Conserved Transcriptome Features Define Prepubertal Primate Spermatogonial Stem Cells as Adark Spermatogonia and Identify Unique Regulators
Abstract
1. Introduction
2. Results
2.1. Single-Cell Transcriptomics Identifies Major Germ and Somatic Cell Types in Prepubertal Human, Baboon, and Macaque Testes
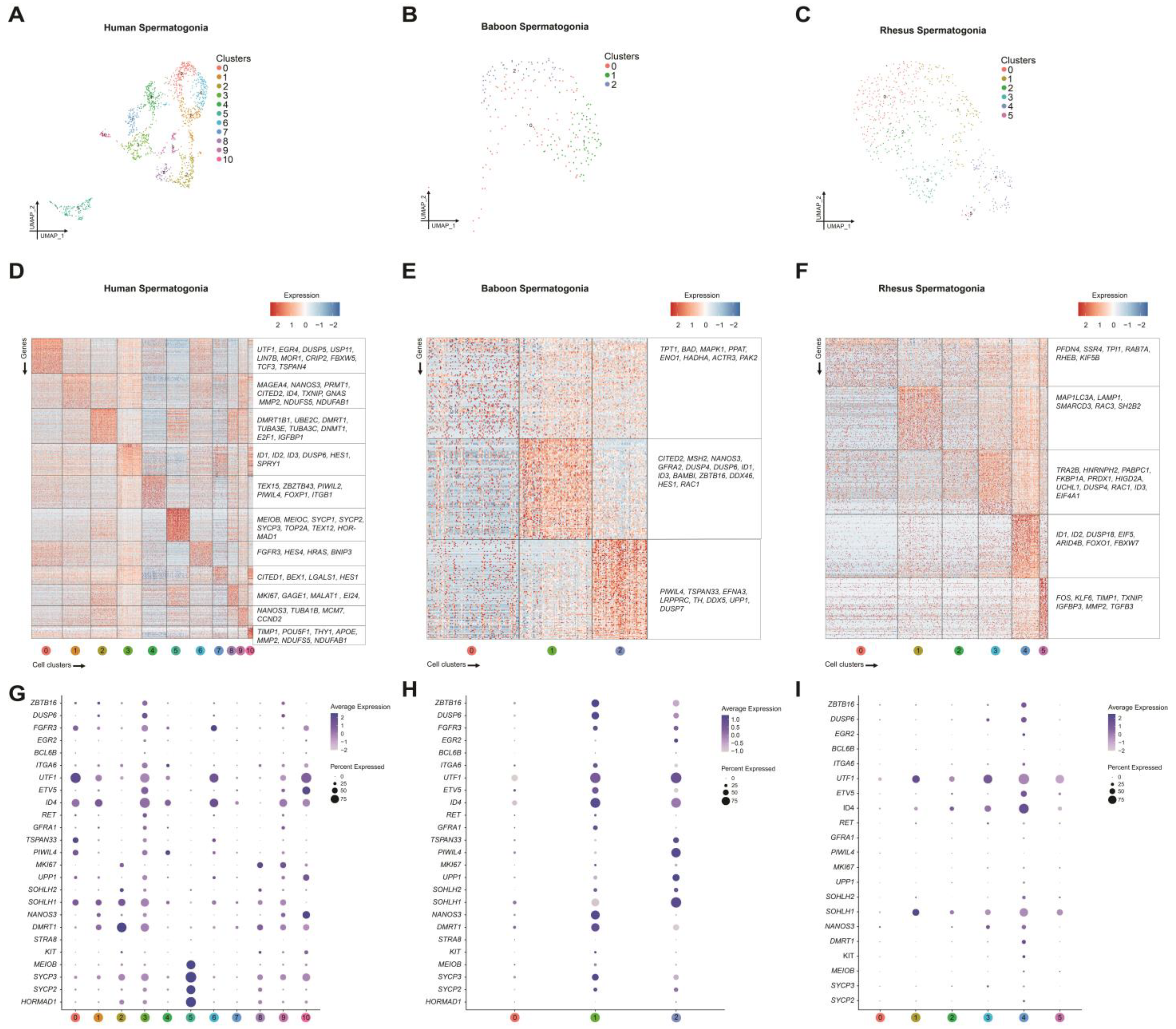
2.2. Focused Analysis of Germ Cells Reveals Discrete Spermatogonial Subtypes in Prepubertal Humans
2.3. Transcriptomic Characterization of Prepubertal Baboon Germ Cells
2.4. Transcriptomic Characterization of Prepubertal Rhesus Macaque Germ Cells
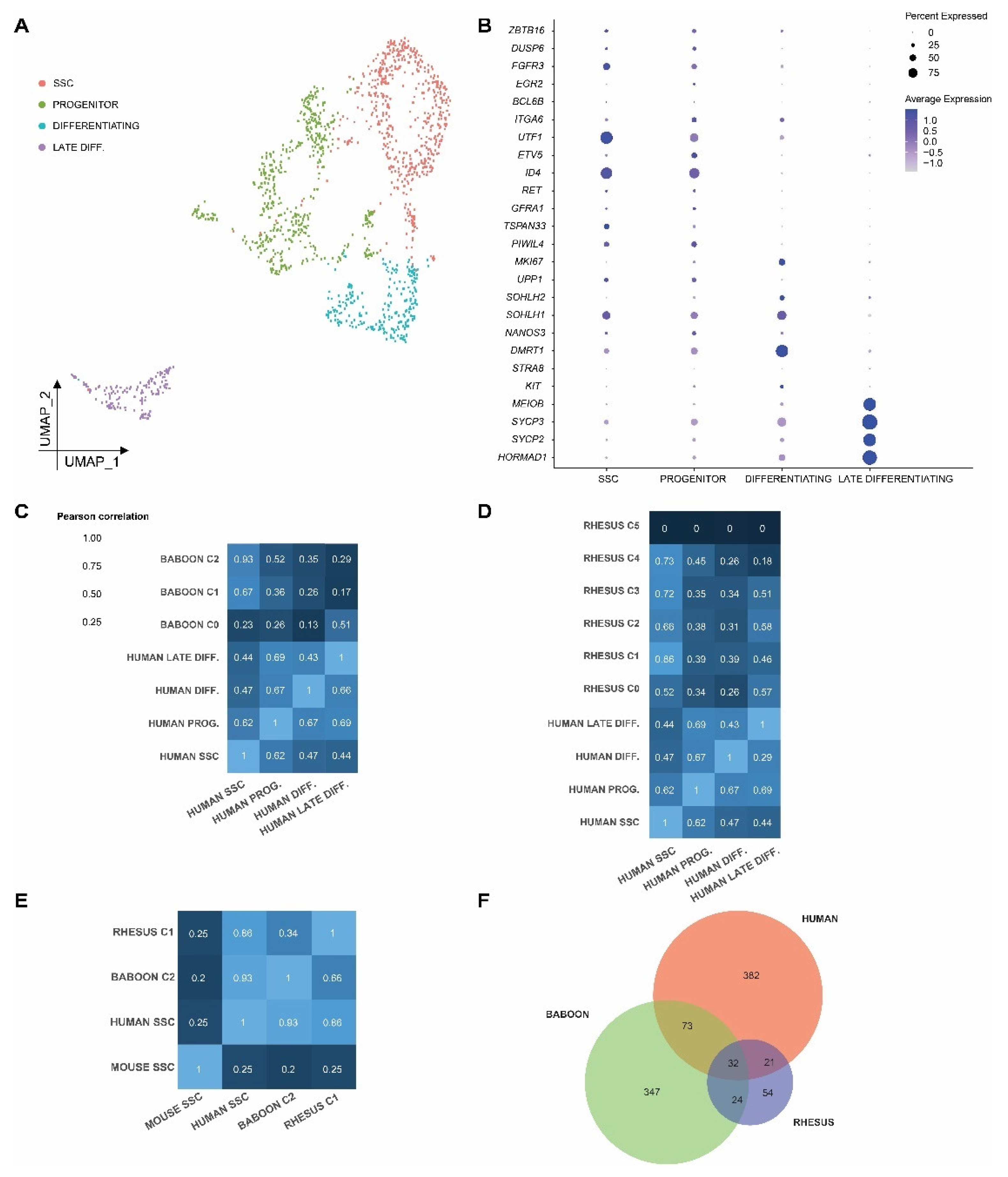
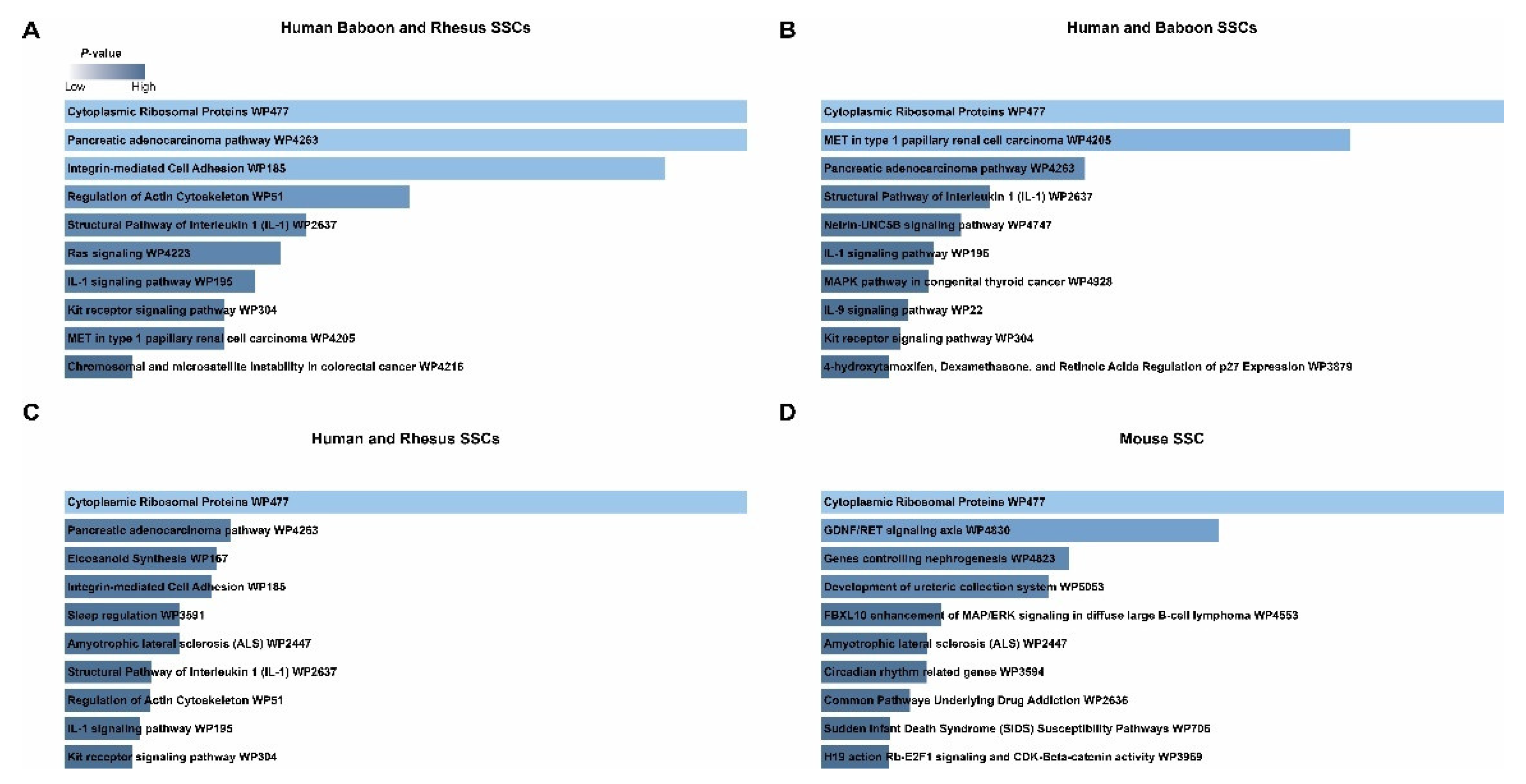
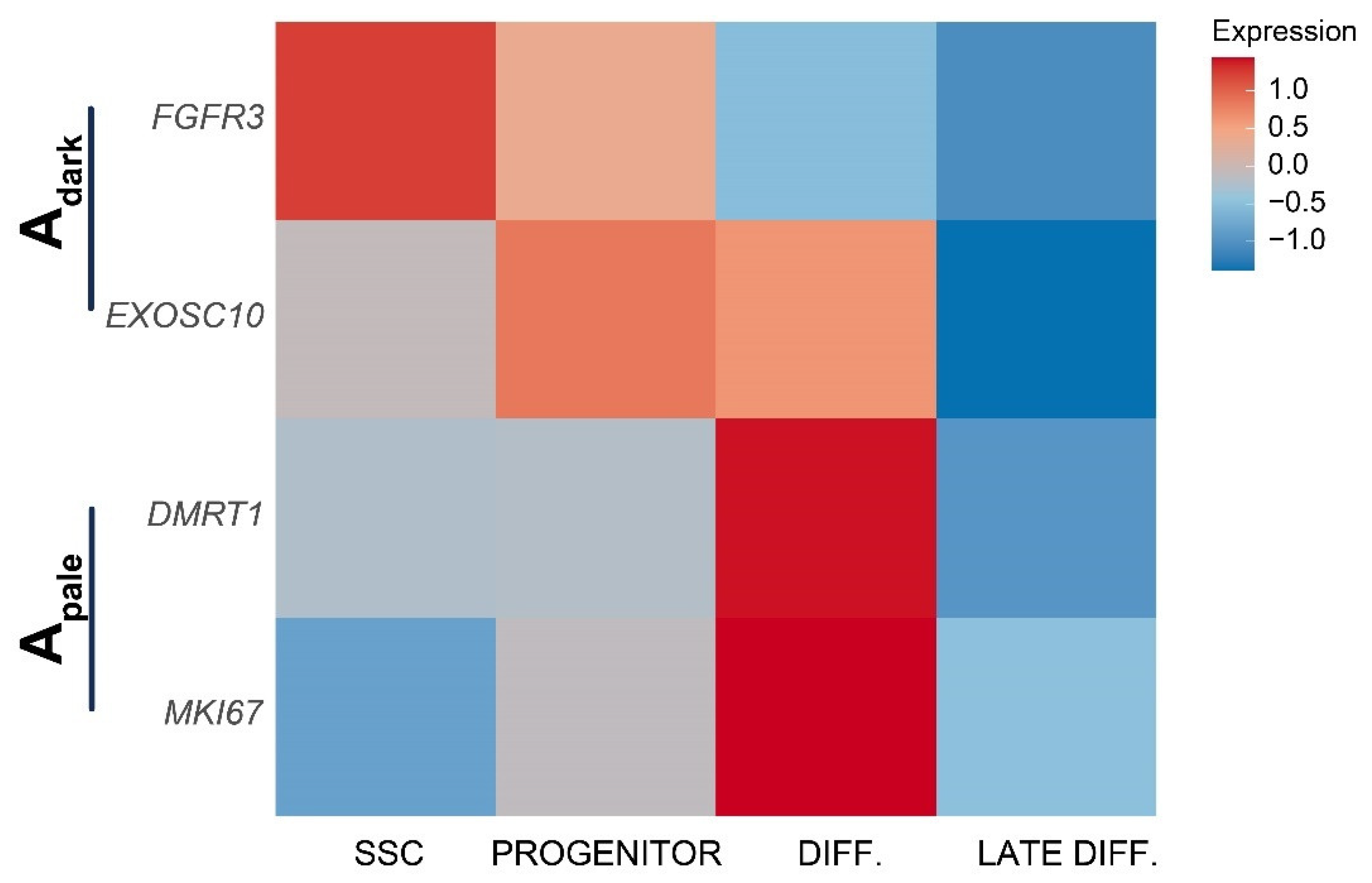
2.5. Cross-Species Comparison of Spermatogonia Reveals Conserved and Divergent Features in Prepubertal Primate and Rodent Spermatogonia
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Single-Cell Suspension Preparation
4.3. Single-Cell RNA-Sequencing of Baboon and Macaque Testis Cells
4.4. Quality Control, Pre-Processing, and Integration of Gene Expression Datasets
4.5. Cross-Species Correlation Analysis of Germ Cells
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lambertini, M.; Del Mastro, L.; Pescio, M.C.; Andersen, C.Y.; Azim, H.A.; Peccatori, F.A.; Costa, M.; Revelli, A.; Salvagno, F.; Gennari, A.; et al. Cancer and fertility preservation: International recommendations from an expert meeting. BMC Med. 2016, 14, 1. [Google Scholar] [CrossRef]
- Khalifeh, F.A.; Sarraf, M.; Dabit, S.T. Full-term delivery following intracytoplasmic sperm injection with spermatozoa extracted from frozen-thawed testicular tissue. Hum. Reprod. 1997, 12, 87–88. [Google Scholar] [CrossRef]
- Fu, L.; Zhou, F.; An, Q.; Zhang, K.; Wang, X.; Xu, J.; Guo, Y.; Lu, W.; Liang, X.; Gu, Y. Sperm Cryopreservation for Male Cancer Patients: More than 10 Years of Experience, in Beijing China. Med. Sci. Monit. 2019, 25, 3256–3261. [Google Scholar] [CrossRef]
- Palermo, G.; Joris, H.; Devroey, P.; Van Steirteghem, A.C. Pregnancies after intracytoplasmic injection of single spermatozoon into an oocyte. Lancet 1992, 340, 17–18. [Google Scholar] [CrossRef]
