Establishing a Proteomics-Based Signature of AKR1C3-Related Genes for Predicting the Prognosis of Prostate Cancer
Abstract
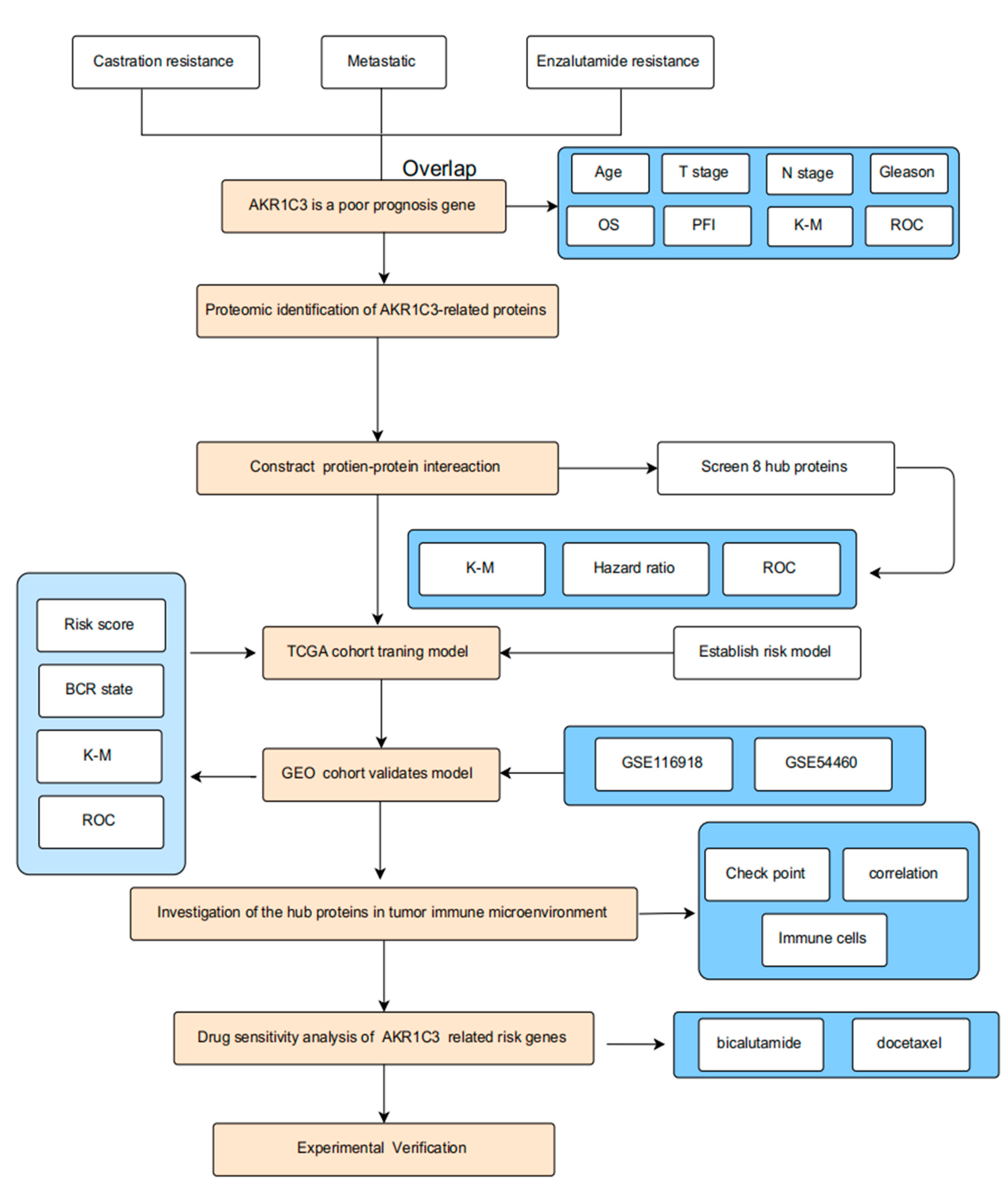
1. Introduction
2. Results
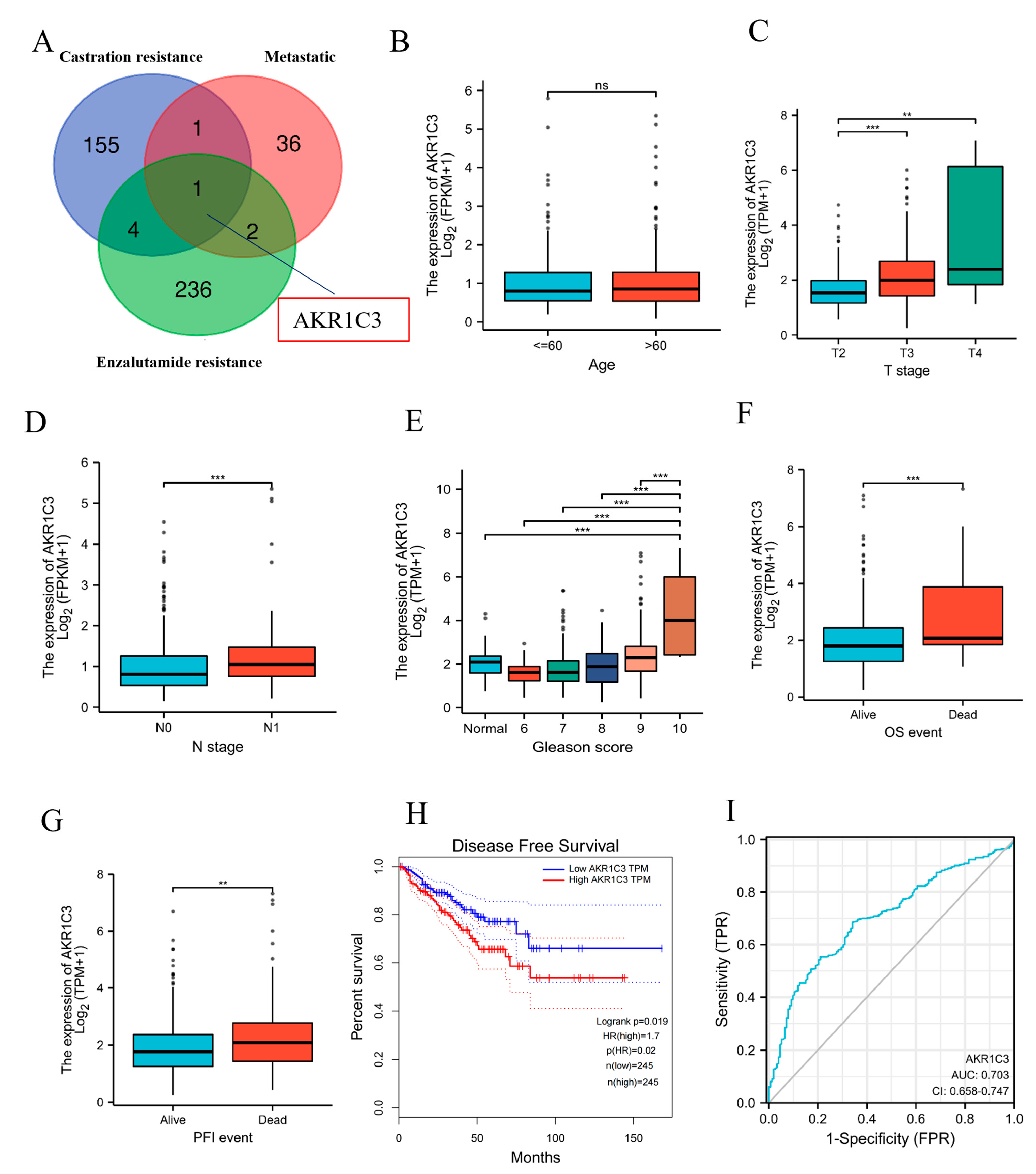
2.1. AKR1C3 as a Poor-Prognosis Gene in PCa Patients
2.2. PPI Network Construction and Risk Gene Selection
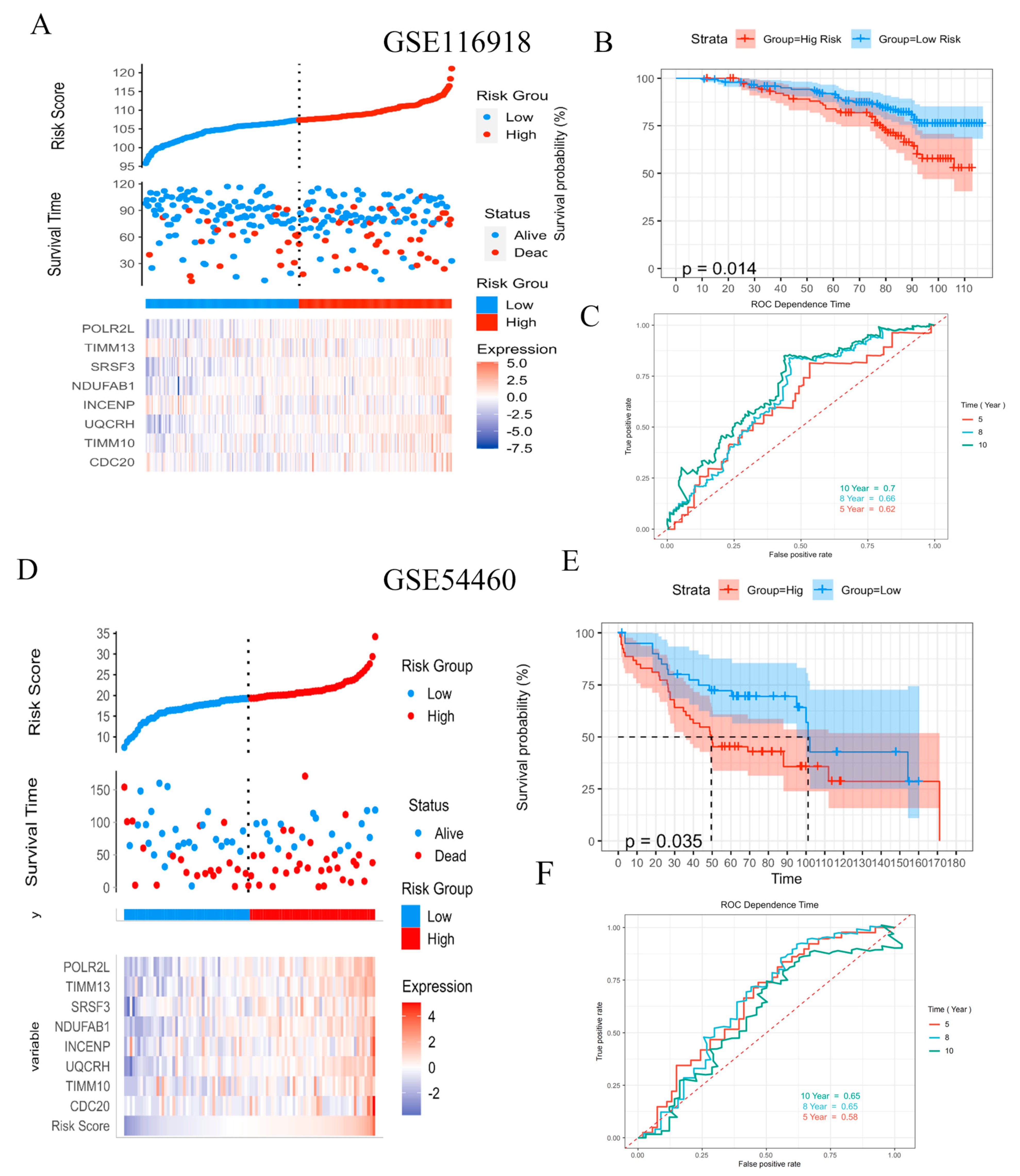
2.3. Construction of Prognostic Risk Model for PCa
2.4. Validation of Prognostic Risk Model for PCa
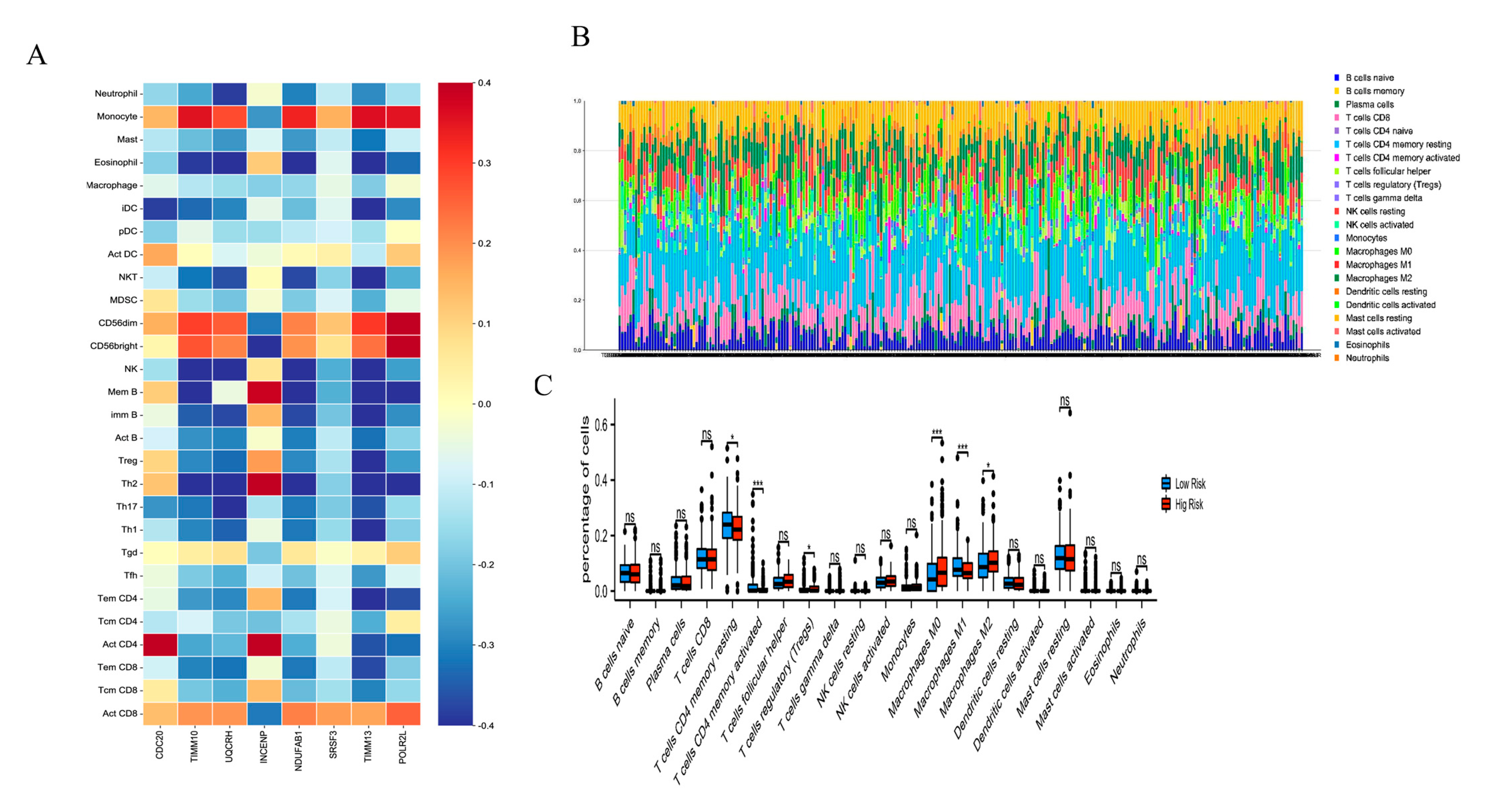
2.5. The Immune Cell Infiltration Analysis of the Risk Score Model in PCa
2.6. Correlation Analysis of the Risk Score Model with Immune Checkpoint Genes
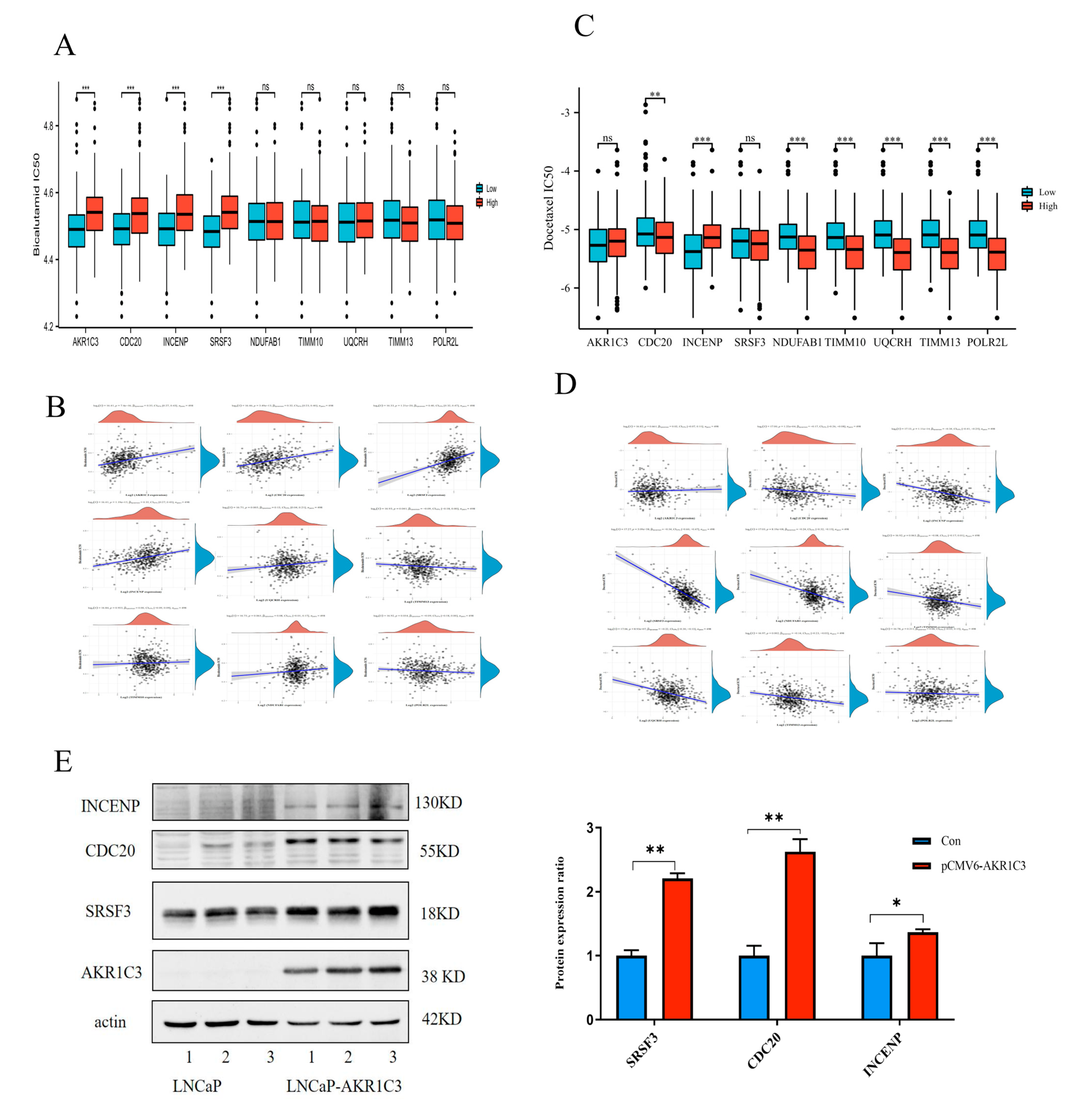
2.7. Drug Sensitivity Evaluation of AKR1C3 and Associated Risk Genes
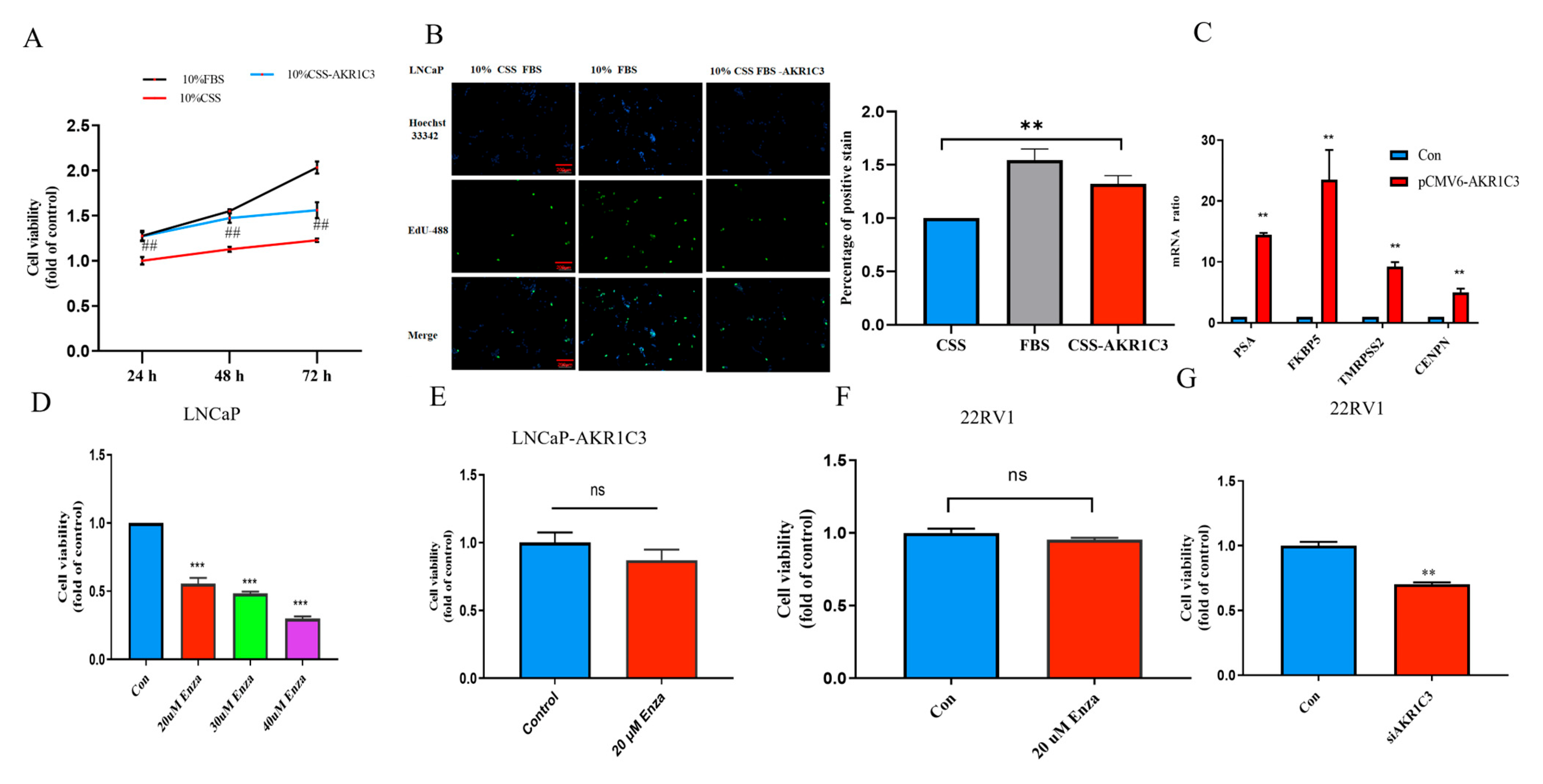
2.8. AKR1C3 Promotes PCa Cell Proliferation and Migration
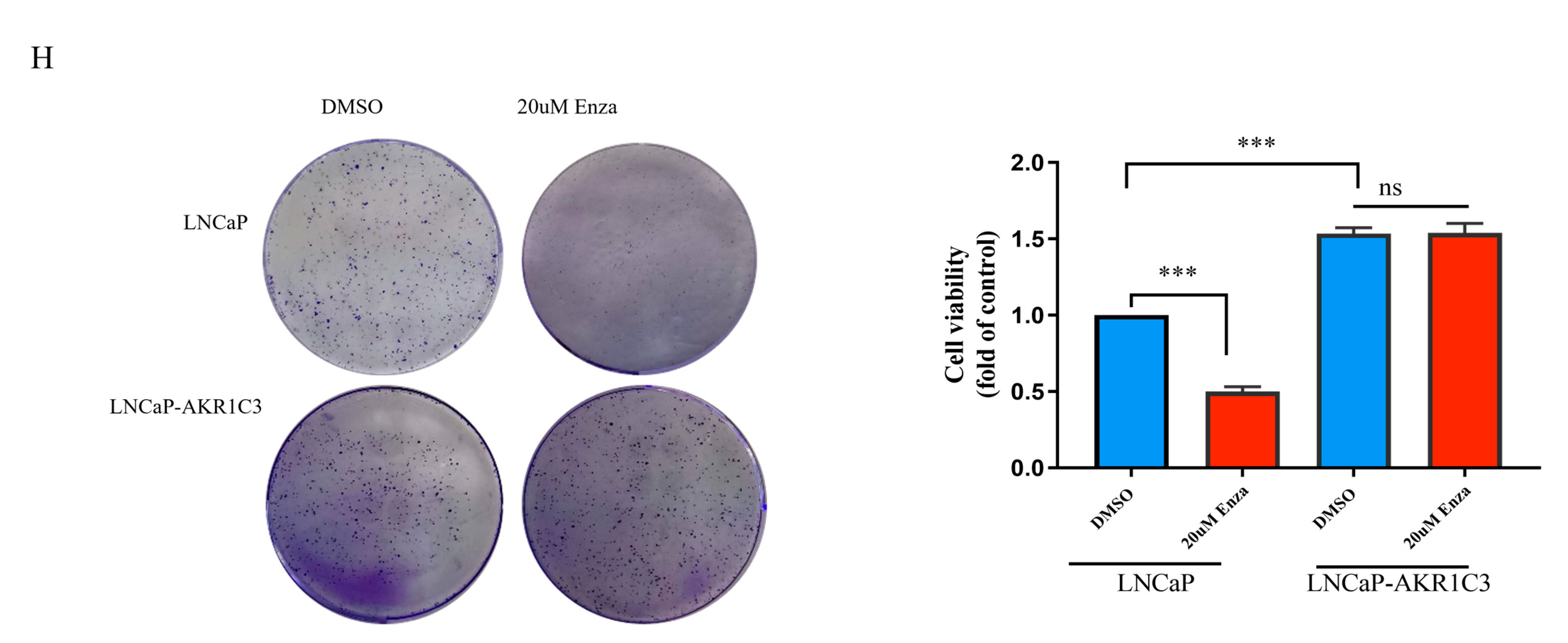
2.9. Overexpression of AKR1C3 Decreased Enzalutamide Sensitivity in P Cells
3. Discussion
4. Materials and Methods
4.1. Data Acquisition and Preprocessing
4.2. Construction of a Prognostic Signature Based on Risk Genes
4.3. Estimation of the Immune Microenvironment Composition
4.4. Drug Sensitivity Analysis
4.5. Cell Culture and Transduction
4.6. MTT
4.7. Colony-Forming Assay
4.8. Wound-Healing and Transwell Migration Assay
4.9. Quantitative Real-Time PCR
4.10. Western Blot Analysis
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Cornford, P.; van den Bergh, R.; Briers, E.; Van den Broeck, T.; Cumberbatch, M.G.; De Santis, M.; Fanti, S.; Fossati, N.; Gandaglia, G.; Gillessen, S.; et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer. Part II-2020 Update: Treatment of Relapsing and Metastatic Prostate Cancer. Eur. Urol. 2021, 79, 263–282. [Google Scholar] [CrossRef] [PubMed]
