Variable Intrinsic Expression of Immunoregulatory Biomarkers in Breast Cancer Cell Lines, Mammospheres, and Co-Cultures
Abstract
1. Introduction
2. Results
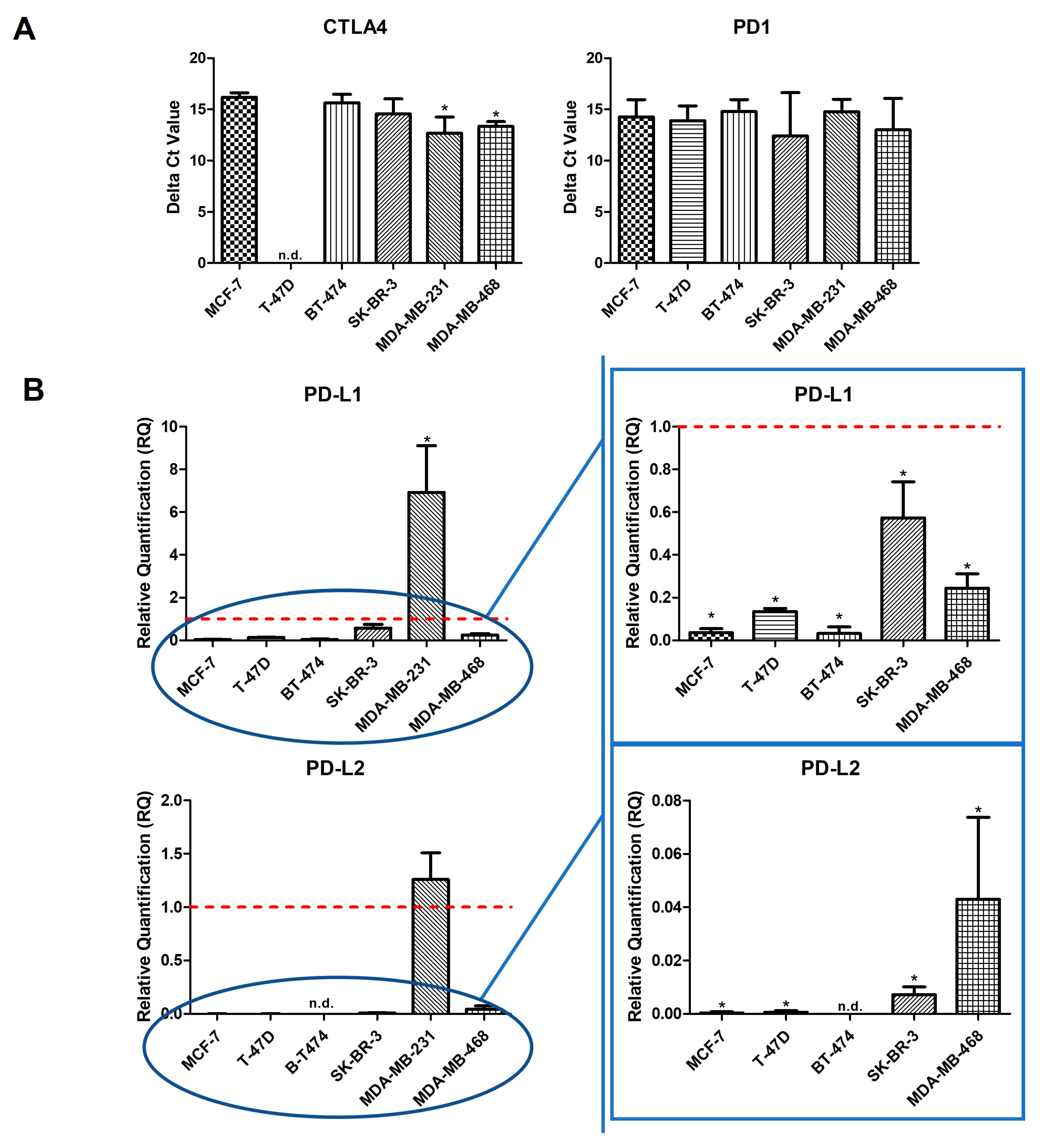
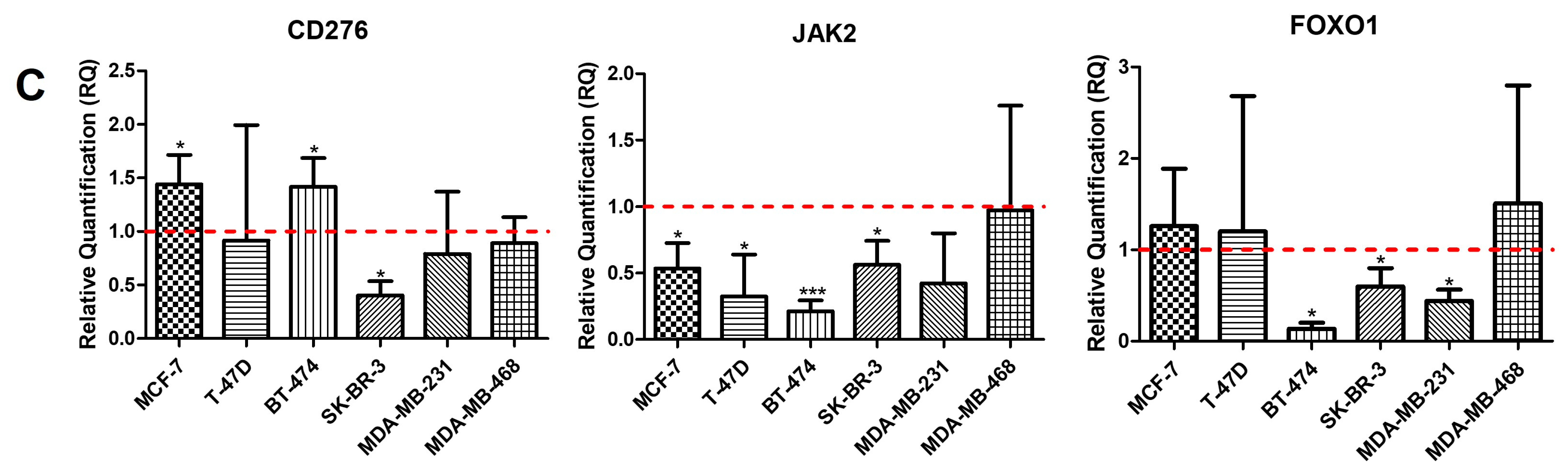
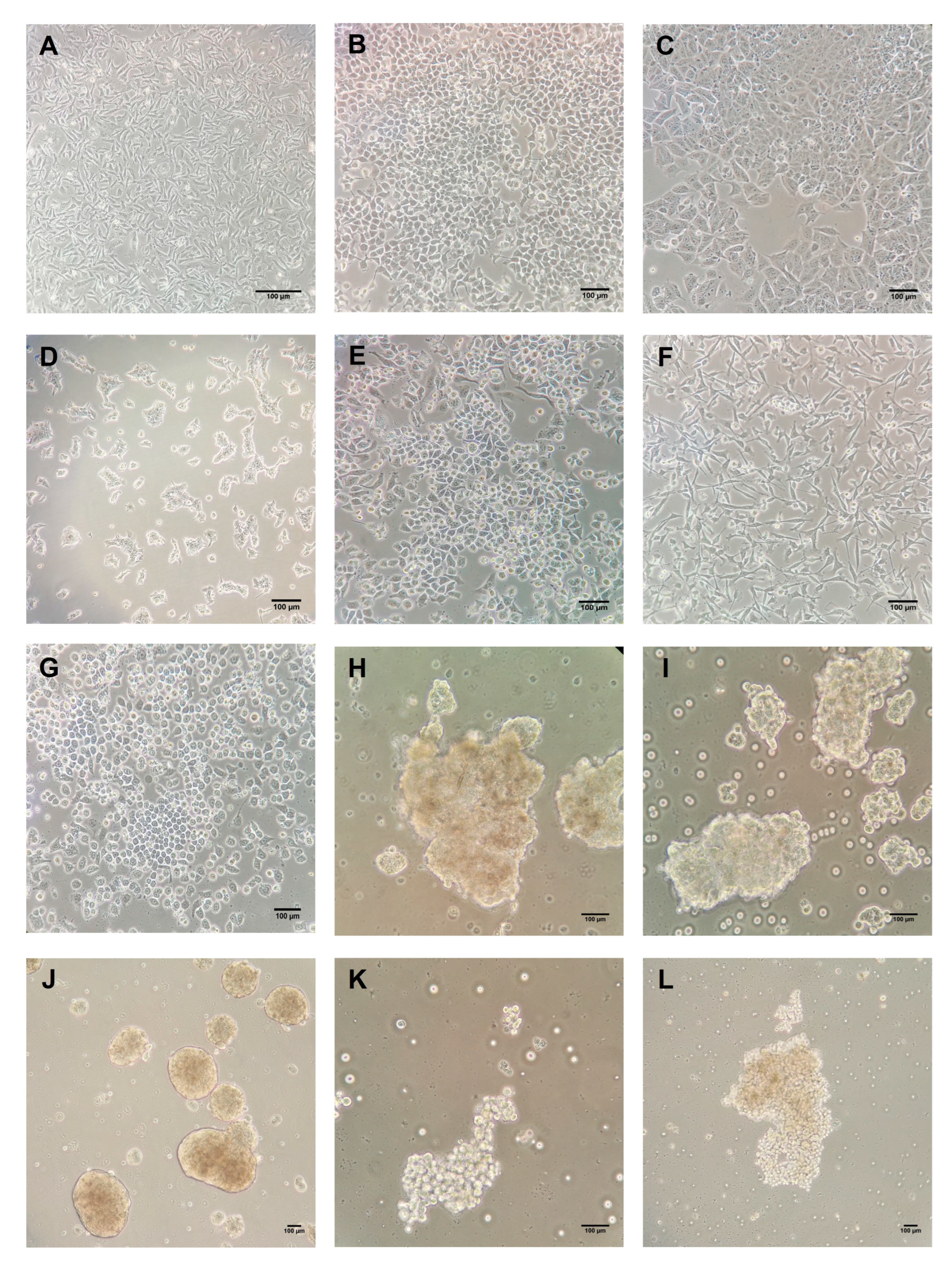
2.1. Intrinsic Gene Expression in BC Cell Lines
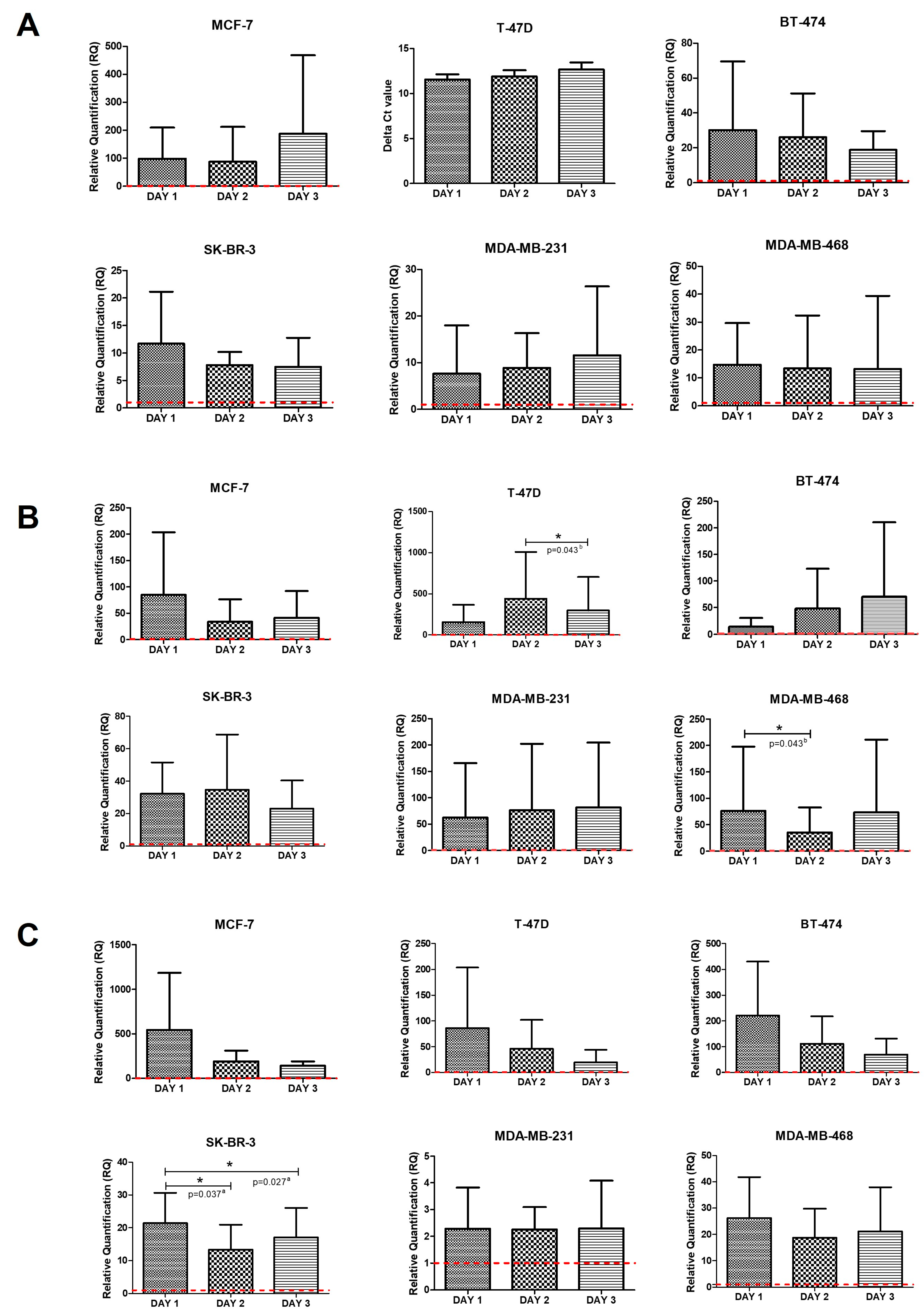
2.2. Intrinsic Gene Expression in Mammospheres
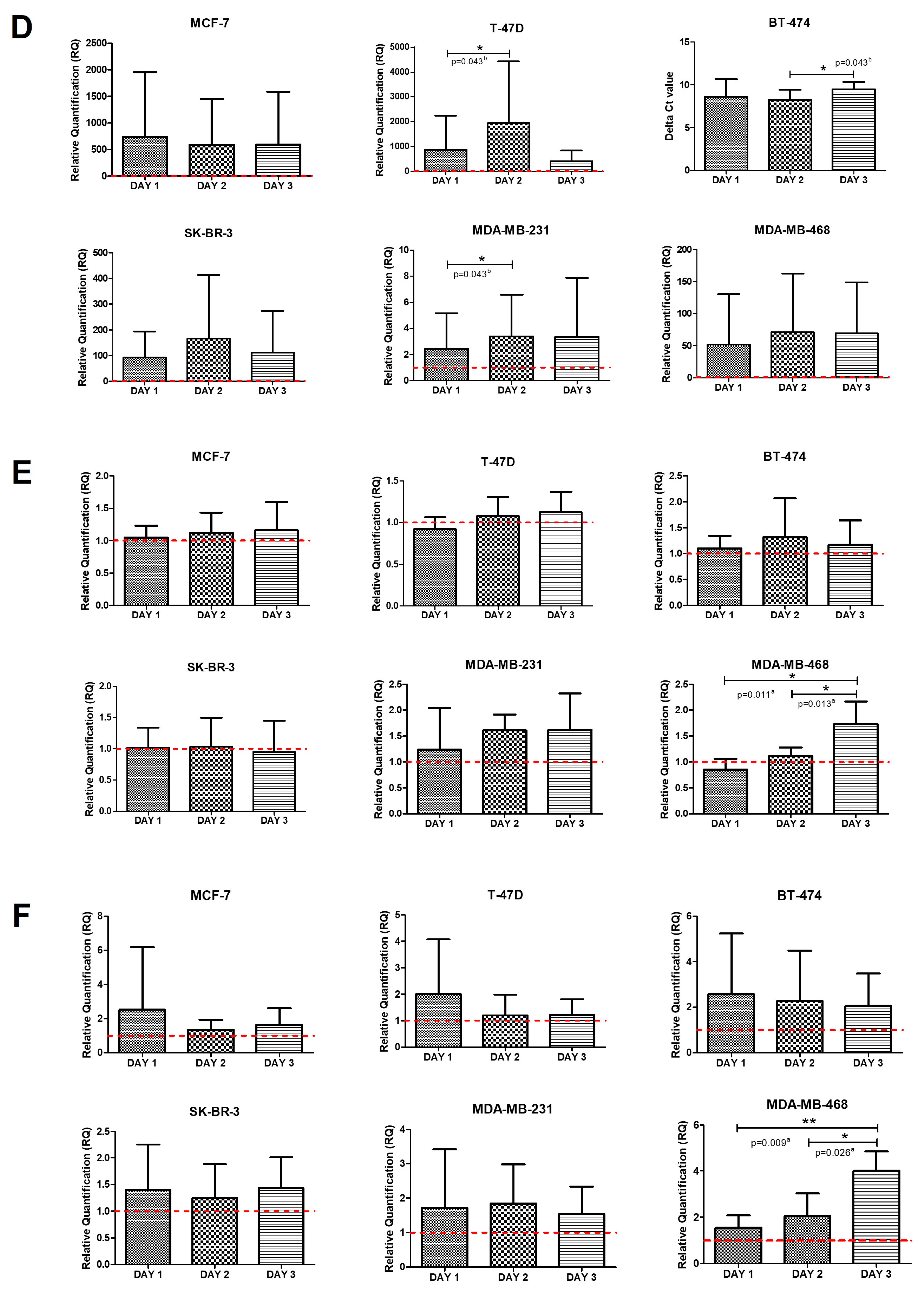
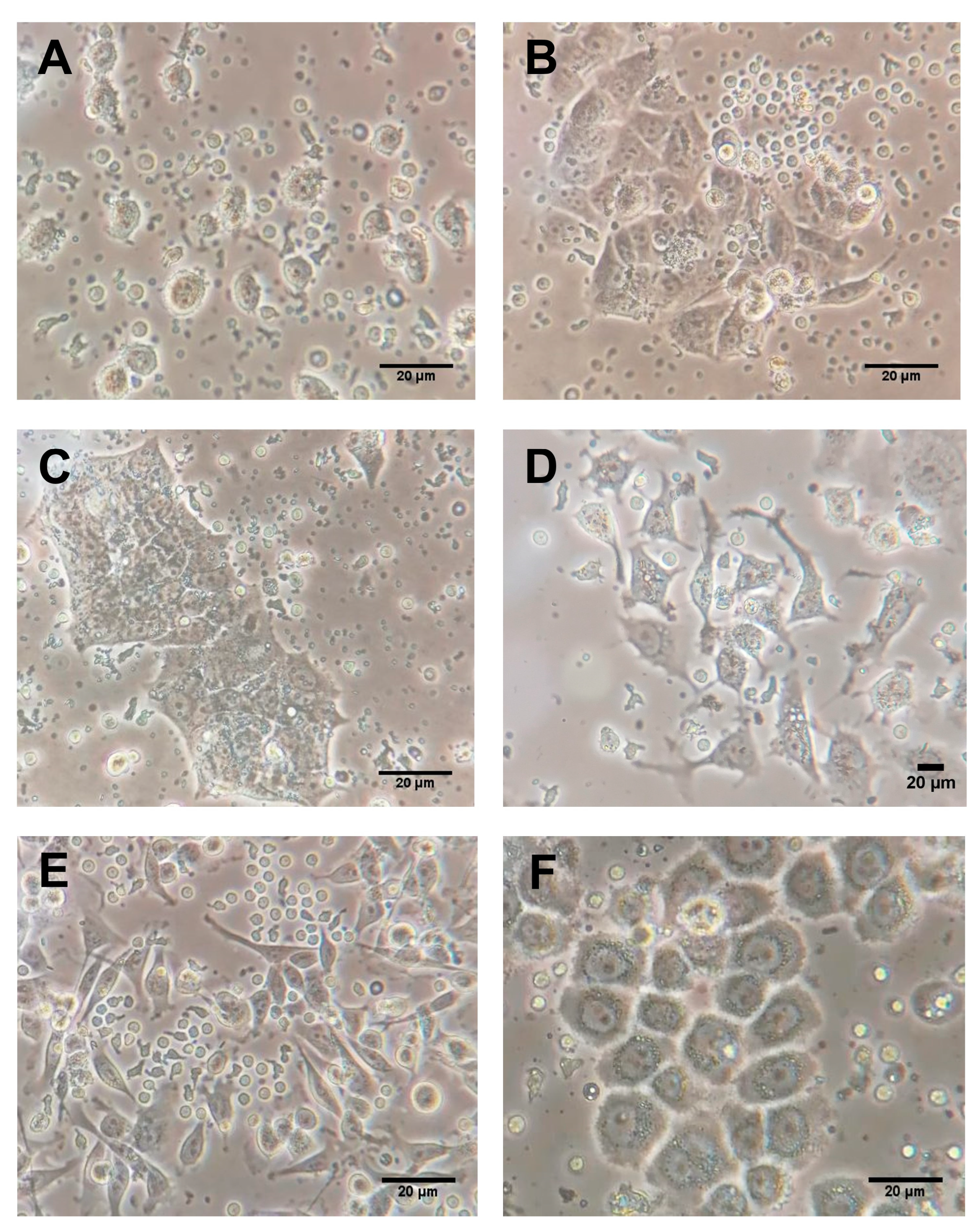
2.3. Intrinsic Gene Expression in Co-Cultures of BC Cell Lines and PBMCs
3. Discussion
4. Materials and Methods
4.1. BC Cell Lines Culture
4.2. Isolation of Human PBMCs
4.3. Co-Culture of PBMCs with BC Cell Lines
4.4. Mammosphere Culture
4.5. RNA Isolation and Complementary DNA Synthesis
4.6. Real-Time Quantitative Polymerase Chain Reaction (qRT-PCR)
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Herschkowitz, J.I.; Simin, K.; Weigman, V.J.; Mikaelian, I.; Usary, J.; Hu, Z.; Rasmussen, K.E.; Jones, L.P.; Assefnia, S.; Chandrasekharan, S. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007, 8, R76. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; Van De Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Sørlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; Van De Rijn, M.; Jeffrey, S.S. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef]
- Yarchoan, M.; Hopkins, A.; Jaffee, E.M. Tumor mutational burden and response rate to PD-1 inhibition. N. Engl. J. Med. 2017, 377, 2500. [Google Scholar] [CrossRef]
- Emens, L.A.; Adams, S.; Barrios, C.H.; Dieras, V.; Iwata, H.; Loi, S.; Rugo, H.S.; Schneeweiss, A.; Winer, E.P.; Patel, S. LBA16 IMpassion130: Final OS analysis from the pivotal phase III study of atezolizumab+ nab-paclitaxel vs. placebo+ nab-paclitaxel in previously untreated locally advanced or metastatic triple-negative breast cancer. Ann. Oncol. 2020, 31, S1148. [Google Scholar] [CrossRef]
- Emens, L.A.; Cruz, C.; Eder, J.P.; Braiteh, F.; Chung, C.; Tolaney, S.M.; Kuter, I.; Nanda, R.; Cassier, P.A.; Delord, J.-P. Long-term clinical outcomes and biomarker analyses of atezolizumab therapy for patients with metastatic triple-negative breast cancer: A phase 1 study. JAMA Oncol. 2019, 5, 74–82. [Google Scholar] [CrossRef]
- Schmid, P.; Adams, S.; Rugo, H.S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Diéras, V.; Hegg, R.; Im, S.-A.; Shaw Wright, G. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N. Engl. J. Med. 2018, 379, 2108–2121. [Google Scholar] [CrossRef]
- Schmid, P.; Rugo, H.S.; Adams, S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Diéras, V.; Henschel, V.; Molinero, L.; Chui, S.Y. Atezolizumab plus nab-paclitaxel as first-line treatment for unresectable, locally advanced or metastatic triple-negative breast cancer (IMpassion130): Updated efficacy results from a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2020, 21, 44–59. [Google Scholar] [CrossRef]
- Cortes, J.; Cescon, D.W.; Rugo, H.S.; Nowecki, Z.; Im, S.-A.; Yusof, M.M.; Gallardo, C.; Lipatov, O.; Barrios, C.H.; Holgado, E. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): A randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet 2020, 396, 1817–1828. [Google Scholar] [CrossRef]
- Schmid, P.; Cortes, J.; Dent, R.; Pusztai, L.; McArthur, H.; Kümmel, S.; Bergh, J.; Denkert, C.; Park, Y.H.; Rina Hui, M.B.; et al. Event-free survival with pembrolizumab in early triple-negative breast cancer. N. Engl. J. Med. 2022, 386, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Miles, D.; Gligorov, J.; André, F.; Cameron, D.; Schneeweiss, A.; Barrios, C.; Xu, B.; Wardley, A.; Kaen, D.; Andrade, L. Primary results from IMpassion131, a double-blind, placebo-controlled, randomised phase III trial of first-line paclitaxel with or without atezolizumab for unresectable locally advanced/metastatic triple-negative breast cancer. Ann. Oncol. 2021, 32, 994–1004. [Google Scholar] [CrossRef] [PubMed]
