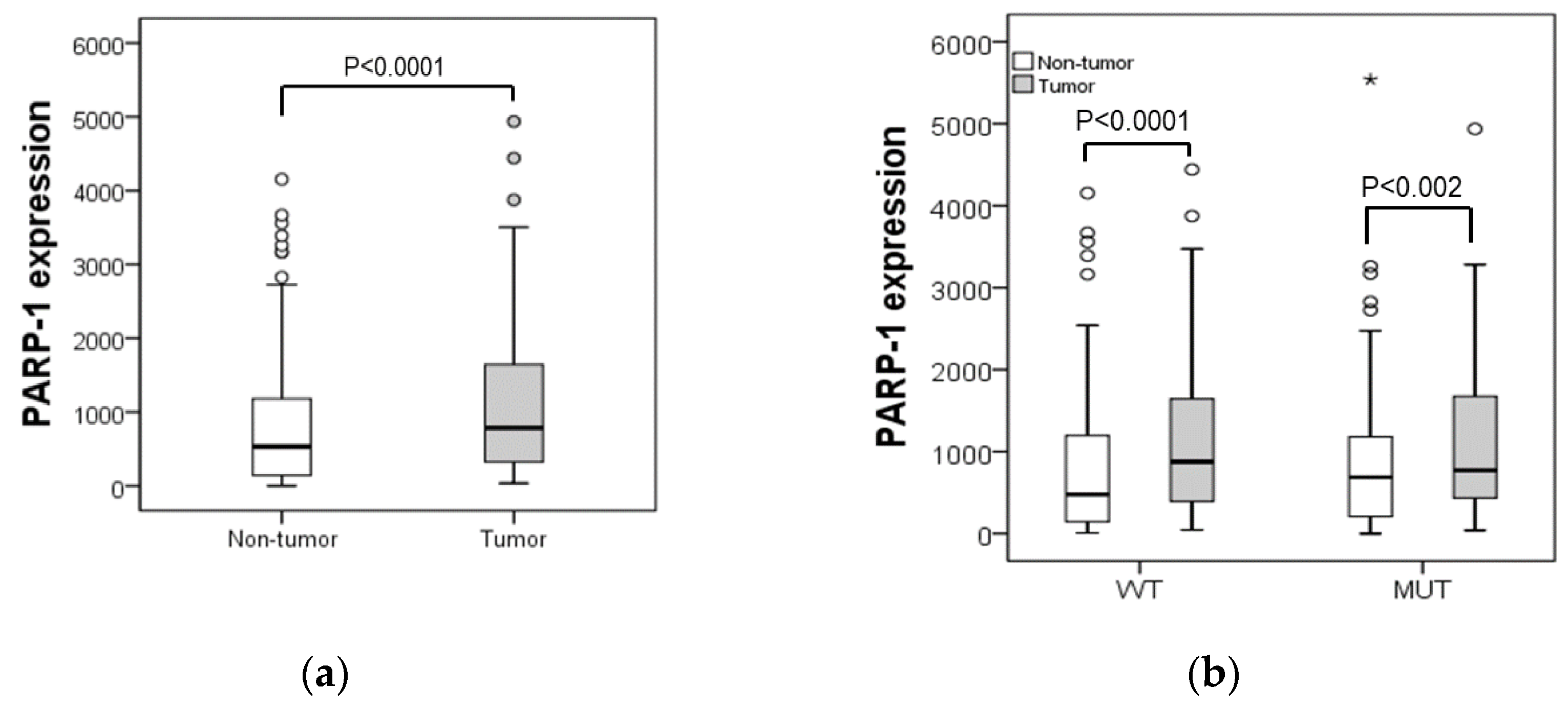
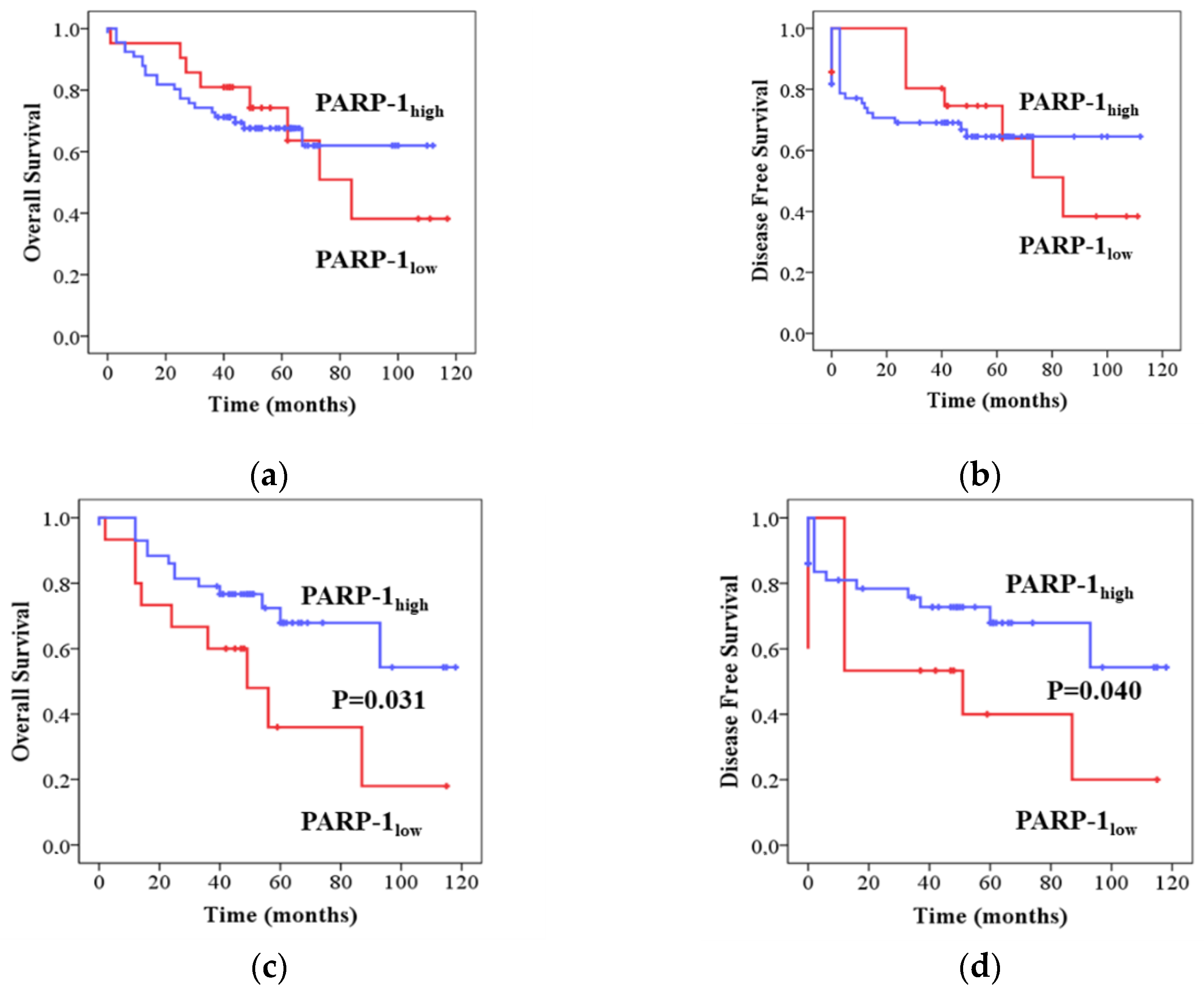
2.1. PARP-1 Correlates with Clinical Outcomes in CRC Patients in a p53 Dependent Manner
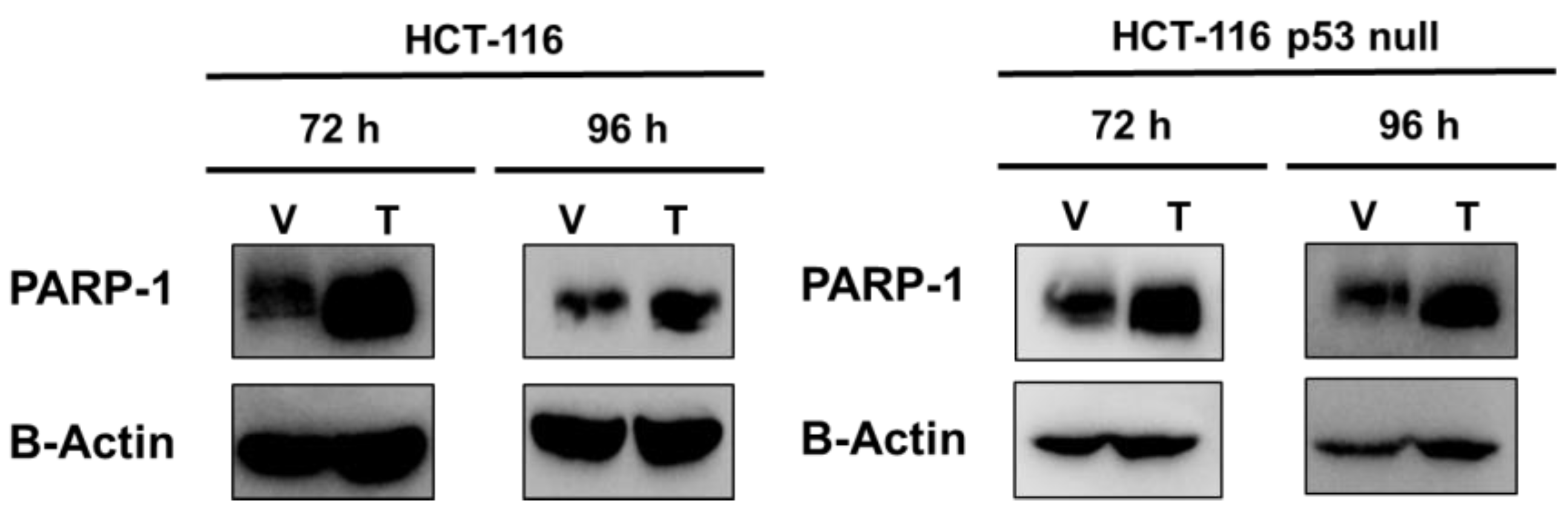
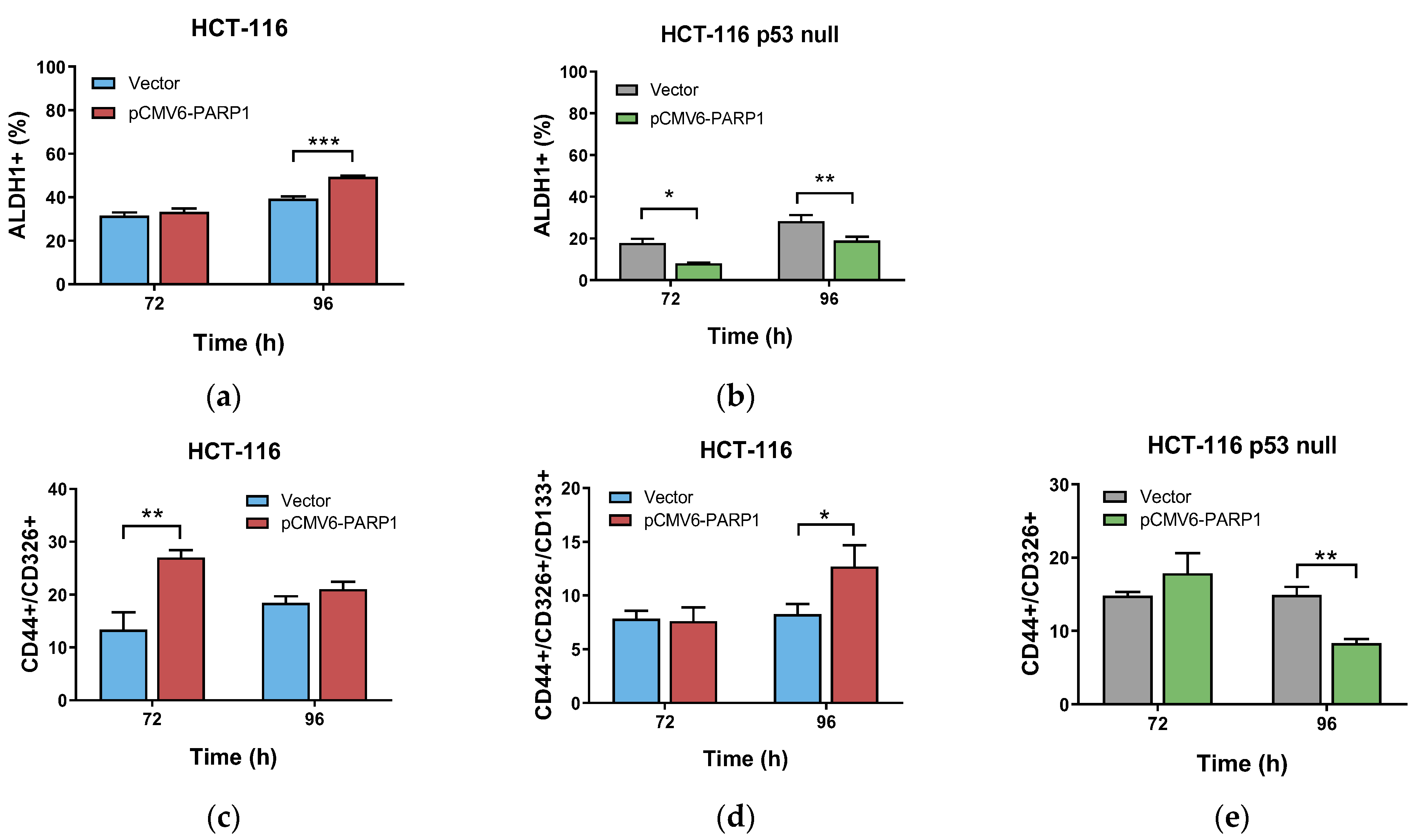
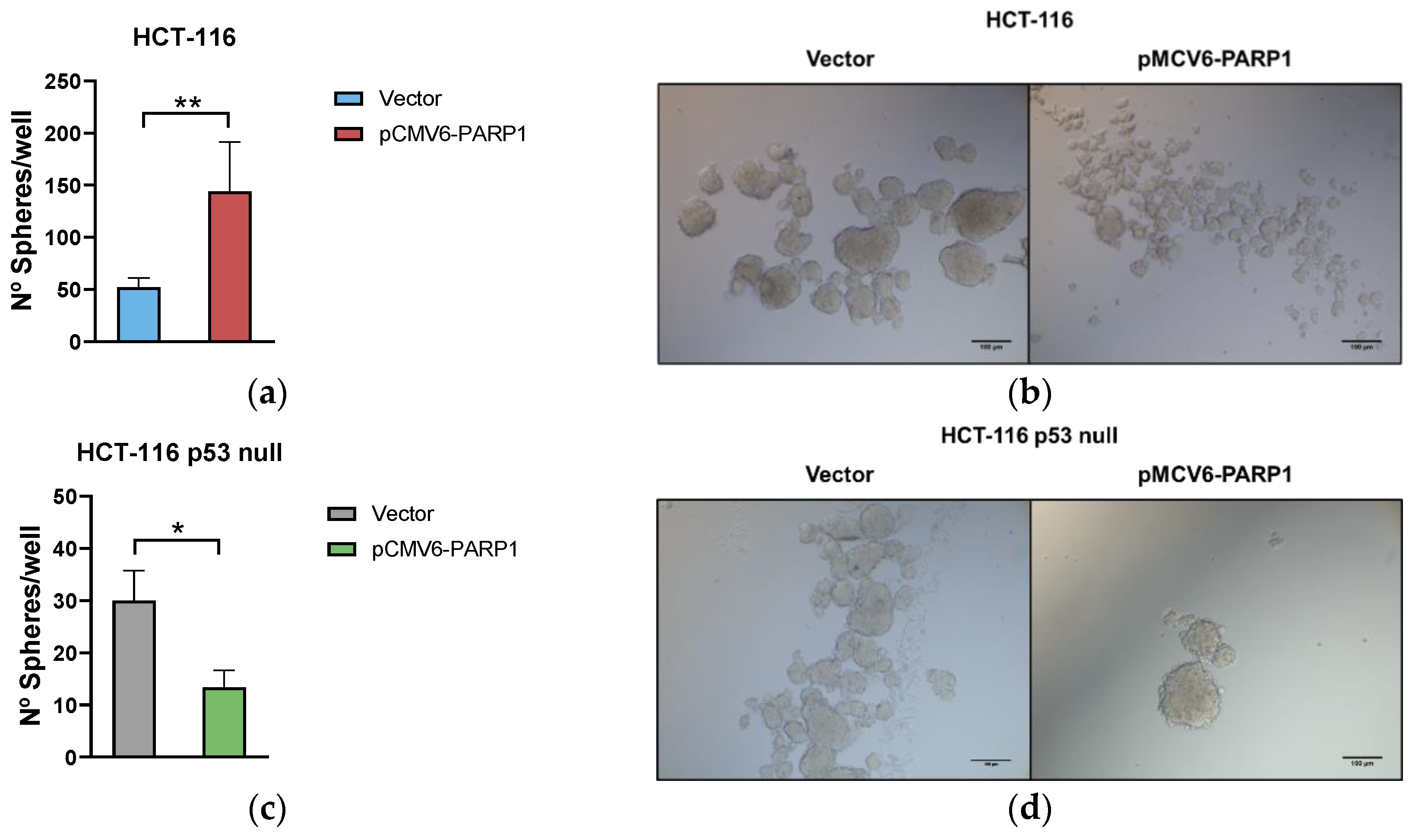
2.2. PARP-1 Regulates Tumourigenic Properties through the Regulation of Cancer Stem-Like Cells Phenotype
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Analysis of p53 Mutations in CRC Samples
4.3. RNA Extraction and First-Strand cDNA Synthesis
4.4. Real-Time PCR (RT-PCR)
4.5. Cell Culture
4.6. Transient Transfection
4.7. Western Blotting
4.8. ALDEFLUOR Assay
4.9. Phenotypic Characterization of CSCs
4.10. Sphere Forming Assay
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Van Der Jeught, K.; Xu, H.C.; Li, Y.J.; Lu, X.B.; Ji, G. Drug resistance and new therapies in colorectal cancer. World J. Gastroenterol. 2018, 24, 3834–3848. [Google Scholar] [CrossRef] [PubMed]
- Das, P.K.; Islam, F.; Lam, A.K. The Roles of Cancer Stem Cells and Therapy Resistance in Colorectal Carcinoma. Cells 2020, 9, 1392. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Kim, Y.K. Cancer Stem Cells as a Potential Target to Overcome Multidrug Resistance. Front. Oncol. 2020, 10, 764. [Google Scholar] [CrossRef]
- Batlle, E.; Clevers, H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124–1134. [Google Scholar] [CrossRef]
- Ray Chaudhuri, A.; Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 610–621. [Google Scholar] [CrossRef]
- Martí, J.M.; Fernández-Cortés, M.; Serrano-Sáenz, S.; Zamudio-Martinez, E.; Delgado-Bellido, D.; Garcia-Diaz, A.; Oliver, F.J. The Multifactorial Role of PARP-1 in Tumor Microenvironment. Cancers 2020, 12, 739. [Google Scholar] [CrossRef]
- Gonçalves, A.; Finetti, P.; Sabatier, R.; Gilabert, M.; Adelaide, J.; Borg, J.P.; Chaffanet, M.; Viens, P.; Birnbaum, D.; Bertucci, F. Poly(ADP-ribose) polymerase-1 mRNA expression in human breast cancer: A meta-analysis. Breast Cancer Res. Treat. 2011, 127, 273–281. [Google Scholar] [CrossRef]
