Label-Free Multiplexed Microfluidic Analysis of Protein Interactions Based on Photonic Crystal Surface Mode Imaging
Abstract
1. Introduction
2. Results and Discussion
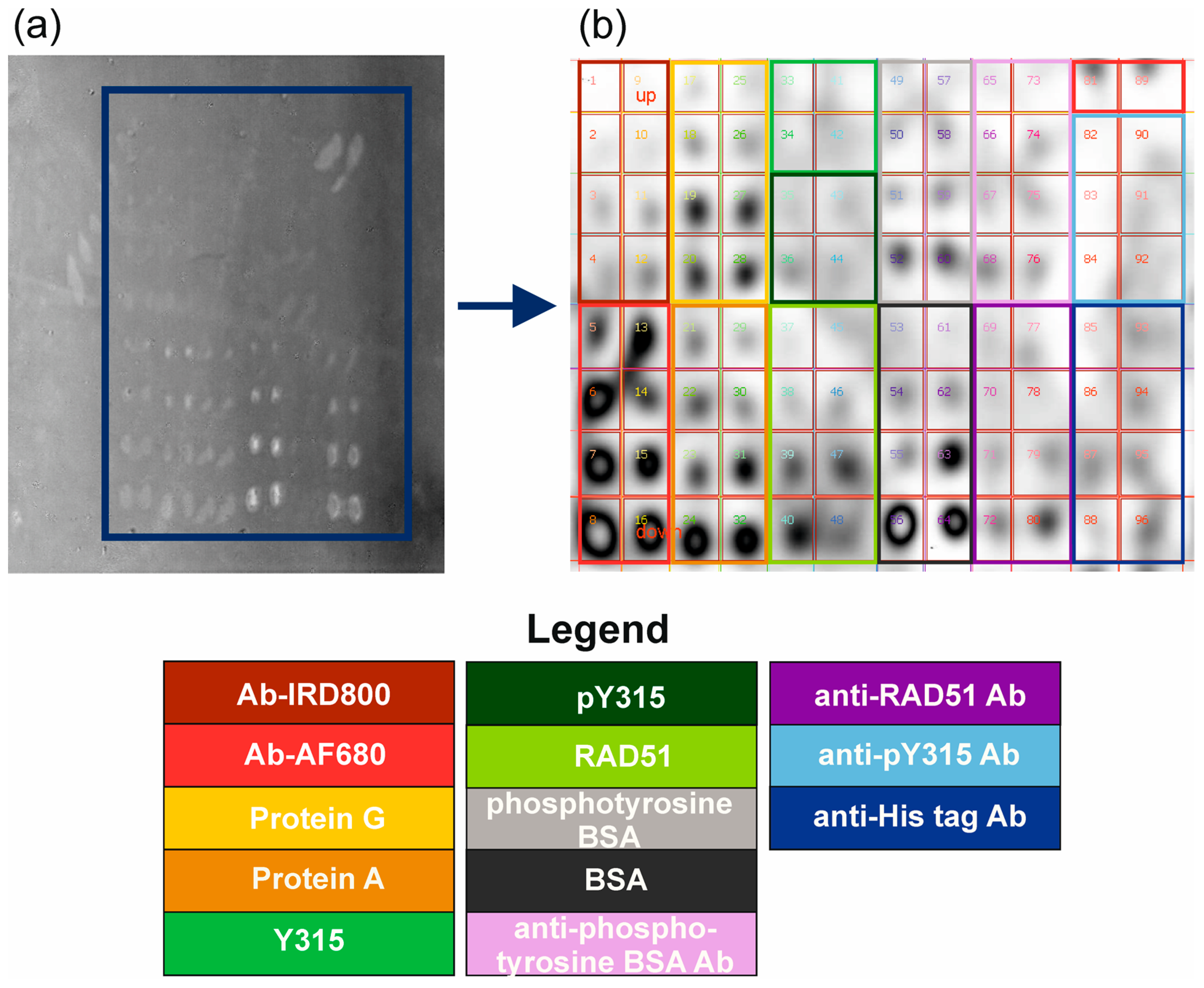
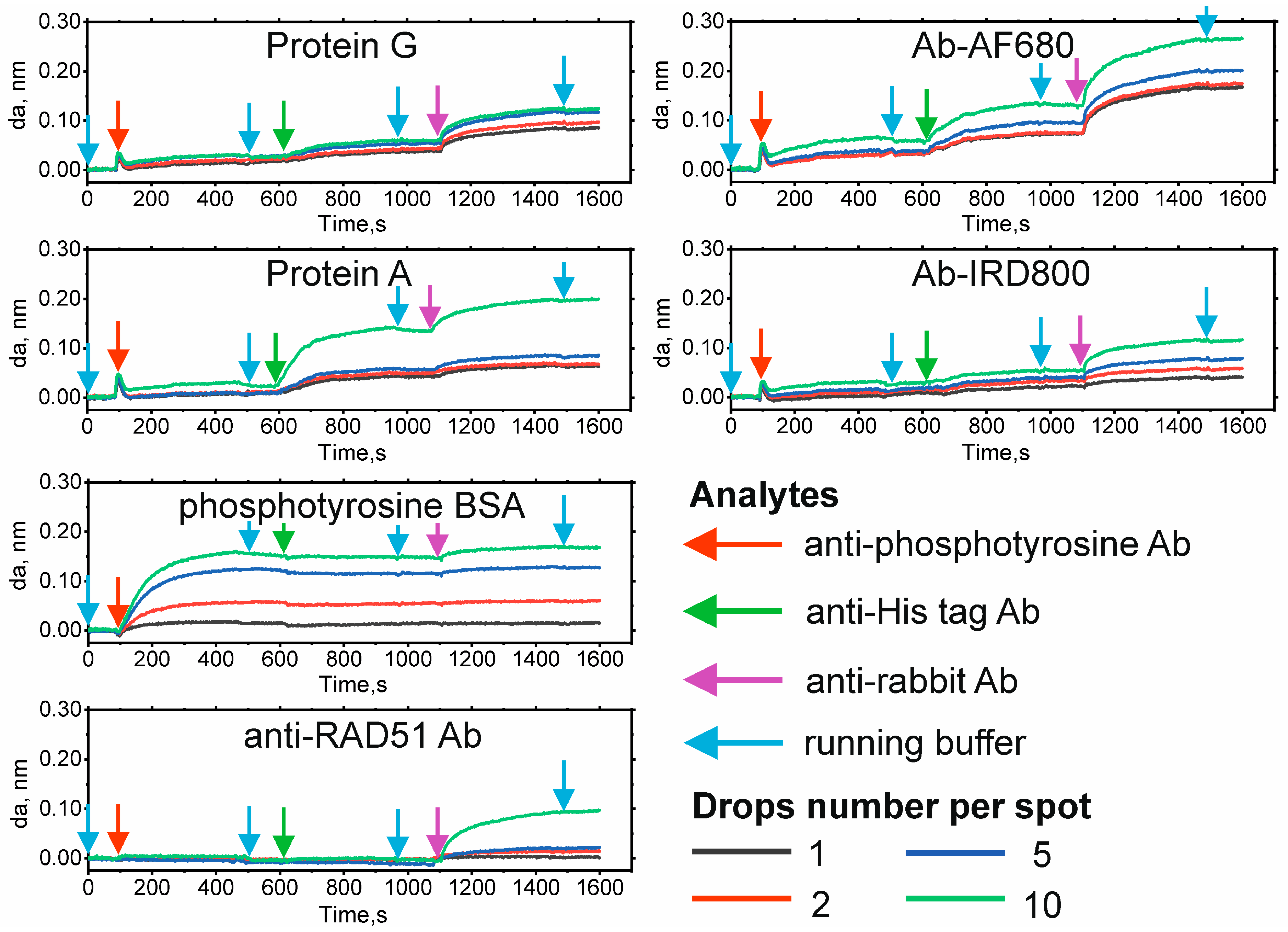
2.1. Chip Biofunctionalization and Possible Protein Interactions
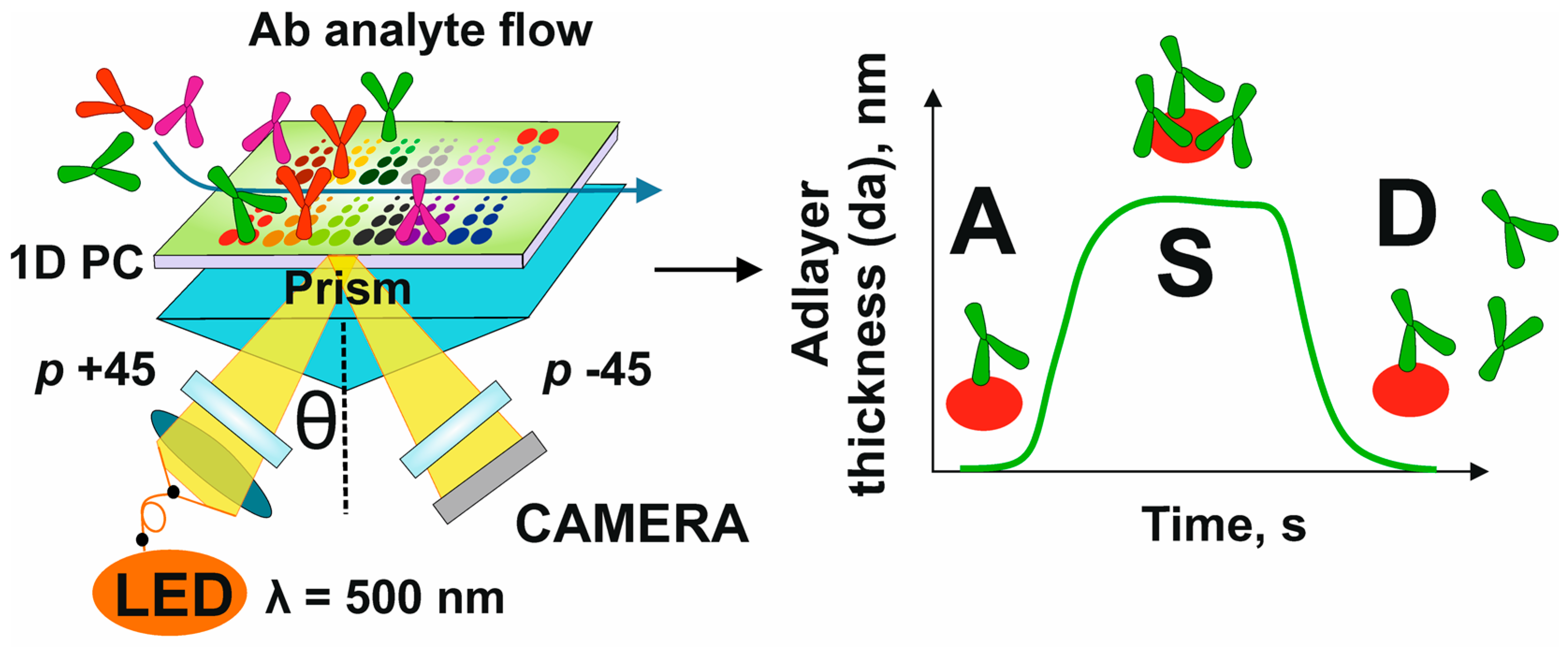
2.2. Photonic Crystal Surface Mode Imaging
3. Materials and Methods
3.1. Materials
3.2. Photonic Crystal Chip Fabrication
3.3. Chip Surface Modification
3.4. Protein Spotting and Chip Biofunctionalization
3.5. Photonic Crystal Surface Mode Imaging
3.6. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shi, L.; Gehin, T.; Chevolot, Y.; Souteyrand, E.; Mangé, A.; Solassol, J.; Laurenceau, E. Anti-heat shock protein autoantibody profiling in breast cancer using customized protein microarray. Anal. Bioanal. Chem. 2016, 408, 1497–1506. [Google Scholar] [CrossRef]
- Matsuda, K.M.; Yoshizaki, A.; Yamaguchi, K.; Fukuda, E.; Okumura, T.; Ogawa, K.; Ono, C.; Norimatsu, Y.; Kotani, H.; Hisamoto, T.; et al. Autoantibody landscape revealed by wet protein array: Sum of autoantibody levels reflects disease status. Front. Immunol. 2022, 13, 893086. [Google Scholar] [CrossRef]
- Jayaraman, V.; Krishna, K.; Yang, Y.; Rajasekaran, K.J.; Ou, Y.; Wang, T.; Bei, K.; Krishnamurthy, H.K.; Rajasekaran, J.J.; Rai, A.J.; et al. An ultra-high-density protein microarray for high throughput single-tier serological detection of Lyme disease. Sci. Rep. 2020, 10, 18085. [Google Scholar] [CrossRef]