- Fyles, A.; Nguyen, L.T.; Neel, B.G.; Sacher, A.; Rottapel, R.; Wang, B.X.; Ohashi, P.S.; Sridhar, S.S.J.O. Phase I study of local radiation and tremelimumab in patients with inoperable locally recurrent or metastatic breast cancer. Oncotarget 2019, 10, 2947. [Google Scholar]
- Santa-Maria, C.A.; Kato, T.; Park, J.-H.; Kiyotani, K.; Rademaker, A.; Shah, A.N.; Gross, L.; Blanco, L.Z.; Jain, S.; Flaum, L.J.O. A pilot study of durvalumab and tremelimumab and immunogenomic dynamics in metastatic breast cancer. Oncotarget 2018, 9, 18985. [Google Scholar] [CrossRef]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef]
- Keir, M.E.; Butte, M.J.; Freeman, G.J.; Sharpe, A.H. PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 2008, 26, 677–704. [Google Scholar] [CrossRef]
- Ni, L.; Dong, C. New B7 family checkpoints in human cancers. Mol. Cancer Ther. 2017, 16, 1203–1211. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, W.; Huang, Y.; Cui, R.; Li, X.; Li, B. Evolving roles for targeting CTLA-4 in cancer immunotherapy. Cell. Physiol. Biochem. 2018, 47, 721–734. [Google Scholar] [CrossRef]
- Contardi, E.; Palmisano, G.L.; Tazzari, P.L.; Martelli, A.M.; Fala, F.; Fabbi, M.; Kato, T.; Lucarelli, E.; Donati, D.; Polito, L. CTLA-4 is constitutively expressed on tumor cells and can trigger apoptosis upon ligand interaction. Int. J. Cancer 2005, 117, 538–550. [Google Scholar] [CrossRef]
- Erfani, N.; Razmkhah, M.; Ghaderi, A. Circulating soluble CTLA4 (sCTLA4) is elevated in patients with breast cancer. Cancer Investig. 2010, 28, 828–832. [Google Scholar] [CrossRef]
- Oaks, M.K.; Hallett, K.M. Cutting edge: A soluble form of CTLA-4 in patients with autoimmune thyroid disease. J. Immunol. 2000, 164, 5015–5018. [Google Scholar] [CrossRef]
- Zhao, Y.; Harrison, D.L.; Song, Y.; Ji, J.; Huang, J.; Hui, E. Antigen-presenting cell-intrinsic PD-1 neutralizes PD-L1 in cis to attenuate PD-1 signaling in T cells. Cell Rep. 2018, 24, 379–390.e376. [Google Scholar] [CrossRef]
- Wang, X.; Yang, X.; Zhang, C.; Wang, Y.; Cheng, T.; Duan, L.; Tong, Z.; Tan, S.; Zhang, H.; Saw, P.E. Tumor cell-intrinsic PD-1 receptor is a tumor suppressor and mediates resistance to PD-1 blockade therapy. Proc. Natl. Acad. Sci. USA 2020, 117, 6640–6650. [Google Scholar] [CrossRef]
- Yao, H.; Wang, H.; Li, C.; Fang, J.-Y.; Xu, J. Cancer cell-intrinsic PD-1 and implications in combinatorial immunotherapy. Front. Immunol. 2018, 9, 1774. [Google Scholar] [CrossRef]
- Li, J.; Chen, L.; Xiong, Y.; Zheng, X.; Xie, Q.; Zhou, Q.; Shi, L.; Wu, C.; Wang, H.; Jiang, J. Knockdown of PD-L1 in human gastric cancer cells inhibits tumor progression and improves the cytotoxic sensitivity to CIK therapy. Cell Physiol. Biochem. 2017, 41, 907–920. [Google Scholar] [CrossRef] [PubMed]
- Ren, T.; Zheng, B.; Huang, Y.; Wang, S.; Bao, X.; Liu, K.; Guo, W. Osteosarcoma cell intrinsic PD-L2 signals promote invasion and metastasis via the RhoA-ROCK-LIMK2 and autophagy pathways. Cell Death Dis. 2019, 10, 261. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Xiong, Y.; Yue, J.; Hanley, S.J.; Watari, H. Tumor-intrinsic PD-L1 signaling in cancer initiation, development and treatment: Beyond immune evasion. Front. Oncol. 2018, 8, 386. [Google Scholar] [CrossRef] [PubMed]
- Almozyan, S.; Colak, D.; Mansour, F.; Alaiya, A.; Al-Harazi, O.; Qattan, A.; Al-Mohanna, F.; Al-Alwan, M.; Ghebeh, H. PD-L1 promotes OCT4 and Nanog expression in breast cancer stem cells by sustaining PI3K/AKT pathway activation. Int. J. Cancer 2017, 141, 1402–1412. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, F.; Zheng, Y.; Wang, X.; Wang, K.; Yu, Y.; Zhao, H. Propofol reduced mammosphere formation of breast cancer stem cells via PD-L1/Nanog in vitro. Oxidative Med. Cell. Longev. 2019, 2019, 9078209. [Google Scholar] [CrossRef] [PubMed]