- Byers, L.A.; Wang, J.; Nilsson, M.B.; Fujimoto, J.; Saintigny, P.; Yordy, J.; Giri, U.; Peyton, M.; Fan, Y.H.; Diao, L.; et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov. 2012, 2, 798–811. [Google Scholar] [CrossRef]
- Ossovskaya, V.; Koo, I.C.; Kaldjian, E.P.; Alvares, C.; Sherman, B.M. Upregulation of poly (ADP-Ribose) polymerase-1 (PARP1) in triple-negative breast cancer and other primary human tumor types. Genes Cancer 2010, 1, 812–821. [Google Scholar] [CrossRef]
- Nosho, K.; Yamamoto, H.; Mikami, M.; Taniguchi, H.; Takahashi, T.; Adachi, Y.; Imamura, A.; Imai, K.; Shinomura, Y. Overexpression of poly(ADP-ribose) polymerase-1 (PARP-1) in the early stage of colorectal carcinogenesis. Eur. J. Cancer 2006, 42, 2374–2381. [Google Scholar] [CrossRef]
- Jarrar, A.; Lotti, F.; DeVecchio, J.; Ferrandon, S.; Gantt, G.; Mace, A.; Karagkounis, G.; Orloff, M.; Venere, M.; Hitomi, M.; et al. Poly(ADP-Ribose) Polymerase Inhibition Sensitizes Colorectal Cancer-Initiating Cells to Chemotherapy. Stem Cells 2019, 37, 42–53. [Google Scholar] [CrossRef]
- Pilié, P.G.; Tang, C.; Mills, G.B.; Yap, T.A. State-of-the-art strategies for targeting the DNA damage response in cancer. Nat. Rev. Clin. Oncol. 2018, 16, 81–104. [Google Scholar] [CrossRef]
- Leichman, L.; Groshen, S.; O’Neil, B.H.; Messersmith, W.; Berlin, J.; Chan, E.; Leichman, C.G.; Cohen, S.J.; Cohen, D.; Lenz, H.-J.; et al. Phase II Study of Olaparib (AZD-2281) After Standard Systemic Therapies for Disseminated Colorectal Cancer. Oncologist 2016, 21, 172–177. [Google Scholar] [CrossRef]
- Dörsam, B.; Seiwert, N.; Foersch, S.; Stroh, S.; Nagel, G.; Begaliew, D.; Diehl, E.; Kraus, A.; McKeague, M.; Minneker, V.; et al. PARP-1 protects against colorectal tumor induction, but promotes inflammation-driven colorectal tumor progression. Proc. Natl. Acad. Sci. USA 2018, 115, E4061–E4070. [Google Scholar] [CrossRef]