- Hamid, A.R.; Pfeiffer, M.J.; Verhaegh, G.W.; Schaafsma, E.; Brandt, A.; Sweep, F.C.; Sedelaar, J.P.; Schalken, J.A. Aldo-keto reductase family 1 member C3 (AKR1C3) is a biomarker and therapeutic target for castration-resistant prostate cancer. Mol. Med. 2013, 18, 1449–1455. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Lou, W.; Zhu, Y.; Yang, J.C.; Nadiminty, N.; Gaikwad, N.W.; Evans, C.P.; Gao, A.C. Intracrine Androgens and AKR1C3 Activation Confer Resistance to Enzalutamide in Prostate Cancer. Cancer Res. 2015, 75, 1413–1422. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Zhao, L.; Zhang, H.; Liu, X.; Zhao, L.; Zhao, X.; Li, Y.; Li, J. AKR1C3 overexpression may serve as a promising biomarker for prostate cancer progression. Diagn. Pathol. 2014, 9, 42. [Google Scholar] [CrossRef]
- Miyazaki, Y.; Teramoto, Y.; Shibuya, S.; Goto, T.; Okasho, K.; Mizuno, K.; Uegaki, M.; Yoshikawa, T.; Akamatsu, S.; Kobayashi, T.; et al. Consecutive Prostate Cancer Specimens Revealed Increased Aldo(-)Keto Reductase Family 1 Member C3 Expression with Progression to Castration-Resistant Prostate Cancer. J. Clin. Med. 2019, 8, 601. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Gu, Y.; Hui, K.; Huang, J.; Xu, S.; Wu, S.; Li, L.; Fan, J.; Wang, X.; Hsieh, J.T.; et al. AKR1C3, a crucial androgenic enzyme in prostate cancer, promotes epithelial-mesenchymal transition and metastasis through activating ERK signaling. Urol. Oncol. Semin. Orig. Investig. 2018, 36, 411–472. [Google Scholar] [CrossRef]