- Tu, X.; Qin, B.; Zhang, Y.; Zhang, C.; Kahila, M.; Nowsheen, S.; Yin, P.; Yuan, J.; Pei, H.; Li, H. PD-L1 (B7-H1) competes with the RNA exosome to regulate the DNA damage response and can be targeted to sensitize to radiation or chemotherapy. Mol. Cell 2019, 74, 1215–1226.e1214. [Google Scholar] [CrossRef] [PubMed]
- Arigami, T.; Narita, N.; Mizuno, R.; Nguyen, L.; Ye, X.; Chung, A.; Giuliano, A.E.; Hoon, D.S. B7–H3 ligand expression by primary breast cancer and associated with regional nodal metastasis. Ann. Surg. 2010, 252, 1044–1051. [Google Scholar] [CrossRef] [PubMed]
- Castellanos, J.R.; Purvis, I.J.; Labak, C.M.; Guda, M.R.; Tsung, A.J.; Velpula, K.K.; Asuthkar, S. B7-H3 role in the immune landscape of cancer. Am. J. Clin. Exp. Immunol. 2017, 6, 66. [Google Scholar] [PubMed]
- Sun, J.; Guo, Y.-D.; Li, X.-N.; Zhang, Y.-Q.; Gu, L.; Wu, P.-P.; Bai, G.-H.; Xiao, Y. B7-H3 expression in breast cancer and upregulation of VEGF through gene silence. OncoTargets Ther. 2014, 7, 1979. [Google Scholar] [CrossRef] [PubMed]
- Flem-Karlsen, K.; Fodstad, Ø.; Nunes-Xavier, C.E. B7-H3 immune checkpoint protein in human cancer. Curr. Med. Chem. 2020, 27, 4062–4086. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, W.; Phillips, J.B.; Arora, R.; McClellan, S.; Li, J.; Kim, J.-H.; Sobol, R.W.; Tan, M. Immunoregulatory protein B7-H3 regulates cancer stem cell enrichment and drug resistance through MVP-mediated MEK activation. Oncogene 2019, 38, 88–102. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, J.; Marzese, D.M.; Wang, X.; Yang, Z.; Li, C.; Zhang, H.; Zhang, J.; Chen, C.C.; Kelly, D.F. B7H3 regulates differentiation and serves as a potential biomarker and theranostic target for human glioblastoma. Lab. Investig. 2019, 99, 1117–1129. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Tao, B.; Chen, Y.; Guo, Z.; Yang, X.; Peng, L.; Xia, X.; Chen, L. B7-H3 regulates glioma growth and cell invasion through a JAK2/STAT3/slug-dependent signaling pathway. OncoTargets Ther. 2020, 13, 2215. [Google Scholar] [CrossRef]
- Chapoval, A.I.; Ni, J.; Lau, J.S.; Wilcox, R.A.; Flies, D.B.; Liu, D.; Dong, H.; Sica, G.L.; Zhu, G.; Tamada, K. B7-H3: A costimulatory molecule for T cell activation and IFN-γ production. Nat. Immunol. 2001, 2, 269–274. [Google Scholar] [CrossRef]
- Prasad, D.V.; Nguyen, T.; Li, Z.; Yang, Y.; Duong, J.; Wang, Y.; Dong, C. Murine B7-H3 is a negative regulator of T cells. J. Immunol. 2004, 173, 2500–2506. [Google Scholar] [CrossRef]
- Picarda, E.; Ohaegbulam, K.C.; Zang, X. Molecular pathways: Targeting B7-H3 (CD276) for human cancer immunotherapy. Clin. Cancer Res. 2016, 22, 3425–3431. [Google Scholar] [CrossRef]
- Hammarén, H.M.; Virtanen, A.T.; Raivola, J.; Silvennoinen, O. The regulation of JAKs in cytokine signaling and its breakdown in disease. Cytokine 2019, 118, 48–63. [Google Scholar] [CrossRef] [PubMed]
- Bellucci, R.; Martin, A.; Bommarito, D.; Wang, K.; Hansen, S.H.; Freeman, G.J.; Ritz, J. Interferon-γ-induced activation of JAK1 and JAK2 suppresses tumor cell susceptibility to NK cells through upregulation of PD-L1 expression. Oncoimmunology 2015, 4, e1008824. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Tang, M.-Y.; Chen, W.; Wang, Z.; Wang, S.-L. High jak2 protein expression predicts a poor prognosis in patients with resectable pancreatic ductal adenocarcinoma. Dis. Mrk. 2020, 2020, 776560. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, T.; Miyamoto, M.; Aoyama, T.; Soyama, H.; Goto, T.; Hirata, J.; Suzuki, A.; Nagaoka, I.; Tsuda, H.; Furuya, K. JAK2/STAT3 pathway as a therapeutic target in ovarian cancers. Oncol. Lett. 2018, 15, 5772–5780. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Sun, W.; Yung, M.M.; Dai, S.; Cai, Y.; Chen, C.-W.; Meng, Y.; Lee, J.B.; Braisted, J.C.; Xu, Y. Autocrine activation of JAK2 by IL-11 promotes platinum drug resistance. Oncogene 2018, 37, 3981–3997. [Google Scholar] [CrossRef]
