COVID-19 Biogenesis and Intracellular Transport
Abstract
1. Introduction
2. Proteins of SARS-CoV-2
| Name of Protein | Function | Reference |
|---|---|---|
| S | The spike S-protein binds to its receptor (ACE2) and perforates (after its cleavage) the double membrane composed of the PM and endosomal membrane. | [31,33,38] |
| E | The envelope (E) protein is crucial for budding and replication | [52,58,59,63,64,65,66] |
| M | The triple-spanning membrane M protein binds to the nucleocapsid and generates the curvature of the host cell membrane. The cytoplasmic domains of M proteins homo-oligomerize. The M–E complex ensures the uniform size of viral particles for viral maturation and mediates virion release. | [62] |
| N | The N protein forms biomolecular condensates with RNA. | [61,63] |
| NSP1 | NSP1 binds to 18S ribosomal RNA in the mRNA entry channel of the ribosome and leads to global inhibition of mRNA translation upon infection. It degrades host mRNA and inhibits IFN signaling. NSP1 suppress IFN-I signaling. | [67,68] |
| NSP2 | NSP2 induces repression of mRNA translation, impairing interferon production. | [69] |
| NSP3 | NSP3/papain-like protease is responsible for cleaving viral polyproteins during replication. NSP3 is the largest protein encoded by the coronavirus. It cleaves viral proteins. NSP3 reverses PARP9/DTX3L-dependent ADP-ribosylation induced by interferon signaling. | [70,71] |
| NSP4 | NSP4 induces pro-inflammatory mitochondrial DNA release in inner membrane-derived vesicles. | [72] |
| NSP5 | NSP5/3C-like protease is responsible for cleaving viral polyproteins during replication. NSP5 exhibits catalytic activity and interferon antagonism. NSP5 activates the NF-κB pathway. | [73,74,75,76] |
| NSP6 | NSP6 participates in the biogenesis of the SARS-CoV-2 replication organelle. It forms double membrane vacuoles (DMVs). NSP6 impairs lysosome acidification to inhibit autophagic flux. NSP6 suppress IFN-I signaling. It triggers NLRP3-dependent pyroptosis. | [14,76,77] |
| NSP7 | They form a dexadecameric complex. They form polymerase complex consisting of the NSP12 catalytic subunit and NSP7–NSP8 cofactors. Processing clamp for RdRp. NSP8 and NSP9 bind to the 7SL RNA in the signal recognition particle and interfere with protein trafficking to the cell membrane upon infection. | [67,78] |
| NSP8 | ||
| NSP9 | RNA binding. NSP8 and NSP9 bind to the 7SL RNA in the signal recognition particle and interfere with protein trafficking to the cell membrane upon infection. | [67] |
| NSP10 | SARS-CoV-2 NSP10/NSP14-ExoN and NSP16 functions as an exoribonuclease with NSP14. | [79] |
| NSP11 | Real function is unclear. NSP11 has a helical propensity. | [80] |
| NSP12 | RNA-dependent RNA-cap2′-O-methyltransfeaze. Viral mRNA capping protecting it from RNases and RNA polymerase. Viral replication and transcription. Forms polymerase complex consisting of the NSP12 catalytic subunit and NSP7-NSP8 cofactors. NSP12 is the viral RNA-dependent RNA polymerase (RdRp) and suppresses host antiviral responses. | [78,81] |
| NSP13 | RNA helicase, 5′-triphosphatase. 2 NSP13, NSP14, and NSP15 function as potent interferon antagonists. | [82,83,84,85,86] |
| NSP14 | NSP13, NSP14, and NSP15 function as potent interferon antagonists. | [86,87,88,89] |
| NSP15 | Endoribonuclease NSP13, NSP14, and NSP15 function as potent interferon antagonists. | [86] |
| NSP16 | 2′-O-methyltransferase NSP16, | [14,90] |
3. Cell Structure of Human Airways
4. ACE2 as a Receptor for SARS-CoV-2
5. ACE2 Localization
6. Role of TMPRSS2
7. Entry of SARS-CoV-2 into Cells
8. Alterations in Cells Induced by SARS-CoV-2
9. Viral Replication Organelle
10. Mechanisms of DMV Formation
11. Function of NSP6 and Mystery of DMV Production
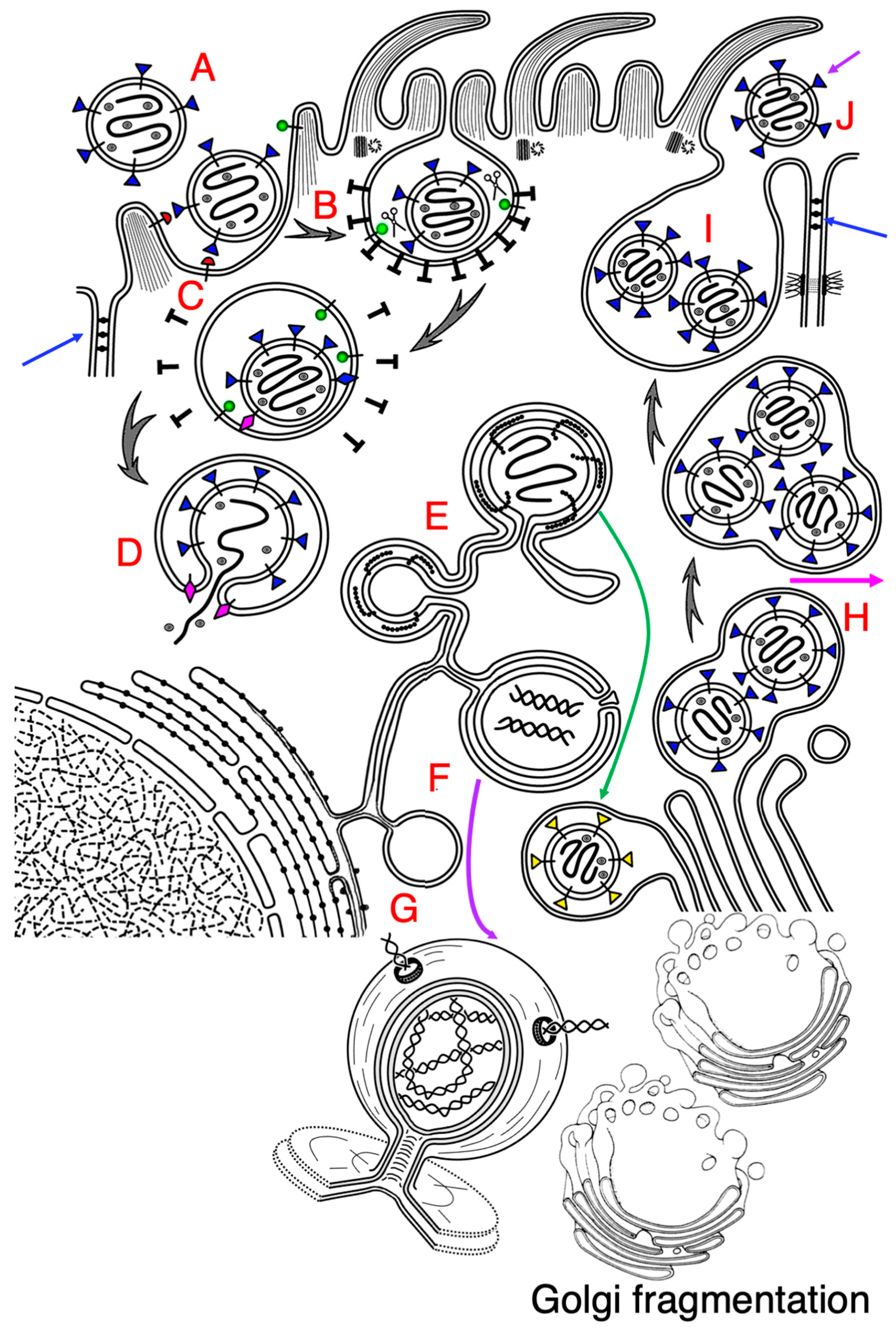
12. Virion Budding and Intracellular Transport
13. Conclusions, Unclear Questions, and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| APM | apical plasma membrane |
| BLPM | basolateral plasma membrane |
| DMV | double membrane vacuole |
| dsRNA | double-stranded form RNA |
| ER | endoplasmic reticulum |
| ERES | ER exit site |
| GC | Golgi complex |
| NSP | nonstructural protein |
| PM | plasma membrane |
| VRO | viral replication organelle. |
| ZER | zippered ER |
References
- Tyrrell, D.A.; Bynoe, M.L. Cultivation of a novel type of common-cold virus in organ cultures. Br. Med. J. 1965, 1, 1467–1470. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science 2020, 368, 1016–1020. [Google Scholar] [CrossRef] [PubMed]
- Azkur, A.K.; Akdis, M.; Azkur, D.; Sokolowska, M.; van de Veen, W.; Brüggen, M.C.; O’Mahony, L.; Gao, Y.; Nadeau, K.; Akdis, C.A. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy 2020, 75, 1564–1581. [Google Scholar] [CrossRef]
- Paules, C.I.; Marston, H.D.; Fauci, A.S. Coronavirus Infections-More Than Just the Common Cold. JAMA 2020, 323, 707–708. [Google Scholar] [CrossRef]
- Wu, A.; Peng, Y.; Huang, B.; Ding, X.; Wang, X.; Niu, P.; Meng, J.; Zhu, Z.; Zhang, Z.; Wang, J.; et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 2020, 27, 325–328. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Klein, S.; Cortese, M.; Winter, S.L.; Wachsmuth-Melm, M.; Neufeldt, C.J.; Cerikan, B.; Stanifer, M.L.; Boulant, S.; Bartenschlager, R.; Chlanda, P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020, 11, 5885. [Google Scholar] [CrossRef]
- Turonová, B.; Sikora, M.; Schürmann, C.; Hagen, W.J.H.; Welsch, S.; Blanc, F.E.C.; Von Bülow, S.; Gecht, M.; Bagola, K.; Hörner, C.; et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science 2020, 370, 203–208. [Google Scholar] [CrossRef]
- Laue, M.; Kauter, A.; Hoffmann, T.; Möller, L.; Michel, J.; Nitsche, A. Morphometry of SARS-CoV and SARS-CoV-2 particles in ultrathin plastic sections of infected Vero cell cultures. Sci. Rep. 2021, 11, 3515. [Google Scholar] [CrossRef]
- Baselga, M.; Moreo, E.; Uranga-Murillo, I.; Arias, M.; Junquera, C. Ultrastructural analysis and three-dimensional reconstruction of cellular structures involved in SARS-CoV-2 spread. Histochem. Cell Biol. 2023, 159, 47–60. [Google Scholar] [CrossRef]
- Ke, Z.; Oton, J.; Qu, K.; Cortese, M.; Zila, V.; McKeane, L.; Nakane, T.; Zivanov, J.; Neufeldt, C.J.; Cerikan, B.; et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature 2020, 588, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Song, Y.; Chen, Y.; Wu, N.; Xu, J.; Sun, C.; Zhang, J.; Weng, T.; Zhang, Z.; Wu, Z.; et al. Molecular Architecture of the SARS-CoV-2 Virus. Cell 2020, 183, 730–738.e13. [Google Scholar] [CrossRef] [PubMed]
- Cortese, M.; Lee, J.Y.; Cerikan, B.; Neufeldt, C.J.; Oorschot, V.M.J.; Köhrer, S.; Hennies, J.; Schieber, N.L.; Ronchi, P.; Mizzon, G.; et al. Integrative Imaging Reveals SARS-CoV-2-Induced Reshaping of Subcellular Morphologies. Cell Host Microbe 2020, 28, 853–866. [Google Scholar] [CrossRef] [PubMed]