- Bianchini, D.; Lorente, D.; Rodriguez-Vida, A.; Omlin, A.; Pezaro, C.; Ferraldeschi, R.; Zivi, A.; Attard, G.; Chowdhury, S.; de Bono, J.S. Antitumour activity of enzalutamide (MDV3100) in patients with metastatic castration-resistant prostate cancer (CRPC) pre-treated with docetaxel and abiraterone. Eur. J. Cancer 2014, 50, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.J. Enzalutamide: A Review in Castration-Resistant Prostate Cancer. Drugs 2018, 78, 1913–1924. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yang, J.C.; Armstrong, C.M.; Lou, W.; Liu, L.; Qiu, X.; Zou, B.; Lombard, A.P.; D’Abronzo, L.S.; Evans, C.P.; et al. AKR1C3 Promotes AR-V7 Protein Stabilization and Confers Resistance to AR-Targeted Therapies in Advanced Prostate Cancer. Mol. Cancer Ther. 2019, 18, 1875–1886. [Google Scholar] [CrossRef] [PubMed]
- Bober, P.; Alexovic, M.; Tomkova, Z.; Kilik, R.; Sabo, J. RHOA and mDia1 Promotes Apoptosis of Breast Cancer Cells Via a High Dose of Doxorubicin Treatment. Open Life Sci. 2019, 14, 619–627. [Google Scholar] [CrossRef]
- Dai, L.; Song, Z.X.; Wei, D.P.; Zhang, J.D.; Liang, J.Q.; Wang, B.B.; Ma, W.T.; Li, L.Y.; Dang, Y.L.; Zhao, L.; et al. CDC20 and PTTG1 are Important Biomarkers and Potential Therapeutic Targets for Metastatic Prostate Cancer. Adv. Ther. 2021, 38, 2973–2989. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Huang, H.; Liu, A.; Li, J.; Liu, C.; Sun, B.; Chen, L.; Gao, Y.; Xu, D.; Su, C. Cell division cycle 20 (CDC20) drives prostate cancer progression via stabilization of beta-catenin in cancer stem-like cells. eBioMedicine 2019, 42, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wu, Z.; Ding, W.; Xiao, C.; Zhang, Y.; Gao, S.; Gao, Y.; Cai, W. SREBP1 siRNA enhance the docetaxel effect based on a bone-cancer dual-targeting biomimetic nanosystem against bone metastatic castration-resistant prostate cancer. Theranostics 2020, 10, 1619–1632. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Gong, Q.; Lin, Z.; Wang, Y.; Li, M.; Wang, L.; Ding, H.; Li, P. Emerging Roles of SRSF3 as a Therapeutic Target for Cancer. Front. Oncol. 2020, 10, 577636. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Sun, Y.; Xu, Q. Inhibition of SRSF3 Alleviates Proliferation and Migration of Gastric Cancer Cells by Regulating the PI3K/AKT/mTOR Signalling Pathway. Folia Biol. 2021, 67, 102–107. [Google Scholar]
- Park, E.R.; Kim, S.B.; Lee, J.S.; Kim, Y.H.; Lee, D.H.; Cho, E.H.; Park, S.H.; Han, C.J.; Kim, B.Y.; Choi, D.W.; et al. The mitochondrial hinge protein, UQCRH, is a novel prognostic factor for hepatocellular carcinoma. Cancer Med. 2017, 6, 749–760. [Google Scholar] [CrossRef]
- Gao, F.; Liu, Q.; Li, G.; Dong, F.; Qiu, M.; Lv, X.; Zhang, S.; Guo, Z. Identification of ubiquinol cytochrome c reductase hinge (UQCRH) as a potential diagnostic biomarker for lung adenocarcinoma. Open Biol. 2016, 6, 150256. [Google Scholar] [CrossRef] [PubMed]
- Adams, R.R.; Eckley, D.M.; Vagnarelli, P.; Wheatley, S.P.; Gerloff, D.L.; Mackay, A.M.; Svingen, P.A.; Kaufmann, S.H.; Earnshaw, W.C. Human INCENP colocalizes with the Aurora-B/AIRK2 kinase on chromosomes and is overexpressed in tumour cells. Chromosoma 2001, 110, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Von Keudell, G.; Suzuki, T.; Dohmae, N.; Nakakido, M.; Piao, L.; Yoshioka, Y.; Nakamura, Y.; Hamamoto, R. PRMT1 promotes mitosis of cancer cells through arginine methylation of INCENP. Oncotarget 2015, 6, 35173–35182. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Chen, B.; Xu, X.; Lin, G. Prospective nested case-control study of feature genes related to leukemic evolution of myelodysplastic syndrome. Mol. Biol. Rep. 2013, 40, 469–476. [Google Scholar] [CrossRef] [PubMed]
- De Antonellis, P.; Carotenuto, M.; Vandenbussche, J.; De Vita, G.; Ferrucci, V.; Medaglia, C.; Boffa, I.; Galiero, A.; Di Somma, S.; Magliulo, D.; et al. Early targets of miR-34a in neuroblastoma. Mol. Cell. Proteom. 2014, 13, 2114–2131. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Li, X.; Zhao, H.; Sun, B.; Liu, A.; Han, X.; Cui, Z.; Yuan, L. Combining multi-dimensional data to identify a key signature (gene and miRNA) of cisplatin-resistant gastric cancer. J. Cell. Biochem. 2018, 119, 6997–7008. [Google Scholar] [CrossRef] [PubMed]
- Triepels, R.; Smeitink, J.; Loeffen, J.; Smeets, R.; Buskens, C.; Trijbels, F.; van den Heuvel, L. The human nuclear-encoded acyl carrier subunit (NDUFAB1) of the mitochondrial complex I in human pathology. J. Inherit. Metab. Dis. 1999, 22, 163–173. [Google Scholar] [CrossRef]
- Luan, S.; Li, P.; Yi, T. Series test of cluster and network analysis for lupus nephritis, before and after IFN-K-immunosuppressive therapy. Nephrology 2018, 23, 997–1006. [Google Scholar] [CrossRef]
- Huang, M.Y.; Jiang, X.M.; Wang, B.L.; Sun, Y.; Lu, J.J. Combination therapy with PD-1/PD-L1 blockade in non-small cell lung cancer: Strategies and mechanisms. Pharmacol. Ther. 2021, 219, 107694. [Google Scholar] [CrossRef]