- Wiman, K.G. p53 talks to PARP: The increasing complexity of p53-induced cell death. Cell Death Differ. 2013, 20, 1438–1439. [Google Scholar] [CrossRef]
- Kanai, M.; Hanashiro, K.; Kim, S.H.; Hanai, S.; Boulares, A.H.; Miwa, M.; Fukasawa, K. Inhibition of Crm1–p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat. Cell Biol. 2007, 9, 1175–1183. [Google Scholar] [CrossRef]
- Fischbach, A.; Krüger, A.; Hampp, S.; Assmann, G.; Rank, L.; Hufnagel, M.; Stöckl, M.T.; Fischer, J.M.F.; Veith, S.; Rossatti, P.; et al. The C-terminal domain of p53 orchestrates the interplay between non-covalent and covalent poly(ADP-ribosyl)ation of p53 by PARP1. Nucleic Acids Res. 2018, 46, 804–822. [Google Scholar] [CrossRef]
- Wesierska-Gadek, J.; Schmid, G. Overexpressed poly(ADP-ribose) polymerase delays the release of rat cells from p53-mediated G(1) checkpoint. J. Cell. Biochem. 2000, 80, 85–103. [Google Scholar] [CrossRef]
- Qiu, W.G.; Polotskaia, A.; Xiao, G.; Di, L.; Zhao, Y.; Hu, W.; Philip, J.; Hendrickson, R.C.; Bargonetti, J. Identification, validation, and targeting of the mutant p53-PARP-MCM chromatin axis in triple negative breast cancer. npj Breast Cancer 2017, 3, 1–9. [Google Scholar] [CrossRef]
- Polotskaia, A.; Xiao, G.; Reynoso, K.; Martin, C.; Qiu, W.G.; Hendrickson, R.C.; Bargonettia, J. Proteome-wide analysis of mutant p53 targets in breast cancer identifies new levels of gain-of-function that influence PARP, PCNA, and MCM4. Proc. Natl. Acad. Sci. USA 2015, 112, E1220–E1229. [Google Scholar] [CrossRef] [PubMed]
- Ríos-Arrabal, S.; Puentes-Pardo, J.D.; Moreno-Sanjuan, S.; Szuba, Á.; Casado, J.; García-Costela, M.; Escudero-Feliu, J.; Verbeni, M.; Cano, C.; González-Puga, C.; et al. Endothelin-1 as a mediator of heme oxygenase-1-induced stemness in colorectal cancer: Influence of p53. J. Pers. Med. 2021, 11, 509. [Google Scholar] [CrossRef] [PubMed]