- Park, S.-Y.; Lee, C.-J.; Choi, J.-H.; Kim, J.-H.; Kim, J.-W.; Kim, J.-Y.; Nam, J.-S. The JAK2/STAT3/CCND2 Axis promotes colorectal Cancer stem cell persistence and radioresistance. J. Exp. Clin. Cancer Res. 2019, 38, 399. [Google Scholar] [CrossRef]
- Kim, J.W.; Gautam, J.; Kim, J.E.; Kim, J.; Kang, K.W. Inhibition of tumor growth and angiogenesis of tamoxifen-resistant breast cancer cells by ruxolitinib, a selective JAK2 inhibitor. Oncol. Lett. 2019, 17, 3981–3989. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, T.; Zou, S.; Jiang, B.; Hua, D. B7-H3 promotes cell migration and invasion through the Jak2/Stat3/MMP9 signaling pathway in colorectal cancer. Mol. Med. Rep. 2015, 12, 5455–5460. [Google Scholar] [CrossRef]
- Kang, F.-B.; Wang, L.; Jia, H.-C.; Li, D.; Li, H.-J.; Zhang, Y.-G.; Sun, D.-X. B7-H3 promotes aggression and invasion of hepatocellular carcinoma by targeting epithelial-to-mesenchymal transition via JAK2/STAT3/Slug signaling pathway. Cancer Cell Int. 2015, 15, 45. [Google Scholar] [CrossRef]
- Obsil, T.; Obsilova, V. Structure/function relationships underlying regulation of FOXO transcription factors. Oncogene 2008, 27, 2263–2275. [Google Scholar] [CrossRef]
- Zhang, X.; Jiang, L.; Liu, H. Forkhead Box Protein O1: Functional Diversity and Post-Translational Modification, a New Therapeutic Target? Drug Des. Dev. Ther. 2021, 15, 1851. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.; Wang, Y.; Xiao, W.; Pujado, S.P.; Xu, F.; Tian, C.; Xiao, E.; Choi, Y.; Graves, D.T. FOXO1 regulates dendritic cell activity through ICAM-1 and CCR7. J. Immunol. 2015, 194, 3745–3755. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-B.; Zhao, Z.-B.; Liu, Q.-Z.; Hu, T.-D.; Long, J.; Yan, K.; Lian, Z.-X. FoxO1 is a regulator of MHC-II expression and anti-tumor effect of tumor-associated macrophages. Oncogene 2018, 37, 1192–1204. [Google Scholar] [CrossRef] [PubMed]
- Bothur, E.; Raifer, H.; Haftmann, C.; Stittrich, A.-B.; Brüstle, A.; Brenner, D.; Bollig, N.; Bieringer, M.; Kang, C.-H.; Reinhard, K. Antigen receptor-mediated depletion of FOXP3 in induced regulatory T-lymphocytes via PTPN2 and FOXO1. Nat. Commun. 2015, 6, 8576. [Google Scholar] [CrossRef]
- Rao, R.R.; Li, Q.; Bupp, M.R.G.; Shrikant, P.A. Transcription factor Foxo1 represses T-bet-mediated effector functions and promotes memory CD8+ T cell differentiation. Immunity 2012, 36, 374–387. [Google Scholar] [CrossRef]
- Wu, Y.; Elshimali, Y.; Sarkissyan, M.; Mohamed, H.; Clayton, S.; Vadgama, J.V. Expression of FOXO1 is associated with GATA3 and Annexin-1 and predicts disease-free survival in breast cancer. Am. J. Cancer Res. 2012, 2, 104. [Google Scholar] [CrossRef]
- Gao, Z.; Liu, R.; Ye, N.; Liu, C.; Li, X.; Guo, X.; Zhang, Z.; Li, X.; Yao, Y.; Jiang, X. FOXO1 inhibits tumor cell migration via regulating cell surface morphology in non-small cell lung cancer cells. Cell. Physiol. Biochem. 2018, 48, 138–148. [Google Scholar] [CrossRef]
- Qiu, X.; Tan, G.; Wen, H.; Lian, L.; Xiao, S. Forkhead box O1 targeting replication factor C subunit 2 expression promotes glioma temozolomide resistance and survival. Ann. Transl. Med. 2021, 9, 692. [Google Scholar] [CrossRef]