- Willsmore, Z.N.; Coumbe, B.; Crescioli, S.; Reci, S.; Gupta, A.; Harris, R.J.; Chenoweth, A.; Chauhan, J.; Bax, H.J.; Mccraw, A.; et al. Combined anti-PD-1 and anti-CTLA-4 checkpoint blockade: Treatment of melanoma and immune mechanisms of action. Eur. J. Immunol. 2021, 51, 544–556. [Google Scholar] [CrossRef]
- Au, L.; Hatipoglu, E.; Robert, D.M.M.; Litchfield, K.; Beattie, G.; Rowan, A.; Schnidrig, D.; Thompson, R.; Byrne, F.; Horswell, S.; et al. Determinants of anti-PD-1 response and resistance in clear cell renal cell carcinoma. Cancer Cell 2021, 39, 1497–1518. [Google Scholar] [CrossRef]
- Pachynski, R.K.; Morishima, C.; Szmulewitz, R.; Harshman, L.; Appleman, L.; Monk, P.; Bitting, R.L.; Kucuk, O.; Millard, F.; Seigne, J.D.; et al. IL-7 expands lymphocyte populations and enhances immune responses to sipuleucel-T in patients with metastatic castration-resistant prostate cancer (mCRPC). J. Immunother. Cancer 2021, 9, e002903. [Google Scholar] [CrossRef]
- Weng, Y.S.; Tseng, H.Y.; Chen, Y.A.; Shen, P.C.; Al, H.A.; Chen, L.M.; Tung, Y.C.; Hsu, H.L. MCT-1/miR-34a/IL-6/IL-6R signaling axis promotes EMT progression, cancer stemness and M2 macrophage polarization in triple-negative breast cancer. Mol. Cancer 2019, 18, 42. [Google Scholar] [CrossRef]
- Seif, F.; Sharifi, L.; Khoshmirsafa, M.; Mojibi, Y.; Mohsenzadegan, M. A Review of Preclinical Experiments Toward Targeting M2 Macrophages in Prostate Cancer. Curr. Drug Targets 2019, 20, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Maolake, A.; Izumi, K.; Shigehara, K.; Natsagdorj, A.; Iwamoto, H.; Kadomoto, S.; Takezawa, Y.; Machioka, K.; Narimoto, K.; Namiki, M.; et al. Tumor-associated macrophages promote prostate cancer migration through activation of the CCL22-CCR4 axis. Oncotarget 2017, 8, 9739–9751. [Google Scholar] [CrossRef] [PubMed]