- Pauly, F.; Dexlin-Mellby, L.; Ek, S.; Ohlin, M.; Olsson, N.; Jirström, K.; Dictor, M.; Schoenmakers, S.; Borrebaeck, C.A.K.; Wingren, C. Protein expression profiling of formalin-fixed paraffin-embedded tissue using recombinant antibody microarrays. J. Proteome Res. 2013, 12, 5943–5953. [Google Scholar] [CrossRef]
- Heiss, K.; Heidepriem, J.; Fischer, N.; Weber, L.K.; Dahlke, C.; Jaenisch, T.; Loeffler, F.F. Rapid response to pandemic threats: Immunogenic epitope detection of pandemic pathogens for diagnostics and vaccine development using peptide microarrays. J. Proteome Res. 2020, 19, 4339–4354. [Google Scholar] [CrossRef]
- Phung, N.L.; Walter, J.G.; Jonczyk, R.; Seiler, L.K.; Scheper, T.; Blume, C. Development of an aptamer-based lateral flow assay for the detection of C-reactive protein using microarray technology as a prescreening platform. ACS Comb. Sci. 2020, 22, 617–629. [Google Scholar] [CrossRef]
- Yuan, J.; Wang, E.; Fox, B.A. Immune monitoring technology primer: Protein microarray (‘seromics’). J. Immunother. Cancer 2016, 4, 2. [Google Scholar] [CrossRef]
- Verardo, D.; Liljedahl, L.; Richter, C.; Agnarsson, B.; Axelsson, U.; Prinz, C.N.; Höök, F.; Borrebaeck, C.A.K.; Linke, H. Fluorescence signal enhancement in antibody microarrays using lightguiding nanowires. Nanomaterials 2021, 11, 227. [Google Scholar] [CrossRef]
- Liang, C.; Luan, J.; Wang, Z.; Jiang, Q.; Gupta, R.; Cao, S.; Liu, K.-K.; Morrissey, J.J.; Kharasch, E.D.; Naik, R.R.; et al. Gold nanorod size-dependent fluorescence enhancement for ultrasensitive fluoroimmunoassays. ACS Appl. Mater. Interfaces 2021, 13, 11414–11423. [Google Scholar] [CrossRef]