- Dziaman, T.; Ludwiczak, H.; Ciesla, J.M.; Banaszkiewicz, Z.; Winczura, A.; Chmielarczyk, M.; Wisniewska, E.; Marszalek, A.; Tudek, B.; Olinski, R. PARP-1 Expression is Increased in Colon Adenoma and Carcinoma and Correlates with OGG1. PLoS ONE 2014, 9, e115558. [Google Scholar] [CrossRef] [PubMed]
- Liebl, M.C.; Hofmann, T.G. The role of p53 signaling in colorectal cancer. Cancers 2021, 13, 2125. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, C.; Jin, J.; Wang, J.; Huang, J.; Ma, Z.; Huang, X.; He, X.; Zhou, Y.; Xu, Y.; et al. High PARP-1 expression predicts poor survival in acute myeloid leukemia and PARP-1 inhibitor and SAHA-bendamustine hybrid inhibitor combination treatment synergistically enhances anti-tumor effects. eBioMedicine 2018, 38, 47–56. [Google Scholar] [CrossRef]
- Thakur, N.; Yim, K.; Abdul-Ghafar, J.; Seo, K.J.; Chong, Y. High poly(ADP-ribose) polymerase expression does relate to poor survival in solid cancers: A systematic review and meta-analysis. Cancers 2021, 13, 5594. [Google Scholar] [CrossRef]
- Klauschen, F.; von Winterfeld, M.; Stenzinger, A.; Sinn, B.V.; Budczies, J.; Kamphues, C.; Bahra, M.; Wittschieber, D.; Weichert, W.; Striefler, J.; et al. High nuclear poly-(ADP-ribose)-polymerase expression is prognostic of improved survival in pancreatic cancer. Histopathology 2012, 61, 409–416. [Google Scholar] [CrossRef]
- Aiad, H.A.; Kandil, M.A.H.; El-Tahmody, M.A.; Abulkheir, I.L.; Abulkasem, F.M.; Elmansori, A.A.; Aleskandarany, M.A. The prognostic and predictive significance of PARP-1 in locally advanced breast cancer of Egyptian patients receiving neoadjuvant chemotherapy. Appl. Immunohistochem. Mol. Morphol. 2015, 23, 571–579. [Google Scholar] [CrossRef]
- Mantovani, F.; Collavin, L.; Del Sal, G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2018, 26, 199–212. [Google Scholar] [CrossRef]
- Liu, D.P.; Song, H.; Xu, Y. A common gain of function of p53 cancer mutants in inducing genetic instability. Oncogene 2010, 29, 949–956. [Google Scholar] [CrossRef]