- McDougall, R.J.; Gillam, L.; Delany, C.; Jayasinghe, Y. Ethics of fertility preservation for prepubertal children: Should clinicians offer procedures where efficacy is largely unproven? J. Med. Ethics 2018, 44, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Onofre, J.; Baert, Y.; Faes, K.; Goossens, E. Cryopreservation of testicular tissue or testicular cell suspensions: A pivotal step in fertility preservation. Hum. Reprod. Update 2016, 22, 744–761. [Google Scholar] [CrossRef] [PubMed]
- Stukenborg, J.-B.; Alves-Lopes, J.P.; Kurek, M.; Albalushi, H.; Reda, A.; Keros, V.; Töhönen, V.; Bjarnason, R.; Romerius, P.; Sundin, M.; et al. Spermatogonial quantity in human prepubertal testicular tissue collected for fertility preservation prior to potentially sterilizing therapy. Hum. Reprod. 2018, 33, 1677–1683. [Google Scholar] [CrossRef] [PubMed]
- Valli-Pulaski, H.; Peters, K.A.; Gassei, K.; Steimer, S.R.; Sukhwani, M.; Hermann, B.P.; Dwomor, L.; David, S.; Fayomi, A.P.; Munyoki, S.; et al. Testicular tissue cryopreservation: 8 years of experience from a coordinated network of academic centers. Hum. Reprod. 2019, 34, 966–977. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Goodyear, S.M.; Abramowitz, L.K.; Bartolomei, M.S.; Tobias, J.W.; Avarbock, M.R.; Brinster, R.L. Fertile offspring derived from mouse spermatogonial stem cells cryopreserved for more than 14 years. Hum. Reprod. 2012, 27, 1249–1259. [Google Scholar] [CrossRef]
- Fayomi, A.P.; Peters, K.; Sukhwani, M.; Valli-Pulaski, H.; Shetty, G.; Meistrich, M.L.; Houser, L.; Robertson, N.; Roberts, V.; Ramsey, C.; et al. Autologous grafting of cryopreserved prepubertal rhesus testis produces sperm and offspring. Science 2019, 363, 1314–1319. [Google Scholar] [CrossRef]
- Sadri-Ardekani, H.; Mizrak, S.C.; van Daalen, S.K.; Korver, C.M.; Roepers-Gajadien, H.L.; Koruji, M.; Hovingh, S.; de Reijke, T.M.; de la Rosette, J.J.M.C.H.; van der Ween, F.; et al. Propagation of Human Spermatogonial Stem Cells In Vitro. JAMA 2009, 302, 2127–2134. [Google Scholar] [CrossRef]
- Wu, X.; Schmidt, J.A.; Avarbock, M.R.; Tobias, J.W.; Carlson, C.A.; Kolon, T.F.; Ginsberg, J.P.; Brinster, R.L. Prepubertal human spermatogonia and mouse gonocytes share conserved gene expression of germline stem cell regulatory molecules. Proc. Natl. Acad. Sci. USA 2009, 106, 21672–21677. [Google Scholar] [CrossRef] [PubMed]
- Sadri-Ardekani, H.; Akhondi, M.A.; van der Veen, F.; Repping, S.; van Pelt, A.M. In Vitro Propagation of Human Prepubertal Spermatogonial Stem Cells. JAMA 2011, 305, 2416–2418. [Google Scholar] [CrossRef]