- Santerre, M.; Arjona, S.P.; Allen, C.N.; Shcherbik, N.; Sawaya, B.E. Why do SARS-CoV-2 NSPs rush to the ER? J. Neurol. 2021, 268, 2013–2022. [Google Scholar] [CrossRef] [PubMed]
- Batlle, D.; Monteil, V.; Garreta, E.; Hassler, L.; Wysocki, J.; Chandar, V.; Schwartz, R.E.; Mirazimi, A.; Montserrat, N.; Bader, M.; et al. Evidence in favor of the essentiality of human cell membrane-bound ACE2 and against soluble ACE2 for SARS-CoV-2 infectivity. Cell 2022, 185, 1837–1839. [Google Scholar] [CrossRef]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Hou, Y.J.; Okuda, K.; Edwards, C.E.; Martinez, D.R.; Asakura, T.; Dinnon, K.H., 3rd; Kato, T.; Lee, R.E.; Yount, B.L.; Mascenik, T.M.; et al. SARS-CoV-2 Reverse Genetics Reveals a Variable Infection Gradient in the Respiratory Tract. Cell 2020, 182, 429–446.e14. [Google Scholar] [CrossRef]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef]
- Coutard, B.; Valle, C.; de Lamballerie, X.; Canard, B.; Seidah, N.G.; Decroly, E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020, 176, 104742. [Google Scholar] [CrossRef]
- Jennings, B.C.; Kornfeld, S.; Doray, B. A weak COPI binding motif in the cytoplasmic tail of SARS-CoV-2 spike glycoprotein is necessary for its cleavage, glycosylation, and localization. FEBS Lett. 2021, 595, 1758–1767. [Google Scholar] [CrossRef] [PubMed]
- Mousavizadeh, L.; Ghasemi, S. Genotype and phenotype of COVID-19: Their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2021, 54, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Wolff, G.; Limpens, R.W.A.L.; Zevenhoven-Dobbe, J.C.; Laugks, U.; Zheng, S.; de Jong, A.W.M.; Koning, R.I.; Agard, D.A.; Grünewald, K.; Koster, A.J.; et al. A molecular pore spans the double membrane of the coronavirus replication organelle. Science 2020, 369, 1395–1398. [Google Scholar] [CrossRef]
- Kumar, A.; Prasoon, P.; Kumari, C.; Pareek, V.; Faiq, M.A.; Narayan, R.K.; Kulandhasamy, M.; Kant, K. SARS-CoV-2-specific virulence factors in COVID-19. J. Med. Virol. 2021, 93, 1343–1350. [Google Scholar] [CrossRef] [PubMed]
- Beznoussenko, G.V.; Kweon, H.S.; Sesorova, I.S.; Mironov, A.A. Comparison of the Cisterna Maturation-Progression Model with the Kiss-and-Run Model of Intra-Golgi Transport: Role of Cisternal Pores and Cargo Domains. Int. J. Mol. Sci. 2022, 23, 3590. [Google Scholar] [CrossRef] [PubMed]
- Beznoussenko, G.V.; Bejan, A.I.; Parashuraman, S.; Luini, A.; Kweon, H.-S.; Mironov, A.A. The diffusion model of intra-Golgi transport has limited power. Int. J. Mol. Sci. 2023, 24, 1375. [Google Scholar] [CrossRef]
- Mironov, A.A.; Beznoussenko, G.V. Models of Intracellular Transport: Pros and Cons. Front. Cell Dev. Biol. 2019, 7, 146. [Google Scholar] [CrossRef]
- Sesorova, I.S.; Karelina, N.R.; Kazakova, T.E.; Parashuraman, S.; Zdorikova, M.A.; Dimov, I.D.; Seliverstova, E.V.; Beznoussenko, G.V.; Mironov, A.A. Structure of the enterocyte transcytosis compartments during lipid absorption. Histochem. Cell Biol. 2020, 53, 413–429. [Google Scholar] [CrossRef]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Kim, D.; Lee, J.Y.; Yang, J.S.; Kim, J.W.; Kim, V.N.; Chang, H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181, 914–921.e10. [Google Scholar] [CrossRef]
- Gong, Y.; Qin, S.; Dai, L.; Tian, Z. The glycosylation in SARS-CoV-2 and its receptor ACE2. Signal Transduct. Target Ther. 2021, 6, 396. [Google Scholar] [CrossRef]
- Cheng, Y.-W.; Chao, T.-L.; Li, C.-L.; Chiu, M.-F.; Kao, H.-C.; Wang, S.-H.; Pang, Y.H.; Lin, C.H.; Tsai, Y.M.; Lee, W.H.; et al. Furin Inhibitors Block SARS-CoV-2 Spike Protein Cleavage to Suppress Virus Production and Cytopathic Effects. Cell Rep. 2020, 33, 108254. [Google Scholar] [CrossRef] [PubMed]
- Hopfer, H.; Herzig, M.C.; Gosert, R.; Menter, T.; Hench, J.; Tzankov, A.; Hirsch, H.H.; Miller, S.E. Hunting coronavirus by transmission electron microscopy—A guide to SARS-CoV-2-associated ultrastructural pathology in COVID-19 tissues. Histopathology 2021, 78, 358–370. [Google Scholar] [CrossRef] [PubMed]
- Peacock, T.P.; Goldhill, D.H.; Zhou, J.; Baillon, L.; Frise, R.; Swann, O.C.; Kugathasan, R.; Penn, R.; Brown, J.C.; Sanchez-David, R.Y.; et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021, 6, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Pinto, A.L.; Rai, R.K.; Brown, J.C.; Griffin, P.; Edgar, J.R.; Shah, A.; Singanayagam, A.; Hogg, C.; Barclay, W.S.; Futter, C.E.; et al. Ultrastructural insight into SARS-CoV-2 entry and budding in human airway epithelium. Nat. Commun. 2022, 13, 1609. [Google Scholar] [CrossRef] [PubMed]
- Greaney, A.M.; Raredon, M.S.B.; Kochugaeva, M.P.; Niklason, L.E.; Levchenko, A. SARS-CoV-2 leverages airway epithelial protective mechanism for viral infection. bioRxiv 2022. [Google Scholar] [CrossRef]
- Khailany, R.A.; Safdar, M.; Ozaslan, M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020, 19, 100682. [Google Scholar] [CrossRef]
- Stout, A.E.; André, N.M.; Jaimes, J.A.; Millet, J.K.; Whittaker, G.R. Coronaviruses in cats and other companion animals: Where does SARS-CoV-2/COVID-19 fit? Vet. Microbiol. 2020, 247, 108777. [Google Scholar] [CrossRef]
- Baena, V.; Conrad, R.; Friday, P.; Fitzgerald, E.; Kim, T.; Bernbaum, J.; Berensmann, H.; Harned, A.; Nagashima, K.; Narayan, K. FIB-SEM as a Volume Electron Microscopy Approach to Study Cellular Architectures in SARS-CoV-2 and Other Viral Infections: A Practical Primer for a Virologist. Viruses 2021, 13, 611. [Google Scholar] [CrossRef]
- Watanabe, Y.; Allen, J.D.; Wrapp, D.; McLellan, J.S.; Crispin, M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 2020, 369, 330–333. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed]
- Jaimes, J.A.; André, N.M.; Chappie, J.S.; Millet, J.K.; Whittaker, G.R. Phylogenetic Analysis and Structural Modeling of SARS-CoV- 2 Spike Protein Reveals an Evolutionary Distinct and Proteolytically Sensitive Activation Loop. J. Mol. Biol. 2020, 432, 3309–3325. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.Y.; et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 2020, 181, 894–904. [Google Scholar] [CrossRef]
- Meng, B.; Datir, R.; Choi, J. SARS-CoV-2 spike N-terminal domain modulates TMPRSS2-dependent viral entry and fusogenicity. Cell Rep. 2022, 40, 111220. [Google Scholar] [CrossRef]
- Bizzotto, J.; Sanchis, P.; Abbate, M.; Lage-Vickers, S.; Lavignolle, R.; Toro, A.; Olszevicki, S.; Sabater, A.; Cascardo, F.; Vazquez, E.; et al. SARS-CoV-2 Infection Boosts MX1 Antiviral Effector in COVID-19 Patients. iScience 2020, 23, 101585. [Google Scholar] [CrossRef]