- Huang, P.; Chen, G.; Jin, W.; Mao, K.; Wan, H.; He, Y. Molecular Mechanisms of Parthanatos and Its Role in Diverse Diseases. Int. J. Mol. Sci. 2022, 23, 7292. [Google Scholar] [CrossRef]
- Friedmann-Morvinski, D.; Verma, I.M. Dedifferentiation and reprogramming: Origins of cancer stem cells. EMBO Rep. 2014, 15, 244. [Google Scholar] [CrossRef]
- Gilabert, M.; Launay, S.; Ginestier, C.; Bertucci, F.; Audebert, S.; Pophillat, M.; Toiron, Y.; Baudelet, E.; Finetti, P.; Noguchi, T.; et al. Poly(ADP-Ribose) Polymerase 1 (PARP1) Overexpression in Human Breast Cancer Stem Cells and Resistance to Olaparib. PLoS ONE 2014, 9, e104302. [Google Scholar] [CrossRef]
- Valencia-González, H.A.; Ruíz, G.; Ortiz-Sánchez, E.; García-Carrancá, A. Cancer stem cells from tumor cell lines activate the DNA damage response pathway after ionizing radiation more efficiently than noncancer stem cells. Stem Cells Int. 2019, 2019, 7038953. [Google Scholar] [CrossRef]
- Venere, M.; Hamerlik, P.; Wu, Q.; Rasmussen, R.D.; Song, L.A.; Vasanji, A.; Tenley, N.; Flavahan, W.A.; Hjelmeland, A.B.; Bartek, J.; et al. Therapeutic targeting of constitutive PARP activation compromises stem cell phenotype and survival of glioblastoma-initiating cells. Cell Death Differ. 2014, 21, 258. [Google Scholar] [CrossRef]
- Vitale, I.; Manic, G.; De Maria, R.; Kroemer, G.; Galluzzi, L. DNA Damage in Stem Cells. Mol. Cell 2017, 66, 306–319. [Google Scholar] [CrossRef]
- Zhao, B.; Zhang, W.D.; Duan, Y.L.; Lu, Y.Q.; Cun, Y.X.; Li, C.H.; Guo, K.; Nie, W.H.; Li, L.; Zhang, R.; et al. Filia Is an ESC-Specific Regulator of DNA Damage Response and Safeguards Genomic Stability. Cell Stem Cell 2015, 16, 684–698. [Google Scholar] [CrossRef]
- Li, H.; Liu, Z.Y.; Wu, N.; Chen, Y.C.; Cheng, Q.; Wang, J. PARP inhibitor resistance: The underlying mechanisms and clinical implications. Mol. Cancer 2020, 19, 107. [Google Scholar] [CrossRef]
- Tang, D.G. Understanding cancer stem cell heterogeneity and plasticity. Cell Res. 2012, 22, 457–472. [Google Scholar] [CrossRef]