- Chen, B.; Wang, Y.-B.; Zhang, Z.-L.; Xia, W.-L.; Wang, H.-X.; Xiang, Z.-Q.; Hu, K.; Han, Y.-F.; Wang, Y.-X.; Huang, Y.-R.; et al. Xeno-free culture of human spermatogonial stem cells supported by human embryonic stem cell-derived fibroblast-like cells. Asian J. Androl. 2009, 11, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Mirzapour, T.; Movahedin, M.; Ibrahim, T.A.T.; Koruji, M.; Haron, A.W.; Nowroozi, M.R.; Rafieian, S.H. Effects of basic fibroblast growth factor and leukaemia inhibitory factor on proliferation and short-term culture of human spermatogonial stem cells. Andrologia 2012, 44 (Suppl. S1), 41–55. [Google Scholar] [CrossRef] [PubMed]
- Koruji, M.; Shahverdi, A.; Janan, A.; Piryaei, A.; Lakpour, M.R.; Sedighi, M.A.G. Proliferation of small number of human spermatogonial stem cells obtained from azoospermic patients. J. Assist. Reprod. Genet. 2012, 29, 957–967. [Google Scholar] [CrossRef]
- Goharbakhsh, L.; Mohazzab, A.; Salehkhou, S.; Heidari, M.; Zarnani, A.H.; Parivar, K.; Akhondi, M.M. Isolation and Culture of Human Spermatogonial Stem Cells Derived from Testis Biopsy. Avicenna J. Med. Biotechnol. 2013, 5, 54–61. [Google Scholar]
- Guo, Y.; Liu, L.; Sun, M.; Hai, Y.; Li, Z.; He, Z. Expansion and long-term culture of human spermatogonial stem cells via the activation of SMAD3 and AKT pathways. Exp. Biol. Med. 2015, 240, 1112–1122. [Google Scholar] [CrossRef]
- Luo, J.; Megee, S.; Dobrinski, I. Asymmetric Distribution of UCH-L1 in Spermatogonia Is Associated with Maintenance and Differentiation of Spermatogonial Stem Cells. J. Cell. Physiol. 2009, 220, 460–468. [Google Scholar] [CrossRef]
- Clermont, Y.; Hermo, L. Spermatogonial stem cells in the albino rat. Am. J. Anat. 1975, 142, 159–175. [Google Scholar] [CrossRef]
- Bartmanska, J.; Clermont, Y. Renewal of type A spermatogonia in adult rats. Cell Tissue Kinet. 1983, 16, 135–143. [Google Scholar] [PubMed]
- Clermont, Y.; Bustos-Obregon, E. Re-examination of spermatogonial renewal in the rat by means of seminiferous tubules mounted “in toto”. Am. J. Anat. 1968, 122, 237–247. [Google Scholar] [CrossRef]
- Clermont, Y. Kinetics of spermatogenesis in mammals: Seminiferous epithelium cycle and spermatogonial renewal. Physiol. Rev. 1972, 52, 198–236. [Google Scholar] [CrossRef]