- Sauer, U. Analytical protein microarrays: Advancements towards clinical applications. Sensors 2017, 17, 256. [Google Scholar] [CrossRef]
- Vinogradov, A.P.; Dorofeenko, A.V.; Pukhov, A.A.; Lisyansky, A.A. Exciting surface plasmon polaritons in the Kretschmann configuration by a light beam. Phys. Rev. B 2018, 97, 235407. [Google Scholar] [CrossRef]
- Špačková, B.; Lynn, N.S.; Slabý, J.; Šípová, H.; Homola, J. A route to superior performance of a nanoplasmonic biosensor: Consideration of both photonic and mass transport aspects. ACS Photonics 2018, 5, 1019–1025. [Google Scholar] [CrossRef]
- Miyazaki, C.M.; Shimizu, F.M.; Ferreira, M. Surface plasmon resonance (SPR) for sensors and biosensors. Nanocharacterization Techniques. In Micro and Nano Technologies; Da Róz, A.L., Ferreira, M., de Lima Leite, F., Oliveira, O.N., Eds.; William Andrew Publishing: Norwich, UK, 2017; pp. 183–200. [Google Scholar]
- Fasoli, J.B.; Corn, R.M. Surface enzyme chemistries for ultrasensitive microarray biosensing with SPR imaging. Langmuir 2015, 31, 9527–9536. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, S.; Yin, L.; Yang, Y.; Guan, Y.; Wang, W.; Xu, H.; Tao, N. Quantification of epidermal growth factor receptor expression level and binding kinetics on cell surfaces by surface plasmon resonance imaging. Anal. Chem. 2015, 87, 9960–9965. [Google Scholar] [CrossRef]
- Nelson, B.P.; Grimsrud, T.E.; Liles, M.R.; Goodman, R.M.; Corn, R.M. Surface plasmon resonance imaging measurements of DNA and RNA hybridization adsorption onto DNA microarrays. Anal. Chem. 2001, 73, 1–7. [Google Scholar] [CrossRef]