- Qin, C.; Ji, Z.; Zhai, E.; Xu, K.; Zhang, Y.; Li, Q.; Jing, H.; Wang, X.; Song, X. PARP inhibitor olaparib enhances the efficacy of radiotherapy on XRCC2-deficient colorectal cancer cells. Cell Death Dis. 2022, 13, 1–8. [Google Scholar] [CrossRef]
- Quiñonero, F.; Mesas, C.; Muñoz-Gámez, J.A.; Jiménez-Luna, C.; Perazzoli, G.; Prados, J.; Melguizo, C.; Ortiz, R. PARP1 inhibition by Olaparib reduces the lethality of pancreatic cancer cells and increases their sensitivity to Gemcitabine. Biomed. Pharmacother. 2022, 155, 113669. [Google Scholar] [CrossRef] [PubMed]
- Long, W.; Zhao, W.; Ning, B.; Huang, J.; Chu, J.; Li, L.; Ma, Q.; Xing, C.; Wang, H.Y.; Liu, Q.; et al. PHF20 collaborates with PARP1 to promote stemness and aggressiveness of neuroblastoma cells through activation of SOX2 and OCT4. J. Mol. Cell Biol. 2018, 10, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Bellio, C.; DiGloria, C.; Foster, R.; James, K.; Konstantinopoulos, P.A.; Growdon, W.B.; Rueda, B.R. PARP inhibition induces enrichment of DNA repair-proficient CD133 and CD117 positive ovarian cancer stem cells. Mol. Cancer Res. 2019, 17, 431–445. [Google Scholar] [CrossRef] [PubMed]
- Smeby, J.; Kryeziu, K.; Berg, K.C.G.; Eilertsen, I.A.; Eide, P.W.; Johannessen, B.; Guren, M.G.; Nesbakken, A.; Bruun, J.; Lothe, R.A.; et al. Molecular correlates of sensitivity to PARP inhibition beyond homologous recombination deficiency in pre-clinical models of colorectal cancer point to wild-type TP53 activity. eBioMedicine 2020, 59, 102923. [Google Scholar] [CrossRef]
- Casado, J.; Iñigo-Chaves, A.; Jiménez-Ruiz, S.M.; Ríos-Arrabal, S.; Carazo-Gallego, Á.; González-Puga, C.; Núñez, M.I.; Ruíz-Extremera, Á.; Salmerón, J.; León, J. AA-NAT, MT1 and MT2 Correlates with Cancer Stem-Like Cell Markers in Colorectal Cancer: Study of the Influence of Stage and p53 Status of Tumors. Int. J. Mol. Sci. 2017, 18, 1251. [Google Scholar] [CrossRef]