- Clermont, Y. The cycle of the seminiferous epithelium in man. Am. J. Anat. 1963, 112, 35–51. [Google Scholar] [CrossRef]
- Oakberg, E.F. Spermatogonial stem-cell renewal in the mouse. Anat. Rec. 1971, 169, 515–531. [Google Scholar] [CrossRef] [PubMed]
- Lord, T.; Oatley, J.M. A revised Asingle model to explain stem cell dynamics in the mouse male germline. Reproduction 2017, 154, R55–R64. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Sharma, M.; Nabeshima, Y.-I.; Braun, R.E.; Yoshida, S. Functional Hierarchy and Reversibility Within the Murine Spermatogenic Stem Cell Compartment. Science 2010, 328, 62–67. [Google Scholar] [CrossRef]
- Hara, K.; Nakagawa, T.; Enomoto, H.; Suzuki, M.; Yamamoto, M.; Simons, B.D.; Yoshida, S. Mouse Spermatogenic Stem Cells Continually Interconvert between Equipotent Singly Isolated and Syncytial States. Cell Stem Cell 2014, 14, 658–672. [Google Scholar] [CrossRef]
- Huckins, C. The spermatogonial stem cell population in adult rats. I. Their morphology, proliferation and maturation. Anat. Rec. 1971, 169, 533–557. [Google Scholar] [CrossRef]
- Tagelenbosch, R.A.; de Rooij, D.G. A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F1 hybrid mouse. Mutat. Res. Mol. Mech. Mutagen. 1993, 290, 193–200. [Google Scholar] [CrossRef]
- Nagano, M.C. Homing Efficiency and Proliferation Kinetics of Male Germ Line Stem Cells Following Transplantation in Mice. Biol. Reprod. 2003, 69, 701–707. [Google Scholar] [CrossRef]
- Oatley, M.; Chan, F.; Kaucher, A.; Oatley, J. Functional and molecular features of the ID4+ germline stem cell population in mouse testes. Genes Dev. 2014, 28, 1351–1362. [Google Scholar] [CrossRef]
- Hermann, B.P.; Cheng, K.; Singh, A.; Roa-De La Cruz, L.; Mutoji, K.N.; Chen, I.-C.; Gildersleeve, H.; Lehle, J.D.; Mayo, M.; Westernströer, B.; et al. The Mammalian Spermatogenesis Single-Cell Transcriptome, from Spermatogonial Stem Cells to Spermatids. Cell Rep. 2018, 25, 1650–1667.e8. [Google Scholar] [CrossRef] [PubMed]
- Helsel, A.R.; Yang, Q.-E.; Oatley, M.J.; Lord, T.; Sablitzky, F.; Oatley, J.M. ID4 levels dictate the stem cell state in mouse spermatogonia. Development 2017, 144, 624–634. [Google Scholar] [CrossRef]
- Clermont, Y. Renewal of spermatogonia in man. Am. J. Anat. 1966, 118, 509–524. [Google Scholar] [CrossRef]