- Zhang, H.; Xie, C.; Yue, J.; Jiang, Z.; Zhou, R.; Xie, R.; Wang, Y.; Wu, S. Cancer-associated fibroblasts mediated chemoresistance by a FOXO1/TGFβ1 signaling loop in esophageal squamous cell carcinoma. Mol. Carcinog. 2017, 56, 1150–1163. [Google Scholar] [CrossRef]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef]
- Chen, X.; Shao, Q.; Hao, S.; Zhao, Z.; Wang, Y.; Guo, X.; He, Y.; Gao, W.; Mao, H. CTLA-4 positive breast cancer cells suppress dendritic cells maturation and function. Oncotarget 2017, 8, 13703. [Google Scholar] [CrossRef]
- Navarrete-Bernal, M.G.; Cervantes-Badillo, M.G.; Martínez-Herrera, J.F.; Lara-Torres, C.O.; Gerson-Cwilich, R.; Zentella-Dehesa, A.; Ibarra-Sánchez, M.d.J.; Esparza-López, J.; Montesinos, J.J.; Cortés-Morales, V.A. Biological landscape of triple negative breast cancers expressing CTLA-4. Front. Oncol. 2020, 10, 1206. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Shi, Y.; Kim, B.C.; Jo, M.H.; Cruz, L.O.; Gou, Z.; Ha, T.; Lu, L.-F.; Reich, D.H.; Chen, Y. Force-dependent trans-endocytosis by breast cancer cells depletes costimulatory receptor CD80 and attenuates T cell activation. Biosens. Bioelectron. 2020, 165, 112389. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, M.; Cao, J.; Cai, G.; Li, X.; Liu, Y.; Chen, W. CTLA-4 promotes lymphoma progression through tumor stem cell enrichment and immunosuppression. Open Life Sci. 2021, 16, 909–919. [Google Scholar] [CrossRef]
- Zhang, B.; Dang, J.; Ba, D.; Wang, C.; Han, J.; Zheng, F. Potential function of CTLA-4 in the tumourigenic capacity of melanoma stem cells. Oncol. Lett. 2018, 16, 6163–6170. [Google Scholar] [CrossRef] [PubMed]
- Kleffel, S.; Posch, C.; Barthel, S.R.; Mueller, H.; Schlapbach, C.; Guenova, E.; Elco, C.P.; Lee, N.; Juneja, V.R.; Zhan, Q. Melanoma cell-intrinsic PD-1 receptor functions promote tumor growth. Cell 2015, 162, 1242–1256. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Yan, Y.; Dong, J.; Duan, L. PD-1 expression on uveal melanoma induces tumor proliferation and predicts poor patient survival. Int. J. Biol. Mrk. 2020, 35, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, X.; Liu, S.; Guo, L.; Zhang, B.; Zhang, J.; Ye, Q. PD-1 checkpoint blockade in combination with an mTOR inhibitor restrains hepatocellular carcinoma growth induced by hepatoma cell-intrinsic PD-1. Hepatology 2017, 66, 1920–1933. [Google Scholar] [CrossRef]
- Osta, E.; Gupta, H.B.; Zhang, D.; Kornepati, A.; Clark, C.A.; Curiel, T.J. Tumor cell-intrinsic programmed death protein 1 expression and induction in human cancer cell lines. Am Assoc. Immnol. 2018, 200, 178.33. [Google Scholar] [CrossRef]
- Pu, N.; Gao, S.; Yin, H.; Li, J.-a.; Wu, W.; Fang, Y.; Zhang, L.; Rong, Y.; Xu, X.; Wang, D. Cell-intrinsic PD-1 promotes proliferation in pancreatic cancer by targeting CYR61/CTGF via the hippo pathway. Cancer Lett. 2019, 460, 42–53. [Google Scholar] [CrossRef]
- Du, S.; McCall, N.; Park, K.; Guan, Q.; Fontina, P.; Ertel, A.; Zhan, T.; Dicker, A.P.; Lu, B. Blockade of tumor-expressed PD-1 promotes lung cancer growth. Oncoimmunology 2018, 7, e1408747. [Google Scholar] [CrossRef]
- Rom-Jurek, E.-M.; Kirchhammer, N.; Ugocsai, P.; Ortmann, O.; Wege, A.K.; Brockhoff, G. Regulation of programmed death ligand 1 (PD-L1) expression in breast cancer cell lines in vitro and in immunodeficient and humanized tumor mice. Int. J. Mol. Sci. 2018, 19, 563. [Google Scholar] [CrossRef] [PubMed]
- Zerdes, I.; Wallerius, M.; Sifakis, E.G.; Wallmann, T.; Betts, S.; Bartish, M.; Tsesmetzis, N.; Tobin, N.P.; Coucoravas, C.; Bergh, J. STAT3 activity promotes programmed-death ligand 1 expression and suppresses immune responses in breast cancer. Cancers 2019, 11, 1479. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Wang, X.; Ye, S.; Li, Y.; Chen, M.; Wang, S.; Qin, T.; Zhang, C.; Li, Y.; Long, Q. NPM1 upregulates the transcription of PD-L1 and suppresses T cell activity in triple-negative breast cancer. Nat. Commun. 2020, 11, 1669. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wu, Y.; Zhang, S.; Ma, P.; Jin, X.; Wang, Z.; Yao, M.; Zhang, E.; Tao, B.; Qin, Y. A tumor-specific super-enhancer drives immune evasion by guiding synchronous expression of PD-L1 and PD-L2. Cell Rep. 2019, 29, 3435–3447.e3434. [Google Scholar] [CrossRef]