- Scarano, S.; Mariani, S.; Minunni, M. SPR-based affinity biosensors as innovative analytical devices. J. Lightwave Technol. 2015, 33, 3374–3384. [Google Scholar] [CrossRef]
- Singh, P. SPR biosensors: Historical perspectives and current challenges. Sens. Actuators B 2016, 229, 110–130. [Google Scholar] [CrossRef]
- Konopsky, V.N. Long-range surface plasmons on duplex metal nanolayers. Photonics Nanostructures Fundam. Appl. 2020, 39, 100788. [Google Scholar] [CrossRef]
- Shinn, M.; Robertson, W.M. Surface plasmon-like sensor based on surface electromagnetic waves in a photonic band-gap material. Sens. Actuators B 2005, 105, 360–364. [Google Scholar] [CrossRef]
- Bellingeri, M.; Chiasera, A.; Kriegel, I.; Scotognella, F. Optical properties of periodic, quasi-periodic, and disordered one-dimensional photonic structures. Opt. Mater. 2017, 72, 403–421. [Google Scholar] [CrossRef]
- Konopsky, V.; Mitko, T.; Aldarov, K.; Alieva, E.; Basmanov, D.; Moskalets, A.; Matveeva, A.; Morozova, O.; Klinov, D. Photonic Crystal surface mode imaging for multiplexed and high-throughput label-free biosensing. Biosens. Bioelectron. 2020, 168, 112575. [Google Scholar] [CrossRef]
- Konopsky, V.N.; Alieva, E.V. Photonic crystal surface waves for optical biosensors. Anal. Chem. 2007, 79, 4729–4735. [Google Scholar] [CrossRef]
- Petrova, I.; Konopsky, V.; Nabiev, I.; Sukhanova, A. label-free flow multiplex biosensing via photonic crystal surface mode detection. Sci. Rep. 2019, 9, 8745. [Google Scholar] [CrossRef]
- Konopsky, V.N.; Karakouz, T.; Alieva, E.V.; Vicario, C.; Sekatskii, S.K.; Dietler, G. Photonic crystal biosensor based on optical surface waves. Sensors 2013, 13, 2566–2578. [Google Scholar] [CrossRef]