- Clermont, Y.; Leblond, C.P. Differentiation and renewal of spermatogonia in the monkey, Macacus rhesus. Am. J. Anat. 1959, 104, 237–273. [Google Scholar] [CrossRef] [PubMed]
- Clermont, Y. Two classes of spermatogonial stem cells in the monkey (Cercopithecus aethiops). Am. J. Anat. 1969, 126, 57–71. [Google Scholar] [CrossRef]
- Clermont, Y. Spermatogenesis in man. A study of the spermatogonial population. Fertil. Steril. 1966, 17, 705–721. [Google Scholar] [CrossRef]
- Ehmcke, J.; Wistuba, J.; Schlatt, S. Spermatogonial stem cells: Questions, models and perspectives. Hum. Reprod. Update 2006, 12, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Fouquet, J.P.; Dadoune, J.P. Renewal of Spermatogonia in the Monkey (Macaca Fascilularis). Biol. Reprod. 1986, 35, 199–207. [Google Scholar] [CrossRef]
- Hermann, B.P.; Sukhwani, M.; Hansel, M.C.; Orwig, K.E. Spermatogonial stem cells in higher primates: Are there differences from those in rodents? Reproduction 2010, 139, 479–493. [Google Scholar] [CrossRef] [PubMed]
- Sohni, A.; Tan, K.; Song, H.-W.; Burow, D.; De Rooij, D.G.; Laurent, L.; Hsieh, T.-C.; Rabah, R.; Hammoud, S.S.; Vicini, E.; et al. The Neonatal and Adult Human Testis Defined at the Single-Cell Level. Cell Rep. 2019, 26, 1501–1517.e4. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Nie, X.; Giebler, M.; Mlcochova, H.; Wang, Y.; Grow, E.J.; DonorConnect; Kim, R.; Tharmalingam, M.; Matilionyte, G.; et al. The Dynamic Transcriptional Cell Atlas of Testis Development during Human Puberty. Cell Stem Cell 2020, 26, 262–276.e4. [Google Scholar] [CrossRef]
- Guo, J.; Grow, E.J.; Mlcochova, H.; Maher, G.J.; Lindskog, C.; Nie, X.; Guo, Y.; Takei, Y.; Yun, J.; Cai, L.; et al. The adult human testis transcriptional cell atlas. Cell Res. 2018, 28, 1141–1157. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Yao, C.; Xing, X.; Jing, T.; Li, P.; Zhu, Z.; Yang, C.; Zhai, J.; Tian, R.; Chen, H.; et al. Single-cell analysis of developing and azoospermia human testicles reveals central role of Sertoli cells. Nat. Commun. 2020, 11, 5683. [Google Scholar] [CrossRef]
- Wang, M.; Liu, X.; Chang, G.; Chen, Y.; An, G.; Yan, L.; Gao, S.; Xu, Y.; Cui, Y.; Dong, J.; et al. Single-Cell RNA Sequencing Analysis Reveals Sequential Cell Fate Transition during Human Spermatogenesis. Cell Stem Cell 2018, 23, 599–614.e4. [Google Scholar] [CrossRef]