- Seyran, M.; Takayama, K.; Uversky, V.N.; Lundstrom, K.; Palù, G.; Sherchan, S.P.; Attrish, D.; Rezaei, N.; Aljabali, A.A.A.; Ghosh, S.; et al. The structural basis of accelerated host cell entry by SARS-CoV-2†. FEBS J. 2021, 288, 5010–5020. [Google Scholar] [CrossRef]
- Fantini, J.; Di Scala, C.; Chahinian, H.; Yahi, N. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int. J. Antimicrob. Agents 2020, 55, 105960. [Google Scholar] [CrossRef]
- Peng, R.; Wu, L.A.; Wang, Q.; Qi, J.; Gao, G.F. Cell entry by SARS-CoV-2. Trends Biochem. Sci. 2021, 46, 848–860. [Google Scholar] [CrossRef] [PubMed]
- Maiuolo, J.; Mollace, R.; Gliozzi, M.; Musolino, V.; Carresi, C.; Paone, S.; Scicchitano, M.; Macrì, R.; Nucera, S.; Bosco, F.; et al. The Contribution of Endothelial Dysfunction in Systemic Injury Subsequent to SARS-CoV-2 Infection. Int. J. Mol. Sci. 2020, 21, 9309. [Google Scholar] [CrossRef] [PubMed]
- Kočar, E.; Režen, T.; Rozman, D. Cholesterol, lipoproteins, and COVID-19: Basic concepts and clinical applications. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2021, 1866, 158849. [Google Scholar] [CrossRef]
- Henderson, R.; Edwards, R.J.; Mansouri, K.; Janowska, K.; Stalls, V.; Gobeil, S.M.C.; Kopp, M.; Li, D.; Parks, R.; Hsu, A.L.; et al. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat. Struct. Mol. Biol. 2020, 27, 925–933. [Google Scholar] [CrossRef]
- Riggioni, C.; Comberiati, P.; Giovannini, M.; Agache, I.; Akdis, M.; Alves-Correia, M.; Antó, J.M.; Arcolaci, A.; Azkur, A.K.; Azkur, D.; et al. A compendium answering 150 questions on COVID-19 and SARS-CoV-2. Allergy 2020, 75, 2503–2541. [Google Scholar] [CrossRef]
- Panigrahi, S.; Goswami, T.; Ferrari, B.; Antonelli, C.J.; Bazdar, D.A.; Gilmore, H.; Freeman, M.L.; Lederman, M.M.; Sieg, S.F. SARS-CoV-2 Spike Protein Destabilizes Microvascular Homeostasis. Microbiol. Spectr. 2021, 9, e0073521. [Google Scholar] [CrossRef]
- Scialo, F.; Daniele, A.; Amato, F.; Pastore, L.; Matera, M.G.; Cazzola, M.; Castaldo, G.; Bianco, A. ACE2: The Major Cell Entry Receptor for SARS-CoV-2. Lung 2020, 198, 867–877. [Google Scholar] [CrossRef]
- Somberg, N.H.; Wu, W.W.; Medeiros-Silva, J.; Dregni, A.J.; Jo, H.; DeGrado, W.F.; Hong, M. SARS-CoV-2 Envelope Protein Forms Clustered Pentamers in Lipid Bilayers. Biochemistry 2022, 61, 2280–2294. [Google Scholar] [CrossRef]
- Yuan, Z.; Hu, B.; Xiao, H.; Tan, X.; Li, Y.; Tang, K.; Zhang, Y.; Cai, K.; Ding, B. The E3 Ubiquitin Ligase RNF5 Facilitates SARS-CoV-2 Membrane Protein-Mediated Virion Release. mBio 2022, 13, e0316821. [Google Scholar] [CrossRef] [PubMed]
- Malik, Y.A. Properties of Coronavirus and SARS-CoV-2. Malays. J. Pathol. 2020, 42, 3–11. [Google Scholar] [PubMed]
- Iserman, C.; Roden, C.A.; Boerneke, M.A.; Sealfon, R.S.G.; McLaughlin, G.A.; Jungreis, I.; Fritch, E.J.; Hou, Y.J.; Ekena, J.; Weidmann, C.A.; et al. Genomic RNA Elements Drive Phase Separation of the SARS-CoV-2 Nucleocapsid. Mol. Cell 2020, 80, 1078–1091.e1076. [Google Scholar] [CrossRef] [PubMed]
- Long, S.W.; Olsen, R.J.; Christensen, P.A.; Bernard, D.W.; Davis, J.J.; Shukla, M.; Nguyen, M.; Saavedra, M.O.; Yerramilli, P.; Pruitt, L.; et al. Molecular Architecture of Early Dissemination and Massive Second Wave of the SARS-CoV-2 Virus in a Major Metropolitan Area. mBio 2020, 11, e02707-20. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Ye, Q.; Singh, D.; Cao, Y.; Diedrich, J.K.; Yates, J.R., 3rd; Villa, E.; Cleveland, D.W.; Corbett, K.D. The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Nat. Commun. 2021, 12, 502. [Google Scholar] [CrossRef] [PubMed]
- Chau, B.-A.; Chen, V.; Cochrane, A.W.; Parent, L.J.; Mouland, A.J. Liquid- Liquid phase separation of nucleocapsid proteins during SARS-CoV-2 and HIV-1 replication. Cell Rep. 2023, 42, 111968. [Google Scholar] [CrossRef]
- Savastano, A.; Ibáñez de Opakua, A.; Rankovic, M.; Zweckstetter, M. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nat. Commun. 2020, 11, 6041. [Google Scholar] [CrossRef]
- Huang, C.; Tan, Z.; Zhao, K.; Zou, W.; Wang, H.; Gao, H.; Sun, S.; Bu, D.; Chai, W.; Li, Y. The effect of N-glycosylation of SARS-CoV-2 spike protein on the virus interaction with the host cell ACE2 receptor. iScience 2021, 24, 103272. [Google Scholar] [CrossRef]
- Banerjee, A.K.; Blanco, M.R.; Bruce, E.A.; Honson, D.D.; Chen, L.M.; Chow, A.; Bhat, P.; Ollikainen, N.; Quinodoz, S.A.; Loney, C.; et al. SARS-CoV-2 Disrupts Splicing, Translation, and Protein Trafficking to Suppress Host Defenses. Cell 2020, 183, 1325–1339.e21. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.J.; Leong, M.W.; Rustagi, A.; Beck, A.; Zeng, L.; Holmes, S.; Qi, L.S.; Blish, C.A. SARS-CoV-2 escapes direct NK cell killing through Nsp1-mediated downregulation of ligands for NKG2D. Cell Rep. 2022, 41, 111892. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Choi, J.H.; Dai, D.L.; Luo, J.; Ladak, R.J.; Li, Q.; Wang, Y.; Zhang, C.; Wiebe, S.; Liu, A.C.H.; et al. SARS-CoV-2 impairs interferon production via NSP2-induced repression of mRNA translation. Proc. Natl. Acad. Sci. USA 2022, 119, e2204539119. [Google Scholar] [CrossRef] [PubMed]
- Lavigne, M.; Helynck, O.; Rigolet, P.; Boudria-Souilah, R.; Nowakowski, M.; Baron, B.; Brülé, S.; Hoos, S.; Raynal, B.; Guittat, L.; et al. SARS-CoV-2 Nsp3 unique domain SUD interacts with guanine quadruplexes and G4-ligands inhibit this interaction. Nucleic Acids Res. 2021, 49, 7695–7712. [Google Scholar] [CrossRef] [PubMed]
- Russo, L.C.; Tomasin, R.; Matos, I.A.; Manucci, A.C.; Sowa, S.T.; Dale, K.; Caldecott, K.W.; Lehtiö, L.; Schechtman, D.; Meotti, F.C.; et al. The SARS-CoV-2 Nsp3 macrodomain reverses PARP9/DTX3L-dependent ADP-ribosylation induced by interferon signaling. J. Biol. Chem. 2021, 297, 101041. [Google Scholar] [CrossRef] [PubMed]
- Faizan, M.I.; Chaudhuri, R.; Sagar, S.; Albogami, S.; Chaudhary, N.; Azmi, I.; Akhtar, A.; Ali, S.M.; Kumar, R.; Iqbal, J.; et al. NSP4 and ORF9b of SARS-CoV-2 Induce Pro-Inflammatory Mitochondrial DNA Release in Inner Membrane-Derived Vesicles. Cells 2022, 11, 2969. [Google Scholar] [CrossRef] [PubMed]
- Moustaqil, M.; Ollivier, E.; Chiu, H.P.; Van Tol, S.; Rudolffi-Soto, P.; Stevens, C.; Bhumkar, A.; Hunter, D.J.B.; Freiberg, A.N.; Jacques, D.; et al. SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): Implications for disease presentation across species. Emerg. Microbes Infect. 2021, 10, 178–195. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Deng, J.; Han, L.; Zhuang, M.W.; Xu, Y.; Zhang, J.; Nan, M.-L.; Xiao, Y.; Zhan, P.; Liu, X.; et al. SARS-CoV-2 NSP5 and N protein counteract the RIG-I signaling pathway by suppressing the formation of stress granules. Signal Transduct. Target. Ther. 2022, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Qiao, J.; You, Q.; Zong, S.; Peng, Q.; Liu, Y.; Hu, S.; Liu, W.; Li, S.; Shu, X.; et al. Front SARS-CoV-2 Nsp5 Activates NF-κB Pathway by Upregulating SUMOylation of MAVS. Front. Immunol. 2021, 12, 750969. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, Z.; Guo, J.; Xu, S.; Zhou, J.; Chen, Q.; Tong, X.; Wang, D.; Peng, G.; Fang, L.; et al. SARS-CoV-2 nsp5 Exhibits Stronger Catalytic Activity and Interferon Antagonism than Its SARS-CoV Ortholog. J. Virol. 2022, 96, e0003722. [Google Scholar] [CrossRef]