- Yang, L.; Cai, Y.; Zhang, D.; Sun, J.; Xu, C.; Zhao, W.; Jiang, W.; Pan, C. miR-195/miR-497 regulate CD274 expression of immune regulatory ligands in triple-negative breast cancer. J. Breast Cancer 2018, 21, 371–381. [Google Scholar] [CrossRef]
- Fu, Y.; Liu, C.J.; Kobayashi, D.K.; Johanns, T.M.; Bowman-Kirigin, J.A.; Schaettler, M.O.; Mao, D.D.; Bender, D.; Kelley, D.G.; Uppaluri, R. GATA2 regulates constitutive PD-L1 and PD-L2 expression in brain tumors. Sci. Rep. 2020, 10, 9027. [Google Scholar] [CrossRef]
- Wu, Y.; Chen, M.; Wu, P.; Chen, C.; Xu, Z.P.; Gu, W. Increased PD-L1 expression in breast and colon cancer stem cells. Clin. Exp. Pharmacol. Physiol. 2017, 44, 602–604. [Google Scholar] [CrossRef]
- Azadi, S.; Es, H.A.; Bazaz, S.R.; Thiery, J.P.; Asadnia, M.; Warkiani, M.E. Upregulation of PD-L1 expression in breast cancer cells through the formation of 3D multicellular cancer aggregates under different chemical and mechanical conditions. Biochim. Et Biophys. Acta (BBA)-Mol. Cell Res. 2019, 1866, 118526. [Google Scholar] [CrossRef]
- Mansour, F.A.; Al-Mazrou, A.; Al-Mohanna, F.; Al-Alwan, M.; Ghebeh, H. PD-L1 is overexpressed on breast cancer stem cells through notch3/mTOR axis. Oncoimmunology 2020, 9, 1729299. [Google Scholar] [CrossRef]
- Lim, S.; Liu, H.; Da Silva, L.M.; Arora, R.; Liu, Z.; Phillips, J.B.; Schmitt, D.C.; Vu, T.; McClellan, S.; Lin, Y. Immunoregulatory protein B7-H3 reprograms glucose metabolism in cancer cells by ROS-mediated stabilization of HIF1α. Cancer Res. 2016, 76, 2231–2242. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Tekle, C.; Chen, Y.-W.; Kristian, A.; Zhao, Y.; Zhou, M.; Liu, Z.; Ding, Y.; Wang, B.; Mælandsmo, G.M. B7-H3 silencing increases paclitaxel sensitivity by abrogating Jak2/Stat3 phosphorylation. Mol. Cancer Ther. 2011, 10, 960–971. [Google Scholar] [CrossRef] [PubMed]
- Nunes-Xavier, C.E.; Karlsen, K.F.; Tekle, C.; Pedersen, C.; Øyjord, T.; Hongisto, V.; Nesland, J.M.; Tan, M.; Sahlberg, K.K.; Fodstad, Ø. Decreased expression of B7-H3 reduces the glycolytic capacity and sensitizes breast cancer cells to AKT/mTOR inhibitors. Oncotarget 2016, 7, 6891. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Zhang, T.; Liu, F.; Sun, Z.; Shi, H.; Hua, D.; Yang, C. The co-stimulatory molecule B7-H3 promotes the epithelial-mesenchymal transition in colorectal cancer. Oncotarget 2016, 7, 31755. [Google Scholar] [CrossRef] [PubMed]
- Lemke, D.; Pfenning, P.-N.; Sahm, F.; Klein, A.-C.; Kempf, T.; Warnken, U.; Schnölzer, M.; Tudoran, R.; Weller, M.; Platten, M. Costimulatory protein 4IgB7H3 drives the malignant phenotype of glioblastoma by mediating immune escape and invasiveness. Clin. Cancer Res. 2012, 18, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, Y.; Jia, L.; koo Kim, J.; Li, J.; Deng, P.; Zhang, W.; Krebsbach, P.H.; Wang, C.-Y. CD276 expression enables squamous cell carcinoma stem cells to evade immune surveillance. Cell Stem Cell 2021, 28, 1597–1613. [Google Scholar] [CrossRef]
- Shi, H.; Yang, Y. Identification of inhibitory immune checkpoints and relevant regulatory pathways in breast cancer stem cells. Cancer Med. 2021, 11, 3794–3807. [Google Scholar] [CrossRef]
- Balko, J.M.; Schwarz, L.J.; Luo, N.; Estrada, M.V.; Giltnane, J.M.; Dávila-González, D.; Wang, K.; Sánchez, V.; Dean, P.T.; Combs, S.E. Triple-negative breast cancers with amplification of JAK2 at the 9p24 locus demonstrate JAK2-specific dependence. Sci. Transl. Med. 2016, 8, 334ra53. [Google Scholar] [CrossRef]