- Guo, J.; Sosa, E.; Chitiashvili, T.; Nie, X.; Rojas, E.J.; Oliver, E.; Plath, K.; Hotaling, J.M.; Stukenborg, J.-B.; Clark, A.T.; et al. Single-cell analysis of the developing human testis reveals somatic niche cell specification and fetal germline stem cell establishment. Cell Stem Cell 2021, 28, 764–778.e4. [Google Scholar] [CrossRef]
- Voigt, A.L.; Dardari, R.; Su, L.; Lara, N.L.M.; Sinha, S.; Jaffer, A.; Munyoki, S.K.; Alpaugh, W.; Dufour, A.; Biernaskie, J.; et al. Metabolic transitions define spermatogonial stem cell maturation. Hum. Reprod. 2022, 37, 2095–2112. [Google Scholar] [CrossRef]
- Di Persio, S.; Tekath, T.; Siebert-Kuss, L.M.; Cremers, J.-F.; Wistuba, J.; Li, X.; zu Hörste, G.M.; Drexler, H.C.; Wyrwoll, M.J.; Tüttelmann, F.; et al. Single-cell RNA-seq unravels alterations of the human spermatogonial stem cell compartment in patients with impaired spermatogenesis. Cell Rep. Med. 2021, 2, 100395. [Google Scholar] [CrossRef]
- Suzuki, S.; Diaz, V.D.; Hermann, B.P. What has single-cell RNA-seq taught us about mammalian spermatogenesis? Biol. Reprod. 2019, 101, 617–634. [Google Scholar] [CrossRef]
- Robertson, M.J.; Kent, K.; Tharp, N.; Nozawa, K.; Dean, L.; Mathew, M.; Grimm, S.L.; Yu, Z.; Légaré, C.; Fujihara, Y.; et al. Large-scale discovery of male reproductive tract-specific genes through analysis of RNA-seq datasets. BMC Biol. 2020, 18, 103. [Google Scholar] [CrossRef]
- von Kopylow, K.; Staege, H.; Spiess, A.-N.; Schulze, W.; Will, H.; Primig, M.; Kirchhoff, C. Differential marker protein expression specifies rarefaction zone-containing human Adark spermatogonia. Reproduction 2012, 143, 45–57. [Google Scholar] [CrossRef]
- von Kopylow, K.; Staege, H.; Schulze, W.; Will, H.; Kirchhoff, C. Fibroblast growth factor receptor 3 is highly expressed in rarely dividing human type A spermatogonia. Histochem. Cell Biol. 2012, 138, 759–772. [Google Scholar] [CrossRef] [PubMed]
- Baert, Y.; Braye, A.; Struijk, R.B.; van Pelt, A.M.; Goossens, E. Cryopreservation of testicular tissue before long-term testicular cell culture does not alter in vitro cell dynamics. Fertil. Steril. 2015, 104, 1244–1252.e4. [Google Scholar] [CrossRef] [PubMed]
- Bélanger, L.-F.; Roy, S.; Tremblay, M.; Brott, B.; Steff, A.-M.; Mourad, W.; Hugo, P.; Erikson, R.; Charron, J. Mek2 Is Dispensable for Mouse Growth and Development. Mol. Cell. Biol. 2003, 23, 4778–4787. [Google Scholar] [CrossRef] [PubMed]