- Ricciardi, S.; Guarino, A.M.; Giaquinto, L.; Polishchuk, E.V.; Santoro, M.; Di Tullio, G.; Wilson, C.; Panariello, F.; Soares, V.C.; Dias, S.S.G.; et al. The role of NSP6 in the biogenesis of the SARS-CoV-2 replication organelle. Nature 2022, 606, 761–768. [Google Scholar] [CrossRef]
- Peng, Q.; Peng, R.; Yuan, B.; Zhao, J.; Wang, M.; Wang, X.; Wang, Q.; Sun, Y.; Fan, Z.; Qi, J.; et al. Structural and Biochemical Characterization of the nsp12-nsp7-nsp8 Core Polymerase Complex from SARS-CoV-2. Cell Rep. 2020, 31, 107774. [Google Scholar] [CrossRef]
- Lin, S.; Chen, H.; Chen, Z.; Yang, F.; Ye, F.; Zheng, Y.; Yang, J.; Lin, X.; Sun, H.; Wang, L.; et al. Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-ExoN domain reveals an exoribonuclease with both structural and functional integrity. Nucleic Acids Res. 2021, 49, 5382–5392. [Google Scholar] [CrossRef]
- Gadhave, K.; Kumar, P.; Kumar, A.; Bhardwaj, T.; Garg, N.; Giri, R. Conformational dynamics of 13 amino acids long NSP11 of SARS-CoV-2 under membrane mimetics and different solvent conditions. Microb. Pathog. 2021, 158, 105041. [Google Scholar] [CrossRef]
- Wang, W.; Zhou, Z.; Xiao, X.; Tian, Z.; Dong, X.; Wang, C.; Li, L.; Ren, L.; Lei, X.; Xiang, Z.; et al. SARS-CoV-2 nsp12 attenuates type I interferon production by inhibiting IRF3 nuclear translocation. Cell Mol. Immunol. 2021, 18, 945–953. [Google Scholar] [CrossRef]
- Cao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, L.; et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020, 368, 779–782. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; West, A.M.V.; Silletti, S.; Corbett, K.D. Architecture and self-assembly of the SARS-CoV-2 nucleocapsid protein. Protein Sci. 2020, 29, 1890–1901. [Google Scholar] [CrossRef] [PubMed]
- Newman, J.A.; Douangamath, A.; Yadzani, S.; Yosaatmadja, Y.; Aimon, A.; Brandão-Neto, J.; Dunnett, L.; Gorrie-Stone, T.; Skyner, R.; Fearon, D.; et al. Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase. Nat. Commun. 2021, 12, 4848. [Google Scholar] [CrossRef] [PubMed]
- Yuen, C.K.; Lam, J.Y.; Wong, W.M.; Mak, L.F.; Wang, X.; Chu, H.; Cai, J.-P.; Jin, D.-Y.; To, K.K.-W.; Chan, J.F.-W.; et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microbes Infect. 2020, 9, 1418–1428. [Google Scholar] [CrossRef]
- Pillon, M.C.; Frazier, M.N.; Dillard, L.B.; Williams, J.G.; Kocaman, S.; Krahn, J.M.; Perera, L.; Hayne, C.K.; Gordon, J.; Stewart, Z.D.; et al. Cryo-EM structures of the SARS-CoV-2 endoribonuclease Nsp15 reveal insight into nuclease specificity and dynamics. Nat. Commun. 2021, 12, 636. [Google Scholar] [CrossRef]
- Sinha, S.; Sehgal, A.; Sehgal, R. Association of ACE2 receptor and ACEIs/ARBs with disease severity in COVID-19. Drug Discov. Ther. 2020, 14, 161–170. [Google Scholar] [CrossRef]
- Chandra, A.; Gurjar, V.; Qamar, I.; Singh, N. Identification of potential inhibitors of SARS-CoV-2 endoribonuclease (EndoU) from FDA approved drugs: A drug repurposing approach to find therapeutics for COVID-19. J. Biomol. Struct. Dyn. 2020, 39, 4201–4211. [Google Scholar] [CrossRef]
- Russ, A.; Wittmann, S.; Tsukamoto, Y.; Herrmann, A.; Deutschmann, J.; Lagisquet, J.; Ensser, A.; Kato, H.; Gramberg, T. Nsp16 shields SARS-CoV-2 from efficient MDA5 sensing and IFIT1-mediated restriction. EMBO Rep. 2022, 23, e55648. [Google Scholar] [CrossRef]
- Travaglini, K.L.; Nabhan, A.N.; Penland, L.; Sinha, R.; Gillich, A.; Sit, R.V.; Chang, S.; Conley, S.D.; Mori, Y.; Seita, J.; et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 2020, 587, 619–625. [Google Scholar] [CrossRef]
- Clausen, T.M.; Sandoval, D.R.; Spliid, C.B.; Pihl, J.; Perrett, H.R.; Painter, C.D.; Narayanan, A.; Majowicz, S.A.; Kwong, E.M.; McVicar, R.N.; et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell 2020, 183, 1043–1057.e15. [Google Scholar] [CrossRef]
- Kopf, M.; Schneider, C.; Nobs, S.P. The development and function of lung-resident macrophages and dendritic cells. Nat. Immunol. 2015, 16, 36–44. [Google Scholar] [CrossRef]
- Barlow, J.L.; McKenzie, A.N.J. Innate Lymphoid Cells of the Lung. Annu. Rev. Physiol. 2019, 81, 429–452. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; He, X.; Ma, L.; Zhuang, Z.; Wang, Y.; Lin, M.; Cai, S.; Wei, L.; Wang, Z.; Zhao, Z.; et al. Suppression of ACE2 SUMOylation protects against SARS-CoV-2 infection through TOLLIP-mediated selective autophagy. Nat. Commun. 2022, 13, 5204. [Google Scholar] [CrossRef] [PubMed]
- Heinl, E.S.; Lorenz, S.; Schmidt, B.; Laqtom, N.N.M.; Mazzulli, J.R.; Francelle, L.; Yu, T.W.; Greenberg, B.; Storch, S.; Tegtmeier, I.; et al. CLN7/MFSD8 may be an important factor for SARS-CoV-2 cell entry. iScience 2022, 25, 105082. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045.e9. [Google Scholar] [CrossRef]
- Batah, S.S.; Fabro, A.T. Pulmonary pathology of ARDS in COVID-19: A pathological review for clinicians. Respir. Med. 2021, 176, 106239. [Google Scholar] [CrossRef]
- Guo, X.; Cao, J.; Cai, J.P.; Wu, J.; Huang, J.; Asthana, P.; Wong, S.K.K.; Ye, Z.-W.; Gurung, S.; Zhang, Y.; et al. Control of SARS-CoV-2 infection by MT1-MMP-mediated shedding of ACE2. Nat. Commun. 2022, 13, 7907. [Google Scholar] [CrossRef]
- Camargo, S.M.R.; Vuille-Dit-Bille, R.N.; Meier, C.F.; Verrey, F. ACE2 and gut amino acid transport. Clin. Sci. 2020, 134, 2823–2833. [Google Scholar] [CrossRef] [PubMed]
- Evans, P.C.; Rainger, G.E.; Mason, J.C.; Guzik, T.J.; Osto, E.; Stamataki, Z.; Neil, D.; Hoefer, I.E.; Fragiadaki, M.; Waltenberger, J.; et al. Endothelial dysfunction in COVID-19: A position paper of the ESC Working Group for Atherosclerosis and Vascular Biology, and the ESC Council of Basic Cardiovascular Science. Cardiovasc. Res. 2020, 116, 2177–2184. [Google Scholar] [CrossRef] [PubMed]
- Gheblawi, M.; Wang, K.; Viveiros, A.; Nguyen, Q.; Zhong, J.C.; Turner, A.J.; Raizada, M.K.; Grant, M.B.; Oudit, G.Y. Angiotensin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System: Celebrating the 20th Anniversary of the Discovery of ACE2. Circ. Res. 2020, 126, 1456–1474. [Google Scholar] [CrossRef] [PubMed]
- Donoghue, M.; Hsieh, F.; Baronas, E.; Godbout, K.; Gosselin, M.; Stagliano, N.; Donovan, M.; Woolf, B.; Robison, K.; Jeyaseelan, R.; et al. A novel angiotensin-converting enzyme- related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ. Res. 2000, 87, E1–E9. [Google Scholar] [CrossRef]