- Chen, M.; Pockaj, B.; Andreozzi, M.; Barrett, M.T.; Krishna, S.; Eaton, S.; Niu, R.; Anderson, K.S. JAK2 and PD-L1 amplification enhance the dynamic expression of PD-L1 in triple-negative breast cancer. Clin. Breast Cancer 2018, 18, e1205–e1215. [Google Scholar] [CrossRef]
- Doheny, D.; Sirkisoon, S.; Carpenter, R.L.; Aguayo, N.R.; Regua, A.T.; Anguelov, M.; Manore, S.G.; Arrigo, A.; Jalboush, S.A.; Wong, G.L. Combined inhibition of JAK2-STAT3 and SMO-GLI1/tGLI1 pathways suppresses breast cancer stem cells, tumor growth, and metastasis. Oncogene 2020, 39, 6589–6605. [Google Scholar] [CrossRef]
- Zhao, Q.; Liu, Y.; Wang, T.; Yang, Y.; Ni, H.; Liu, H.; Guo, Q.; Xi, T.; Zheng, L. MiR-375 inhibits the stemness of breast cancer cells by blocking the JAK2/STAT3 signaling. Eur. J. Pharmacol. 2020, 884, 173359. [Google Scholar] [CrossRef]
- To, N.B.; Nguyen, Y.T.-K.; Moon, J.Y.; Ediriweera, M.K.; Cho, S.K. Pentadecanoic Acid, an Odd-Chain Fatty Acid, Suppresses the Stemness of MCF-7/SC Human Breast Cancer Stem-Like Cells through JAK2/STAT3 Signaling. Nutrients 2020, 12, 1663. [Google Scholar] [CrossRef]
- Marotta, L.L.; Almendro, V.; Marusyk, A.; Shipitsin, M.; Schemme, J.; Walker, S.R.; Bloushtain-Qimron, N.; Kim, J.J.; Choudhury, S.A.; Maruyama, R. The JAK2/STAT3 signaling pathway is required for growth of CD44+ CD24–stem cell–like breast cancer cells in human tumors. J. Clin. Investig. 2011, 121, 2723–2735. [Google Scholar] [CrossRef]
- Kim, M.S.; Jeong, J.; Seo, J.; Kim, H.-S.; Kim, S.-J.; Jin, W. Dysregulated JAK2 expression by TrkC promotes metastasis potential, and EMT program of metastatic breast cancer. Sci. Rep. 2016, 6, 33899. [Google Scholar] [CrossRef]
- Kim, J.-H.; Choi, H.S.; Kim, S.-L.; Lee, D.-S. The PAK1-Stat3 signaling pathway activates IL-6 gene transcription and human breast cancer stem cell formation. Cancers 2019, 11, 1527. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.-Y.; Huang, Z.-G.; Tang, Z.-Q.; Xiao, X.-L.; Zeng, J.-J.; Feng, Z.-B. FOXO1 and hsa-microRNA-204-5p affect the biologic behavior of MDA-MB-231 breast cancer cells. Int. J. Clin. Exp. Pathol. 2020, 13, 1146. [Google Scholar] [PubMed]
- Guttilla, I.K.; White, B.A. Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J. Biol. Chem. 2009, 284, 23204–23216. [Google Scholar] [CrossRef]
- Yu, F.; Jin, L.; Yang, G.; Ji, L.; Wang, F.; Lu, Z. Post-transcriptional repression of FOXO1 by QKI results in low levels of FOXO1 expression in breast cancer cells. Oncol. Rep. 2014, 31, 1459–1465. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, T.; Gao, C.; Lv, X.; Liu, K.; Song, H.; Xing, Y.; Xi, T. FOXO1 3′ UTR functions as a ceRNA in repressing the metastases of breast cancer cells via regulating miRNA activity. FEBS Lett. 2014, 588, 3218–3224. [Google Scholar] [CrossRef] [PubMed]
- Vaziri-Gohar, A.; Zheng, Y.; Houston, K.D. IGF-1 receptor modulates FoxO1-mediated tamoxifen response in breast cancer cells. Mol. Cancer Res. 2017, 15, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Han, C.-Y.; Cho, K.-B.; Choi, H.-S.; Han, H.-K.; Kang, K.-W. Role of FoxO1 activation in MDR1 expression in adriamycin-resistant breast cancer cells. Carcinogenesis 2008, 29, 1837–1844. [Google Scholar] [CrossRef]
- Zou, Y.; Lin, X.; Bu, J.; Lin, Z.; Chen, Y.; Qiu, Y.; Mo, H.; Tang, Y.; Fang, W.; Wu, Z. Timeless-stimulated miR-5188-FOXO1/β-catenin-c-Jun feedback loop promotes stemness via ubiquitination of β-catenin in breast cancer. Mol. Ther. 2020, 28, 313–327. [Google Scholar] [CrossRef]
- Truong, T.H.; Dwyer, A.R.; Diep, C.H.; Hu, H.; Hagen, K.M.; Lange, C.A. Phosphorylated progesterone receptor isoforms mediate opposing stem cell and proliferative breast cancer cell fates. Endocrinology 2019, 160, 430–446. [Google Scholar] [CrossRef]