| All 1 | wtp53 2 | mtp53 3 | |||||
|---|---|---|---|---|---|---|---|
| Characteristic | Median ± CL | p | Median ± CL | p | Median ± CL | p | |
| Age (y) *,4 | <72 | 1.63 (0.94–2.73) | 0.654 | 1.81 (1.10–3.03) | 0.157 | 1.46 (0.84–2.46) | 0.489 |
| ≥72 | 1.52 (1.00–2.52) | 1.44 (1.01–2.21) | 1.60 (0.96–2.93) | ||||
| Gender * | Male | 1.60 (0.98–2.89) | 0.323 | 1.71 (1.09–2.93) | 0.184 | 1.50 (0.88–3.20) | 0.403 |
| Female | 1.54 (0.96–2.37) | 1.53 (0.97–1.96) | 1.47 (0.92–2.50) | ||||
| Location * | Colon | 1.62 (0.98–2.57) | 0.430 | 1.71 (1.05–2.44) | 0.325 | 1.51 (0.91–2.81) | 0.540 |
| Rectum | 1.24 (0.86–3.09) | 1.62 (0.86–3.59) | 1.62 (0.98–26.25) | ||||
| DG †& | Well | 1.16 (0.58–2.50) | 0.042 | 1.10 (0.56–1.43) | 0.002 | 1.23 (0.57–4.97) | 0.980 |
| Moderately | 1.77 (1.11–2.57) | 1.78 (1.19–2.63) | 1.65 (0.99–2.52) | ||||
| Poor | 1.54 (0.92–3.28) | 2.32 (0.91–4.55) | 1.27 (0.92–4.80) | ||||
| pTNM Stage * | Stage I + II | 1.69 (1.07–2.66) | 0.501 | 1.69 (1.10–2.65) | 0.785 | 1.70 (0.92–2.74) | 0.445 |
| Stage III + IV | 1.58 (0.96–2.69) | 1.57 (0.92–2.29) | 1.54 (0.88–2.69) | ||||
| Tumour stage * | T1 + T2 | 1.78 (1.07–3.25) | 0.505 | 1.61 (1.17–2.40) | 0.732 | 2.01 (0.57–4.08) | 0.942 |
| T3 + T4 | 1.57 (0.95–2.52) | 1.56 (0.96–2.73) | 1.50 (0.92–2.76) | ||||
| LNM *,# | Absent | 1.42 (0.78–2.48) | 0.503 | 1.58 (0.97–2.52) | 0.850 | 1.46 (0.53–3.33) | 0.383 |
| Present | 1.50 (0.92–2.10) | 1.58 (0.90–2.03) | 1.27 (0.90–2.54) |
| wtp53 1 | mtp53 2 | ||||
|---|---|---|---|---|---|
| Variables | HR [95 % CI] | p | HR [95 % CI] | p | |
| PARP-1 * | Low | 1 | 1 | ||
| High | 1.30 [0.57, 3.00] | 0.532 | 0.36 [0.15, 0.88] | 0.025 | |
| Age | ≤72 | 1 | 1 | ||
| >72 | 2.27 [0.91, 5.64] | 0.078 | 2.32 [0.93, 5.79] | 0.072 | |
| Gender | Female | 1 | 1 | ||
| Male | 1.54 [0.70, 3.38] | 0.286 | 1.21 [0.45, 3.26] | 0.709 | |
| TNM | Stage I + II | 1 | 1 | ||
| Stage III + IV | 1.86 [0.83, 4.16] | 0.130 | 1.72 [0.59, 5.04] | 0.322 | |
| C 3 and/or R 4 | No | 1 | 1 | ||