- Wu, C.T.; Lidsky, P.V.; Xiao, Y.; Cheng, R.; Lee, I.T.; Nakayama, T.; Jiang, S.; He, W.; Demeter, J.; Knight, M.G.; et al. SARS-CoV-2 replication in airway epithelia requires motile cilia and microvillar reprogramming. Cell 2022, 186, 112–130.e20. [Google Scholar] [CrossRef]
- Bezzerri, V.; Gentili, V.; Api, M.; Finotti, A.; Papi, C.; Tamanini, A.; Boni, C.; Baldisseri, E.; Olioso, D.; Duca, M.; et al. SARS-CoV-2 viral entry and replication is impaired in Cystic Fibrosis airways due to ACE2 downregulation. Nat. Commun. 2023, 14, 132. [Google Scholar] [CrossRef] [PubMed]
- Kuba, K.; Imai, Y.; Penninger, J.M. Multiple functions of angiotensin- converting enzyme 2 and its relevance in cardiovascular diseases. Circ. J. 2013, 77, 301–308. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef]
- Tipnis, S.R.; Hooper, N.M.; Hyde, R.; Karran, E.; Christie, G.; Turner, A.J. A human homolog of angiotensin-converting enzyme: Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000, 275, 33238–33243. [Google Scholar] [CrossRef]
- Matuck, B.F.; Dolhnikoff, M.; Duarte-Neto, A.N.; Maia, G.; Gomes, S.C.; Sendyk, D.I.; Zarpellon, A.; Andrade, N.P.; Monteiro, R.A.; Pinho, J.R.R.; et al. Salivary glands are a target for SARS-CoV-2: A source for saliva contamination. J. Pathol. 2021, 254, 239–243. [Google Scholar] [CrossRef]
- Brann, D.H.; Tsukahara, T.; Weinreb, C.; Lipovsek, M.; Van den Berge, K.; Gong, B.; Chance, R.; Macaulay, I.C.; Chou, H.-J.; Fletcher, R.B.; et al. Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia. Sci. Adv. 2020, 6, eabc5801. [Google Scholar] [CrossRef]
- Fodoulian, L.; Tuberosa, J.; Rossier, D.; Boillat, M.; Kan, C.; Pauli, V.; Egervari, K.; Lobrinus, J.A.; Landis, B.N.; Carleton, A.; et al. SARS-CoV-2 receptors and entry genes are expressed in the human olfactory neuroepithelium and brain. iScience 2020, 23, 101839. [Google Scholar] [CrossRef] [PubMed]
- Beyerstedt, S.; Casaro, E.B.; Rangel, É.B. COVID-19: Angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 905–919. [Google Scholar] [CrossRef] [PubMed]
- de Melo, G.D.; Lazarini, F.; Levallois, S.; Hautefort, C.; Michel, V.; Larrous, F.; Verillaud, B.; Aparicio, C.; Wagner, S.; Gheusi, G.; et al. COVID-19-related anosmia is associated with viral persistence and inflammation in human olfactory epithelium and brain infection in hamsters. Sci. Transl. Med. 2021, 13, eabf8396. [Google Scholar] [CrossRef] [PubMed]
- Lamers, M.M.; Beumer, J.; van der Vaart, J.; Knoops, K.; Puschhof, J.; Breugem, T.I.; Ravelli, R.B.G.; van Schayck, J.P.; Mykytyn, A.Z.; Duimel, H.Q.; et al. SARS-CoV-2 productively infects human gut enterocytes. Science 2020, 369, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Fontela, C.; Dowling, W.E.; Funnell, S.G.P.; Gsell, P.-G.; Riveros-Balta, A.X. Animal models for COVID-19. Nature 2020, 586, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Sesorova, I.S.; Dimov, I.D.; Kashin, A.D.; Sesorov, V.V.; Karelina, N.R.; Zdorikova, M.A.; Beznoussenko, G.V.; Mironov, A.A. Cellular and sub-cellular mechanisms of lipid transport from gut to lymph. Tissue Cell 2021, 72, 101529. [Google Scholar] [CrossRef]
- Guney, C.; Akar, F. Epithelial and Endothelial Expressions of ACE2: SARS-CoV-2 Entry Routes. J. Pharm. Pharm. Sci. 2021, 24, 84–93. [Google Scholar] [CrossRef]
- Han, Y.; Duan, X.; Yang, L.; Nilsson-Payant, B.E.; Wang, P.; Duan, F.; Tang, X.; Yaron, T.M.; Zhang, T.; Uhl, S.; et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 2021, 589, 270–275. [Google Scholar] [CrossRef]
- Ebisudani, T.; Sugimoto, S.; Haga, K.; Mitsuishi, A.; Takai-Todaka, R.; Fujii, M.; Toshimitsu, K.; Hamamoto, J.; Sugihara, K.; Hishida, T.; et al. Direct derivation of human alveolospheres for SARS-CoV-2 infection modeling and drug screening. Cell Rep. 2021, 35, 109218. [Google Scholar] [CrossRef]
- Khan, M.; Yoo, S.J.; Clijsters, M.; Backaert, W.; Vanstapel, A.; Speleman, K.; Lietaer, C.; Choi, S.; Hether, T.D.; Marcelis, L.; et al. Visualizing in deceased COVID-19 patients how SARS-CoV-2 attacks the respiratory and olfactory mucosae but spares the olfactory bulb. Cell 2021, 184, 5932–5949.e15. [Google Scholar] [CrossRef]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef] [PubMed]
- Meinhardt, J.; Radke, J.; Dittmayer, C.; Franz, J.; Thomas, C.; Mothes, R.; Laue, M.; Schneider, J.; Brünink, S.; Greuel, S.; et al. Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat. Neurosci. 2021, 24, 168–175. [Google Scholar] [CrossRef]
- Beckman, D.; Bonillas, A.; Diniz, G.B.; Ott, S.; Roh, J.W.; Elizaldi, S.R.; Schmidt, B.A.; Sammak, R.L.; Van Rompay, K.K.; Iyer, S.S.; et al. SARS-CoV-2 infects neurons and induces neuroinflammation in a non-human primate model of COVID-19. Cell Rep. 2022, 41, 111573. [Google Scholar] [CrossRef] [PubMed]
- Stein, S.R.; Ramelli, S.C.; Grazioli, A.; Chung, J.Y.; Singh, M.; Yinda, C.K.; Winkler, C.W.; Sun, J.; Dickey, J.M.; Ylaya, K.; et al. SARS-CoV-2 infection and persistence in the human body and brain at autopsy. Nature 2022, 612, 758–763. [Google Scholar] [CrossRef]
- Theoharides, T.C. Could SARS-CoV-2 Spike Protein Be Responsible for Long-COVID Syndrome? Mol. Neurobiol. 2022, 5, 1850–1861. [Google Scholar] [CrossRef]
- Sungnak, W.; Huang, N.; Bécavin, C.; Berg, M.; Queen, R.; Litvinukova, M.; Talavera-Lopez, C.; Maatz, H.; Reichart, D.; Sampaziotis, F.; et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020, 26, 681–687. [Google Scholar] [CrossRef]
- Lee, I.T.; Nakayama, T.; Wu, C.T.; Goltsev, Y.; Jiang, S.; Gall, P.A.; Liao, C.-K.; Shih, L.-C.; Schürch, C.M.; McIlwain, D.R.; et al. ACE2 localizes to the respiratory cilia and is not increased by ACE inhibitors or ARBs. Nat. Commun. 2020, 11, 5453. [Google Scholar] [CrossRef]
- Bunyavanich, S.; Do, A.; Vicencio, A. Nasal gene expression of angiotensin-converting enzyme 2 in children and adults. JAMA 2020, 323, 2427. [Google Scholar] [CrossRef]
- Schneider, J.L.; Rowe, J.H.; Garcia-de-Alba, C.; Kim, C.F.; Sharpe, A.H.; Haigis, M.C. The aging lung: Physiology, disease, and immunity. Cell 2021, 184, 1990–2019. [Google Scholar] [CrossRef]
- Lima, R.S.; Rocha, L.P.C.; Moreira, P.R. Genetic and epigenetic control of ACE2 expression and its possible role in COVID-19. Cell Biochem. Funct. 2021, 39, 713–726. [Google Scholar] [CrossRef]
- Sarver, D.C.; Wong, G.W. Obesity alters Ace2 and Tmprss2 expression in lung, trachea, and esophagus in a sex-dependent manner: Implications for COVID-19. Biochem. Biophys. Res. Commun. 2021, 538, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Kearns, F.L.; Sandoval, D.R.; Casalino, L.; Clausen, T.M.; Rosenfeld, M.A.; Spliid, C.B.; Amaro, R.E.; Esko, J.D. Spike-heparan sulfate interactions in SARS-CoV-2 infection. Curr. Opin. Struct. Biol. 2022, 76, 102439. [Google Scholar] [CrossRef] [PubMed]