- Takashima, S.; Kanatsu-Shinohara, M.; Tanaka, T.; Takehashi, M.; Morimoto, H.; Shinohara, T. Rac Mediates Mouse Spermatogonial Stem Cell Homing to Germline Niches by Regulating Transmigration through the Blood-Testis Barrier. Cell Stem Cell 2011, 9, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Haataja, L.; Kaartinen, V.; Groffen, J.; Heisterkamp, N. The Small GTPase Rac3 Interacts with the Integrin-binding Protein CIB and Promotes Integrin αIIbβ3-mediated Adhesion and Spreading. J. Biol. Chem. 2002, 277, 8321–8328. [Google Scholar] [CrossRef] [PubMed]
- Kubota, H.; Brinster, R.L. Culture of Rodent Spermatogonial Stem Cells, Male Germline Stem Cells of the Postnatal Animal. Methods Cell Biol. 2008, 86, 59–84. [Google Scholar] [CrossRef]
- Forbes, A.; Lehman, R. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development 1998, 125, 679–690. [Google Scholar] [CrossRef]
- Lai, F.; Zhou, Y.; Luo, X.; Fox, J.; King, M.L. Nanos1 functions as a translational repressor in the Xenopus germline. Mech. Dev. 2011, 128, 153–163. [Google Scholar] [CrossRef]
- Subramaniam, K.; Seydoux, G. Nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development 1999, 126, 4861–4871. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Tsuda, M.; Saga, Y. Functional redundancy among Nanos proteins and a distinct role of Nanos2 during male germ cell development. Development 2007, 134, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, M.; Sasaoka, Y.; Kiso, M.; Abe, K.; Haraguchi, S.; Kobayashi, S.; Saga, Y. Conserved Role of nanos Proteins in Germ Cell Development. Science 2003, 301, 1239–1241. [Google Scholar] [CrossRef]
- Phillips, B.T.; Gassei, K.; Orwig, K.E. Spermatogonial stem cell regulation and spermatogenesis. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 1663–1678. [Google Scholar] [CrossRef]
- de Rooij, D.G. The nature and dynamics of spermatogonial stem cells. Development 2017, 144, 3022–3030. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Murray, E.; Sinha, A.; Laumas, A.; Li, J.; Lesman, D.; Nie, X.; Hotaling, J.; Guo, J.; Cairns, B.R.; et al. Dissecting mammalian spermatogenesis using spatial transcriptomics. Cell Rep. 2021, 37, 109915. [Google Scholar] [CrossRef]