- Yu, J.-M.; Sun, W.; Wang, Z.-H.; Liang, X.; Hua, F.; Li, K.; Lv, X.-X.; Zhang, X.-W.; Liu, Y.-Y.; Yu, J.-J. TRIB3 supports breast cancer stemness by suppressing FOXO1 degradation and enhancing SOX2 transcription. Nat. Commun. 2019, 10, 5720. [Google Scholar] [CrossRef]
- Hinterneder, J.; Carlstrom, J.; Carlson, A.; Nida, D.; Hopkinton, M. Rapid, No-Wash Measurement of Immune Checkpoint Molecules and Cytokines in Co-Cultures of Immune Cells and Cancer Cell Lines; PerkinElmer Application Note; PerkinElmer: Hopkinton, MA, USA, 2018. [Google Scholar]
- Baruch, E.N.; Ortenberg, R.; Avivi, C.; Anafi, L.; Dick-Necula, D.; Stossel, C.; Moshkovits, Y.; Itzhaki, O.; Besser, M.J.; Schachter, J. Immune co-culture cell microarray–a feasible tool for high-throughput functional investigation of lymphocyte–cancer interactions. OncoImmunology 2020, 9, 1741267. [Google Scholar] [CrossRef]
- Shklovskaya, E.; Rizos, H. spatial and temporal changes in PD-L1 expression in cancer: The role of genetic drivers, tumor microenvironment and resistance to therapy. Int. J. Mol. Sci. 2020, 21, 7139. [Google Scholar] [CrossRef] [PubMed]
- Mo, X.; Zhang, H.; Preston, S.; Martin, K.; Zhou, B.; Vadalia, N.; Gamero, A.M.; Soboloff, J.; Tempera, I.; Zaidi, M.R. Interferon-γ signaling in melanocytes and melanoma cells regulates expression of CTLA-4. Cancer Res. 2018, 78, 436–450. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Fang, Y.C.; Li, J. PD-L1 expression levels on tumor cells affect their immunosuppressive activity. Oncol. Lett. 2019, 18, 5399–5407. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Liu, Q.; Yang, J.; Ren, Q.; Cao, W.; Yang, J.; Yu, Z.; Yu, F.; Wu, Y.; Shi, H. Co-culture of apoptotic breast cancer cells with immature dendritic cells: A novel approach for DC-based vaccination in breast cancer. Braz. J. Med. Biol. Res. 2012, 45, 510–515. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montoyo-Pujol, Y.G.; García-Escolano, M.; Ponce, J.J.; Delgado-García, S.; Martín, T.A.; Ballester, H.; Castellón-Molla, E.; Martínez-Peinado, P.; Pascual-García, S.; Sempere-Ortells, J.M.; et al. Variable Intrinsic Expression of Immunoregulatory Biomarkers in Breast Cancer Cell Lines, Mammospheres, and Co-Cultures. Int. J. Mol. Sci. 2023, 24, 4478. https://doi.org/10.3390/ijms24054478
Montoyo-Pujol YG, García-Escolano M, Ponce JJ, Delgado-García S, Martín TA, Ballester H, Castellón-Molla E, Martínez-Peinado P, Pascual-García S, Sempere-Ortells JM, et al. Variable Intrinsic Expression of Immunoregulatory Biomarkers in Breast Cancer Cell Lines, Mammospheres, and Co-Cultures. International Journal of Molecular Sciences. 2023; 24(5):4478. https://doi.org/10.3390/ijms24054478
Chicago/Turabian StyleMontoyo-Pujol, Yoel Genaro, Marta García-Escolano, José J. Ponce, Silvia Delgado-García, Tina Aurora Martín, Hortensia Ballester, Elena Castellón-Molla, Pascual Martínez-Peinado, Sandra Pascual-García, José Miguel Sempere-Ortells, and et al. 2023. "Variable Intrinsic Expression of Immunoregulatory Biomarkers in Breast Cancer Cell Lines, Mammospheres, and Co-Cultures" International Journal of Molecular Sciences 24, no. 5: 4478. https://doi.org/10.3390/ijms24054478
APA StyleMontoyo-Pujol, Y. G., García-Escolano, M., Ponce, J. J., Delgado-García, S., Martín, T. A., Ballester, H., Castellón-Molla, E., Martínez-Peinado, P., Pascual-García, S., Sempere-Ortells, J. M., & Peiró, G. (2023). Variable Intrinsic Expression of Immunoregulatory Biomarkers in Breast Cancer Cell Lines, Mammospheres, and Co-Cultures. International Journal of Molecular Sciences, 24(5), 4478. https://doi.org/10.3390/ijms24054478