- Nonomura, N.; Takayama, H.; Nakayama, M.; Nakai, Y.; Kawashima, A.; Mukai, M.; Nagahara, A.; Aozasa, K.; Tsujimura, A. Infiltration of tumour-associated macrophages in prostate biopsy specimens is predictive of disease progression after hormonal therapy for prostate cancer. BJU Int. 2011, 107, 1918–1922. [Google Scholar] [CrossRef] [PubMed]
- Karpisheh, V.; Mousavi, S.M.; Naghavi, S.P.; Fathi, M.; Mohammadpour, S.M.; Aghebati-Maleki, L.; Jafari, R.; Majidi, Z.N.; Jadidi-Niaragh, F. The role of regulatory T cells in the pathogenesis and treatment of prostate cancer. Life Sci. 2021, 284, 119132. [Google Scholar] [CrossRef]
- Walker, L.S. Treg and CTLA-4: Two intertwining pathways to immune tolerance. J. Autoimmun. 2013, 45, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Tekguc, M.; Wing, J.B.; Osaki, M.; Long, J.; Sakaguchi, S. Treg-expressed CTLA-4 depletes CD80/CD86 by trogocytosis, releasing free PD-L1 on antigen-presenting cells. Proc. Natl. Acad. Sci. USA 2021, 118, e2023739118. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.D.; Kou, Z.Y.; Wang, J.T.; Mi, D.H. Prognostic implications of ferroptosis-associated gene signature in colon adenocarcinoma. World J. Clin. Cases 2021, 9, 8671–8693. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Gao, L.; Xie, T.; Li, J.; Zhai, T.S.; Xu, Y. Identification and Validation of a Prognostic Signature for Prostate Cancer Based on Ferroptosis-Related Genes. Front. Oncol. 2021, 11, 623313. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Hua, X.; Chen, J.; Xiao, H.; Zhang, L.; Zhou, J.; Liang, C.; Tai, S. Identification of a Costimulatory Molecule-Related Signature for Predicting Prognostic Risk in Prostate Cancer. Front. Genet. 2021, 12, 666300. [Google Scholar] [CrossRef] [PubMed]