- Bestle, D.; Heindl, M.R.; Limburg, H.; Van Lam van, T.; Pilgram, O.; Moulton, H.; Stein, D.A.; Hardes, K.; Eickmann, M.; Dolnik, O.; et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance 2020, 3, e202000786. [Google Scholar] [CrossRef] [PubMed]
- Mykytyn, A.Z.; Breugem, T.I.; Riesebosch, S.; Schipper, D.; van den Doel, P.B.; Rottier, R.J.; Lamers, M.M.; Haagmans, B.L. SARS-CoV-2 entry into human airway organoids is serine protease-mediated and facilitated by the multibasic cleavage site. Elife 2021, 10, e64508. [Google Scholar] [CrossRef]
- Tang, T.; Jaimes, J.A.; Bidon, M.K.; Straus, M.R.; Daniel, S.; Whittaker, G.R. Proteolytic Activation of SARS-CoV-2 Spike at the S1/S2 Boundary: Potential Role of Proteases beyond Furin. ACS Infect. Dis. 2021, 7, 264–272. [Google Scholar] [CrossRef]
- Jaimes, J.A.; Millet, J.K.; Whittaker, G.R. Proteolytic Cleavage of the SARS-CoV-2 Spike Protein and the Role of the Novel S1/S2 Site. iScience 2020, 23, 101212. [Google Scholar] [CrossRef]
- Puelles, V.G.; Lütgehetmann, M.; Lindenmeyer, M.T.; Sperhake, J.P.; Wong, M.N.; Allweiss, L.; Chilla, S.; Heinemann, A.; Wanner, N.; Liu, S.; et al. Multiorgan and Renal Tropism of SARS-CoV-2. N. Engl. J. Med. 2020, 383, 590–592. [Google Scholar] [CrossRef]
- Mendonça, L.; Howe, A.; Gilchrist, J.B.; Sheng, Y.; Sun, D.; Knight, M.L.; Zanetti-Domingues, L.C.; Bateman, B.; Krebs, A.-S.; Chen, L.; et al. Correlative multi-scale cryo-imaging unveils SARS-CoV-2 assembly and egress. Nat. Commun. 2021, 12, 4629. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Wang, S.; Wu, Y.; Hou, W.; Yuan, L.; Shen, C.; Wang, J.; Ye, J.; Zheng, Q.; Ma, J.; et al. Virus-Free and Live-Cell Visualizing SARS-CoV-2 Cell Entry for Studies of Neutralizing Antibodies and Compound Inhibitors. Small Methods 2021, 5, 2001031. [Google Scholar] [CrossRef]
- Saxena, S.K.; Kumar, S.; Ansari, S.; Paweska, J.T.; Maurya, V.K.; Tripathi, A.K.; Abdel-Moneim, A.S. Characterization of the novel SARS-CoV-2 Omicron (B.1.1.529) variant of concern and its global perspective. J. Med. Virol. 2022, 94, 1738–1744. [Google Scholar] [CrossRef]
- Bayati, A.; Kumar, R.; Francis, V.; McPherson, P.S. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. J. Biol. Chem. 2021, 296, 100306. [Google Scholar] [CrossRef] [PubMed]
- Benton, D.J.; Wrobel, A.G.; Xu, P.; Roustan, C.; Martin, S.R.; Rosenthal, P.B.; Skehel, J.J.; Gamblin, S.J. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature 2020, 588, 327–330. [Google Scholar] [CrossRef] [PubMed]
- Baristaite, G.; Gurwitz, D. d-Galactose treatment increases ACE2, TMPRSS2, and FURIN and reduces SERPINA1 mRNA expression in A549 human lung epithelial cells. Drug. Dev. Res. 2022, 83, 622–627. [Google Scholar] [CrossRef] [PubMed]
- Menachery, V.D.; Dinnon, K.H., 3rd; Yount, B.L., Jr.; McAnarney, E.T.; Gralinski, L.E.; Hale, A.; Graham, R.L.; Scobey, T.; Anthony, S.J.; Wang, L.; et al. Trypsin Treatment Unlocks Barrier for Zoonotic Bat Coronavirus Infection. J. Virol. 2020, 94, e01774-19. [Google Scholar] [CrossRef] [PubMed]
- Atik, N.; Wirawan, F.; Amalia, R.; Khairani, A.F.; Pradini, G.W. Differences in endosomal Rab gene expression between positive and negative COVID-19 patients. BMC Res. Notes 2022, 15, 252. [Google Scholar] [CrossRef]
- Szlachcic, W.J.; Dabrowska, A.; Milewska, A.; Ziojla, N.; Blaszczyk, K.; Barreto-Duran, E.; Sanak, M.; Surmiak, M.; Owczarek, K.; Grzanka, D.; et al. SARS-CoV-2 infects an in vitro model of the human developing pancreas through endocytosis. iScience 2022, 25, 104594. [Google Scholar] [CrossRef]
- Lalioti, V.; González-Sanz, S.; Lois-Bermejo, I.; González-Jiménez, P.; Viedma-Poyatos, Á.; Merino, A.; Pajares, M.A.; Pérez-Sala, D. Cell surface detection of vimentin, ACE2 and SARS-CoV-2 Spike proteins reveals selective colocalization at primary cilia. Sci. Rep. 2022, 12, 7063. [Google Scholar] [CrossRef]
- Pak, A.J.; Yu, A.; Ke, Z.; Briggs, J.A.G.; Voth, G.A. Cooperative multivalent receptor binding promotes exposure of the SARS-CoV-2 fusion machinery core. Nat. Commun. 2022, 13, 1002. [Google Scholar] [CrossRef]
- Tien, C.F.; Tsai, W.T.; Chen, C.H.; Chou, H.J.; Zhang, M.M.; Lin, J.J.; Lin, E.-J.; Dai, S.-S.; Ping, Y.-H.; Yu, C.-Y.; et al. Glycosylation and S-palmitoylation regulate SARS-CoV-2 spike protein intracellular trafficking. iScience 2022, 25, 104709. [Google Scholar] [CrossRef]
- Meng, B.; Abdullahi, A.; Ferreira, I.A.T.M.; Goonawardane, N.; Saito, A.; Kimura, I.; Yamasoba, D.; Gerber, P.P.; Fatihi, S.; Rathore, S.; et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 2022, 603, 706–714. [Google Scholar] [CrossRef]
- Belouzard, S.; Millet, J.K.; Licitra, B.N.; Whittaker, G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 2012, 4, 1011–1033. [Google Scholar] [CrossRef] [PubMed]
- Bouhaddou, M.; Memon, D.; Meyer, B.; White, K.M.; Rezelj, V.V.; Correa Marrero, M.; Polacco, B.J.; Melnyk, J.E.; Ulferts, S.; Kaake, R.M.; et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell 2020, 182, 685–712.e19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kennedy, A.; Xing, L.; Bui, S.; Reid, W.; Joppich, J.; Ahat, E.; Rose, M.; Tang, Q.; Tai, A.W.; et al. SARS-CoV-2 triggers Golgi fragmentation via down-regulation of GRASP55 to facilitate viral trafficking. bioRxiv 2022. [Google Scholar] [CrossRef]
- Sun, L.; Li, P.; Ju, X.; Rao, J.; Huang, W.; Ren, L.; Zhang, S.; Xiong, T.; Xu, K.; Zhou, X.; et al. In vivo structural characterization of the SARS-CoV-2 RNA genome identifies host proteins vulnerable to repurposed drugs. Cell 2021, 184, 1865–1883.e20. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.G.; Jureka, A.S.; Silvas, J.A.; Nicolini, A.M.; Chvatal, S.A.; Carlson-Stevermer, J.; Oki, J.; Holden, K.; Basler, C.F. Inhibitors of VPS34 and fatty-acid metabolism suppress SARS-CoV-2 replication. Cell Rep. 2021, 36, 109479. [Google Scholar] [CrossRef]
- Yang, Y.; Zheng, L.; Zheng, X.; Ge, L. Autophagosomal Membrane Origin and Formation. In Advances in Experimental Medicine and Biology; Springer: Singapore, 2021; Volume 1208, pp. 17–42. [Google Scholar] [CrossRef]
- Yang, S.L.; DeFalco, L.; Anderson, D.E.; Zhang, Y.; Aw, J.G.A.; Lim, S.Y.; Lim, X.N.; Tan, K.Y.; Zhang, T.; Chawla, T.; et al. Comprehensive mapping of SARS-CoV-2 interactions in vivo reveals functional virus-host interactions. Nat. Commun. 2021, 12, 5113. [Google Scholar] [CrossRef]
- Miao, G.; Zhao, H.; Li, Y.; Ji, M.; Chen, Y.; Shi, Y.; Bi, Y.; Wang, P.; Zhang, H. ORF3a of the COVID-19 virus SARS-CoV-2 blocks HOPS complex-mediated assembly of the SNARE complex required for autolysosome formation. Dev. Cell 2021, 56, 427–442.e5. [Google Scholar] [CrossRef] [PubMed]