- Hermann, B.P.; Sukhwani, M.; Lin, C.C.; Sheng, Y.; Tomko, J.; Rodriguez, M.; Shuttleworth, J.J.; McFarland, D.; Hobbs, R.M.; Pandolfi, P.P.; et al. Characterization, Cryopreservation, and Ablation of Spermatogonial Stem Cells in Adult Rhesus Macaques. Stem Cells 2007, 25, 2330–2338. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.X.Y.; Terry, J.M.; Belgrader, P.; Ryvkin, P.; Bent, Z.W.; Wilson, R.; Ziraldo, S.B.; Wheeler, T.D.; McDermott, G.P.; Zhu, J.; et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017, 8, 14049. [Google Scholar] [CrossRef]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., III; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e21. [Google Scholar] [CrossRef]
- Wickham, H.; Henry, L. Tidyr: Tidy Messy Data. R Package Version 1.0.0. 2019. Available online: https://cran.r-project.org/package=tidyr (accessed on 11 August 2022).
- Valero-Mora, P.M. Ggplot2: Elegant graphics for data analysis. J. Stat. Softw. 2010, 35, 1–3. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’Ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Pico, A.; Kelder, T.; van Iersel, M.; Hanspers, K.; Conklin, B.R.; Evelo, C. WikiPathways: Pathway Editing for the People. PLoS Biol. 2008, 6, e184. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, A.; Hermann, B.P. Conserved Transcriptome Features Define Prepubertal Primate Spermatogonial Stem Cells as Adark Spermatogonia and Identify Unique Regulators. Int. J. Mol. Sci. 2023, 24, 4755. https://doi.org/10.3390/ijms24054755
Singh A, Hermann BP. Conserved Transcriptome Features Define Prepubertal Primate Spermatogonial Stem Cells as Adark Spermatogonia and Identify Unique Regulators. International Journal of Molecular Sciences. 2023; 24(5):4755. https://doi.org/10.3390/ijms24054755
Chicago/Turabian StyleSingh, Anukriti, and Brian P. Hermann. 2023. "Conserved Transcriptome Features Define Prepubertal Primate Spermatogonial Stem Cells as Adark Spermatogonia and Identify Unique Regulators" International Journal of Molecular Sciences 24, no. 5: 4755. https://doi.org/10.3390/ijms24054755
APA StyleSingh, A., & Hermann, B. P. (2023). Conserved Transcriptome Features Define Prepubertal Primate Spermatogonial Stem Cells as Adark Spermatogonia and Identify Unique Regulators. International Journal of Molecular Sciences, 24(5), 4755. https://doi.org/10.3390/ijms24054755