| Yes | 0.66 [0.28, 1.56] | 0.341 | 0.74 [0.25, 2.62] | 0.602 | |
| All 1 | wtp53 2 | mtp53 3 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PARP-1 | PARP-1 | PARP-1 | ||||||||
| Low | High | p | Low | High | p | Low | High | p | ||
| CD133lowCD44low | 30 (61.2) | 19 (38.8) | <0.001 * | 17 (53.1) | 15 (46.9) | <0.001 * | 13 (76.5) | 4 (23.5) | 0.144 | |
| CD133highCD44high | 13 (22.4) | 45 (77.6) | 6 (14.0) | 37 (86.0) | 7 (46.7) | 8 (53.3) | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Puentes-Pardo, J.D.; Moreno-SanJuan, S.; Casado, J.; Escudero-Feliu, J.; López-Pérez, D.; Sánchez-Uceta, P.; González-Novoa, P.; Gálvez, J.; Carazo, Á.; León, J. PARP-1 Expression Influences Cancer Stem Cell Phenotype in Colorectal Cancer Depending on p53. Int. J. Mol. Sci. 2023, 24, 4787. https://doi.org/10.3390/ijms24054787
Puentes-Pardo JD, Moreno-SanJuan S, Casado J, Escudero-Feliu J, López-Pérez D, Sánchez-Uceta P, González-Novoa P, Gálvez J, Carazo Á, León J. PARP-1 Expression Influences Cancer Stem Cell Phenotype in Colorectal Cancer Depending on p53. International Journal of Molecular Sciences. 2023; 24(5):4787. https://doi.org/10.3390/ijms24054787
Chicago/Turabian StylePuentes-Pardo, Jose D., Sara Moreno-SanJuan, Jorge Casado, Julia Escudero-Feliu, David López-Pérez, Paula Sánchez-Uceta, Paula González-Novoa, Julio Gálvez, Ángel Carazo, and Josefa León. 2023. "PARP-1 Expression Influences Cancer Stem Cell Phenotype in Colorectal Cancer Depending on p53" International Journal of Molecular Sciences 24, no. 5: 4787. https://doi.org/10.3390/ijms24054787
APA StylePuentes-Pardo, J. D., Moreno-SanJuan, S., Casado, J., Escudero-Feliu, J., López-Pérez, D., Sánchez-Uceta, P., González-Novoa, P., Gálvez, J., Carazo, Á., & León, J. (2023). PARP-1 Expression Influences Cancer Stem Cell Phenotype in Colorectal Cancer Depending on p53. International Journal of Molecular Sciences, 24(5), 4787. https://doi.org/10.3390/ijms24054787