- Denisova, G.N.; Dimov, I.D.; Zaitseva, A.V.; Artiux, L.J.; Mironov, A.A.; Karelina, N.R. Overloading of differentiated Caco-2 cells during lipid transcytosis induces glycosylation mistakes in the Golgi complex. Biocell 2021, 45, 773–783. [Google Scholar] [CrossRef]
- Mironov, A.A.; Dimov, I.D.; Beznoussenko, G.V. Role of Intracellular Transport in the Centriole-Dependent Formation of Golgi Ribbon. In Results and Problems in Cell Differentiation; Springer: Cham, Switzerland, 2019; Volume 67, pp. 49–79. [Google Scholar] [CrossRef]
- Mironov, A.A.; Sesorova, I.S.; Dimov, I.D.; Karelina, N.R.; Beznoussenko, G.V. Intracellular transports and atherogenesis. Front. Biosci. 2020, 25, 1230–1258. [Google Scholar] [CrossRef]
- Tojo, K.; Yamamoto, N.; Tamada, N.; Mihara, T.; Abe, M.; Nishii, M.; Takeuchi, I.; Goto, T. Early alveolar epithelial cell necrosis is a potential driver of COVID-19-induced acute respiratory distress syndrome. iScience 2023, 26, 105748. [Google Scholar] [CrossRef]
- Mihelc, E.M.; Baker, S.C.; Lanman, J.K. Coronavirus infection induces progressive restructuring of the endoplasmic reticulum involving the formation and degradation of double membrane vesicles. Virology 2021, 556, 9–22. [Google Scholar] [CrossRef]
- Prydz, K.; Saraste, J. The life cycle and enigmatic egress of coronaviruses. Mol. Microbiol. 2022, 117, 1308–1316. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Inoue, T.; Inoue, M.; Murae, M.; Fukasawa, M.; Kaneko, M.K.; Kato, Y.; Noguchi, K. SARS-CoV-2 Spike Protein Mutation at Cysteine-488 Impairs Its Golgi Localization and Intracellular S1/S2 Processing. Int. J. Mol. Sci. 2022, 23, 15834. [Google Scholar] [CrossRef] [PubMed]
- Snijder, E.J.; Limpens, R.W.A.L.; de Wilde, A.H.; de Jong, A.W.M.; Zevenhoven-Dobbe, J.C.; Maier, H.J.; Faas, F.F.G.A.; Koster, A.J.; Bárcena, M. A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS Biol. 2020, 18, e3000715. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Riva, L.; Pu, Y.; Martin-Sancho, L.; Kanamune, J.; Yamamoto, Y.; Sakai, K.; Gotoh, S.; Miorin, L.; De Jesus, P.D.; et al. MDA5 Governs the Innate Immune Response to SARS-CoV-2 in Lung Epithelial Cells. Cell Rep. 2021, 34, 108628. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Cao, Z.; Xie, X.; Zhang, X.; Chen, J.Y.; Wang, H.; Menachery, V.D.; Rajsbaum, R.; Shi, P.-Y. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 2020, 33, 108234. [Google Scholar] [CrossRef]
- Martin-Sancho, L.; Lewinski, M.K.; Pache, L.; Stoneham, C.A.; Yin, X.; Becker, M.E.; Pratt, D.; Churas, C.; Rosenthal, S.B.; Liu, S.; et al. Functional landscape of SARS-CoV-2 cellular restriction. Mol. Cell 2021, 81, 2656–2668.e8. [Google Scholar] [CrossRef]
- Fong, C.H.; Lu, L.; Chen, L.L.; Yeung, M.L.; Zhang, A.J.; Zhao, H.; Yuen, K.-Y.; To, K.K.-W. Interferon-gamma inhibits influenza A virus cellular attachment by reducing sialic acid cluster size. iScience 2022, 25, 104037. [Google Scholar] [CrossRef]
- Klann, K.; Bojkova, D.; Tascher, G.; Ciesek, S.; Münch, C.; Cinatl, J. Growth Factor Receptor Signaling Inhibition Prevents SARS-CoV-2 Replication. Mol. Cell. 2020, 80, 164–174.e4. [Google Scholar] [CrossRef]
- Baulcombe, D. Diced defence. Nature 2001, 409, 295–296. [Google Scholar] [CrossRef]
- Chu, C.Y.; Rana, T.M. Small RNAs: Regulators and guardians of the genome. J. Cell. Physiol. 2007, 213, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Thoms, M.; Buschauer, R.; Ameismeier, M.; Koepke, L.; Denk, T.; Hirschenberger, M.; Kratzat, H.; Hayn, M.; Mackens-Kiani, T.; Cheng, J.; et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 2020, 369, 1249–1255. [Google Scholar] [CrossRef] [PubMed]
- Sa Ribero, M.; Jouvenet, N.; Dreux, M.; Nisole, S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020, 16, e1008737. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Dong, X.; Ma, R.; Wang, W.; Xiao, X.; Tian, Z.; Wang, C.; Wang, Y.; Li, L.; Ren, L.; et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020, 11, 3810. [Google Scholar] [CrossRef]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef]
- Park, A.; Iwasaki, A. Type I and Type III Interferons—Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell Host Microbe 2020, 27, 870–878. [Google Scholar] [CrossRef]
- Xia, H.; Shi, P.Y. Antagonism of Type I Interferon by Severe Acute Respiratory Syndrome Coronavirus 2. J. Interferon Cytokine Res. 2020, 40, 543–548. [Google Scholar] [CrossRef]
- Shvarev, D.; Schoppe, J.; König, C.; Perz, A.; Füllbrunn, N.; Kiontke, S.; Langemeyer, L.; Januliene, D.; Schnelle, K.; Kümmel, D.; et al. Structure of the HOPS tethering complex, a lysosomal membrane fusion machinery. Elife 2022, 11, e80901. [Google Scholar] [CrossRef]
- Miller, K.; McGrath, M.E.; Hu, Z.; Ariannejad, S.; Weston, S.; Frieman, M.; Jackson, W.T. Coronavirus interactions with the cellular autophagy machinery. Autophagy 2020, 16, 2131–2139. [Google Scholar] [CrossRef]
- Gorshkov, K.; Chen, C.Z.; Bostwick, R.; Rasmussen, L.; Tran, B.N.; Cheng, Y.S.; Xu, M.; Pradhan, M.; Henderson, M.; Zhu, W.; et al. The SARS-CoV-2 Cytopathic Effect Is Blocked by Lysosome Alkalizing Small Molecules. ACS Infect. Dis. 2021, 7, 1389–1408. [Google Scholar] [CrossRef]
- van der Hoeven, B.; Oudshoorn, D.; Koster, A.J.; Snijder, E.J.; Kikkert, M.; Bárcena, M. Biogenesis and architecture of arterivirus replication organelles. Virus Res. 2016, 220, 70–90. [Google Scholar] [CrossRef] [PubMed]
- Littler, D.R.; Gully, B.S.; Colson, R.N.; Rossjohn, J. Crystal Structure of the SARS-CoV-2 Non-structural Protein 9, Nsp9. iScience 2020, 23, 101258. [Google Scholar] [CrossRef] [PubMed]
- Snijder, E.J.; Decroly, E.; Ziebuhr, J. The Nonstructural Proteins Directing Coronavirus RNA Synthesis and Processing. Adv. Virus Res. 2016, 96, 59–126. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef]
- Bárcena, M.; Oostergetel, G.T.; Bartelink, W.; Faas, F.G.; Verkleij, A.; Rottier, P.J.; Koster, A.J.; Bosch, B.J. Cryo-electron tomography of mouse hepatitis virus: Insights into the structure of the coronavirion. Proc. Natl. Acad. Sci. USA 2009, 106, 582–587. [Google Scholar] [CrossRef]
- Cottam, E.M.; Maier, H.J.; Manifava, M.; Vaux, L.C.; Chandra-Schoenfelder, P.; Gerner, W.; Britton, P.; Ktistakis, N.T.; Wileman, T. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy 2011, 7, 1335–1347. [Google Scholar] [CrossRef]
- Saraste, J.; Prydz, K. Assembly and Cellular Exit of Coronaviruses: Hijacking an Unconventional Secretory Pathway from the Pre-Golgi Intermediate Compartment via the Golgi Ribbon to the Extracellular Space. Cells 2021, 10, 503. [Google Scholar] [CrossRef]
- Romero-Brey, I.; Merz, A.; Chiramel, A.; Lee, J.Y.; Chlanda, P.; Haselman, U.; Santarella-Mellwig, R.; Habermann, A.; Hoppe, S.; Kallis, S.; et al. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 2012, 8, e1003056. [Google Scholar] [CrossRef]