- Geeleher, P.; Cox, N.J.; Huang, R.S. Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol. 2014, 15, R47. [Google Scholar] [CrossRef] [PubMed]
- Van de Merbel, A.F.; van der Horst, G.; Buijs, J.T.; van der Pluijm, G. Protocols for Migration and Invasion Studies in Prostate Cancer. Methods Mol. Biol. 2018, 1786, 67–79. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, X.; Li, C.; Ding, J.; Yao, Z.; Zhao, T.; Guo, J.; Wang, Y.; Li, J. Establishing a Proteomics-Based Signature of AKR1C3-Related Genes for Predicting the Prognosis of Prostate Cancer. Int. J. Mol. Sci. 2023, 24, 4513. https://doi.org/10.3390/ijms24054513
Cui X, Li C, Ding J, Yao Z, Zhao T, Guo J, Wang Y, Li J. Establishing a Proteomics-Based Signature of AKR1C3-Related Genes for Predicting the Prognosis of Prostate Cancer. International Journal of Molecular Sciences. 2023; 24(5):4513. https://doi.org/10.3390/ijms24054513
Chicago/Turabian StyleCui, Xiaoli, Changcheng Li, Jipeng Ding, Zhou Yao, Tianyu Zhao, Jiahui Guo, Yaru Wang, and Jing Li. 2023. "Establishing a Proteomics-Based Signature of AKR1C3-Related Genes for Predicting the Prognosis of Prostate Cancer" International Journal of Molecular Sciences 24, no. 5: 4513. https://doi.org/10.3390/ijms24054513
APA StyleCui, X., Li, C., Ding, J., Yao, Z., Zhao, T., Guo, J., Wang, Y., & Li, J. (2023). Establishing a Proteomics-Based Signature of AKR1C3-Related Genes for Predicting the Prognosis of Prostate Cancer. International Journal of Molecular Sciences, 24(5), 4513. https://doi.org/10.3390/ijms24054513