- Romero-Brey, I.; Berger, C.; Kallis, S.; Kolovou, A.; Paul, D.; Lohmann, V.; Bartenschlager, R. NS5A Domain 1 and Polyprotein Cleavage Kinetics Are Critical for Induction of Double-Membrane Vesicles Associated with Hepatitis C Virus Replication. mBio 2015, 6, e00759. [Google Scholar] [CrossRef]
- Romero-Brey, I.; Bartenschlager, R. Viral Infection at High Magnification: 3D Electron Microscopy Methods to Analyze the Architecture of Infected Cells. Viruses 2015, 7, 6316–6345. [Google Scholar] [CrossRef]
- Mironov, A.A.; Mironov, A.; Derganc, J.; Beznoussenko, G.V. Membrane Curvature, Trans-Membrane Area Asymmetry, Budding, Fission and Organelle Geometry. Int. J. Mol. Sci. 2020, 21, 7594. [Google Scholar] [CrossRef]
- Mironov, A.A.; Mironov, A.; Tratnjek, L.; Beznoussenko, G.V.; Kreft, M.E. Mitochondria, autophagy and COVID. Int. J. Mol. Sci. 2023, 24. accepted. [Google Scholar]
- Angelini, M.M.; Akhlaghpour, M.; Neuman, B.W.; Buchmeier, M.J. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio 2013, 4, e00524-13. [Google Scholar] [CrossRef] [PubMed]
- Holloway, G.; Johnson, R.I.; Kang, Y.; Dang, V.T.; Stojanovski, D.; Coulson, B.S. Rotavirus NSP6 localizes to mitochondria via a predicted N-terminal a-helix. J. Gen. Virol. 2015, 96, 3519–3524. [Google Scholar] [CrossRef] [PubMed]
- Benvenuto, D.; Angeletti, S.; Giovanetti, M.; Bianchi, M.; Pascarella, S.; Cauda, R.; Ciccozzi, M.; Cassone, A. Evolutionary analysis of SARS-CoV-2: How mutation of Non-Structural Protein 6 (NSP6) could affect viral autophagy. J. Infect. 2020, 81, e24–e27. [Google Scholar] [CrossRef]
- Morelli, E.; Ginefra, P.; Mastrodonato, V.; Beznoussenko, G.V.; Rusten, T.E.; Bilder, D.; Stenmark, H.; Mironov, A.A.; Vaccari, T. Multiple functions of the SNARE protein Snap29 in autophagy, endocytic, and exocytic trafficking during epithelial formation in Drosophila. Autophagy 2014, 10, 2251–2268. [Google Scholar] [CrossRef]
- Tirincsi, A.; Sicking, M.; Hadzibeganovic, D.; Haßdenteufel, S.; Lang, S. The Molecular Biodiversity of Protein Targeting and Protein Transport Related to the Endoplasmic Reticulum. Int. J. Mol. Sci. 2021, 23, 143. [Google Scholar] [CrossRef]
- Svoboda, D.; Nielson, A.; Werber, A.; Higginson, J. An electron microscopic study of viral hepatitis in mice. Am. J. Pathol. 1962, 41, 205–224. [Google Scholar] [PubMed]
- Deng, Y.; Almshwerqi, Z.A.; Ng, M.M.L.; Kohlwein, S.D. Do viruses subvert cholesterol homeostasis to induce host cubic membranes? Trends Cell Biol. 2010, 20, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Netherton, C.L.; Wileman, T. Virus factories, double membrane vesicles and viroplasm generated in animal cells. Curr. Opin. Virol. 2011, 1, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zandi, R. Biophysical Modeling of SARS-CoV-2 Assembly: Genome Condensation and Budding. Viruses 2022, 14, 2089. [Google Scholar] [CrossRef] [PubMed]
- Bacca, M. Mechanics of diffusion-mediated budding and implications for virus replication and infection. J. R. Soc. Interface 2022, 19, 20220525. [Google Scholar] [CrossRef] [PubMed]
- Rath, S.L.; Tripathy, M.; Mandal, N. How Does Temperature Affect the Dynamics of SARS-CoV-2 M Proteins? Insights from Molecular Dynamics Simulations. J. Membr. Biol. 2022, 255, 341–356. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wu, D.; Nguyen, A.; Li, Y.; Adão, R.C.; Valkov, E.; Patterson, G.H.; Piszczek, G.; Schuck, P. Energetic and structural features of SARS-CoV-2 N-protein co-assemblies with nucleic acids. iScience 2021, 24, 102523. [Google Scholar] [CrossRef]
- Ghosh, S.; Dellibovi-Ragheb, T.A.; Kerviel, A.; Pak, E.; Qiu, Q.; Fisher, M.; Takvorian, P.M.; Bleck, C.; Hsu, V.W.; Fehr, A.R.; et al. β-Coronaviruses Use Lysosomes for Egress Instead of the Biosynthetic Secretory Pathway. Cell 2020, 183, 1520–1535.e14. [Google Scholar] [CrossRef]
- Klumperman, J.; Locker, J.K.; Meijer, A.; Horzinek, M.C.; Geuze, H.J.; Rottier, P.J. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 1994, 68, 6523–6534. [Google Scholar] [CrossRef]
- Saville, J.W.; Berezuk, A.M.; Srivastava, S.S.; Subramaniam, S. Three-Dimensional Visualization of Viral Structure, Entry, and Replication Underlying the Spread of SARS-CoV-2. Chem. Rev. 2022, 122, 14066–14084. [Google Scholar] [CrossRef]
- Wu, H.; Fujioka, Y.; Sakaguchi, S.; Suzuki, Y.; Nakano, T. Three-dimensional reconstruction by electron tomography for the application to ultrastructural analysis of SARS-CoV-2 particles. Med. Mol. Morphol. 2022, 55, 60–67. [Google Scholar] [CrossRef]
- Scherer, K.M.; Mascheroni, L.; Carnell, G.W.; Wunderlich, L.C.S.; Makarchuk, S.; Brockhoff, M.; Mela, I.; Fernandez-Villegas, A.; Barysevich, M.; Stewart, H.; et al. SARS-CoV-2 nucleocapsid protein adheres to replication organelles before viral assembly at the Golgi/ERGIC and lysosome-mediated egress. Sci. Adv. 2022, 8, eabl4895. [Google Scholar] [CrossRef]
- Sesorova, I.S.; Sesorov, V.V.; Soloviev, P.B.; Lakunin, K.Y.; Dimov, I.D.; Mironov, A.A. Role of Endothelial Regeneration and Overloading of Enterocytes with Lipids in Capturing of Lipoproteins by Basement Membrane of Rat Aortic Endothelium. Biomedicines 2022, 10, 2858. [Google Scholar] [CrossRef]
- Nikonova, M.A.; Sesorova, I.S.; Dimov, I.D.; Karelina, N.R.; Mironov, A.A. Effect of the First Feeding on Enterocytes of Newborn Rats. Int. J. Mol. Sci. 2022, 23, 14179. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.L.; Kuan, I.I.; Guo, J.Y.; Hsu, W.C.; Tang, W.C.; Chan, H.J.; Chen, Y.-J.; Chen, B.-C.; Wu, H.-C.; Liao, J.C. SARS-CoV-2 N protein mediates intercellular nucleic acid dispersion, a feature reduced in Omicron. iScience 2023, 26, 105995. [Google Scholar] [CrossRef] [PubMed]
- Beznoussenko, G.V.; Parashuraman, S.; Sesorova, I.S.; Mironov, A.A. Exit of different cargoes from the Golgi complex and their post-Golgi trafficking. Histochem. Cell Biol. 2023, submitted.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mironov, A.A.; Savin, M.A.; Beznoussenko, G.V. COVID-19 Biogenesis and Intracellular Transport. Int. J. Mol. Sci. 2023, 24, 4523. https://doi.org/10.3390/ijms24054523
Mironov AA, Savin MA, Beznoussenko GV. COVID-19 Biogenesis and Intracellular Transport. International Journal of Molecular Sciences. 2023; 24(5):4523. https://doi.org/10.3390/ijms24054523
Chicago/Turabian StyleMironov, Alexander A., Maksim A. Savin, and Galina V. Beznoussenko. 2023. "COVID-19 Biogenesis and Intracellular Transport" International Journal of Molecular Sciences 24, no. 5: 4523. https://doi.org/10.3390/ijms24054523
APA StyleMironov, A. A., Savin, M. A., & Beznoussenko, G. V. (2023). COVID-19 Biogenesis and Intracellular Transport. International Journal of Molecular Sciences, 24(5), 4523. https://doi.org/10.3390/ijms24054523