- Konopsky, V.N.; Alieva, E.V. Photonic crystal surface mode imaging biosensor based on wavelength interrogation of resonance peak. Sens. Actuators B 2018, 276, 271–278. [Google Scholar] [CrossRef]
- Pavlichenko, I.; Broda, E.; Fukuda, Y.; Szendrei, K.; Hatz, A.K.; Scarpa, G.; Lugli, P.; Bräuchle, C.; Lotsch, B.V. Bringing One-Dimensional Photonic Crystals to a New Light: An electrophotonic platform for chemical mass transport visualisation and cell monitoring. Mater. Horiz. 2015, 2, 299–308. [Google Scholar] [CrossRef]
- Romano, S.; Lamberti, A.; Masullo, M.; Penzo, E.; Cabrini, S.; Rendina, I.; Mocella, V. Optical biosensors based on photonic crystals supporting bound states in the continuum. Materials 2018, 11, 526. [Google Scholar] [CrossRef]
- Martínez-Pérez, P.; Gómez-Gómez, M.; Angelova, T.; Griol, A.; Hurtado, J.; Bellieres, L.; García-Rupérez, J. Continuous detection of increasing concentrations of thrombin employing a label-free photonic crystal aptasensor. Micromachines 2020, 11, 464. [Google Scholar] [CrossRef]
- Sinibaldi, A.; Danz, N.; Munzert, P.; Michelotti, F. Hybrid inorganic/organic photonic crystal biochips for cancer biomarkers detection. Opt. Laser Technol. 2018, 102, 227–232. [Google Scholar] [CrossRef]
- Huang, C.-S.; Chaudhery, V.; Pokhriyal, A.; George, S.; Polans, J.; Lu, M.; Tan, R.; Zangar, R.C.; Cunningham, B.T. Multiplexed cancer biomarker detection using quartz-based photonic crystal surfaces. Anal. Chem. 2012, 84, 1126–1133. [Google Scholar] [CrossRef]
- Sinibaldi, A.; Danz, N.; Descrovi, E.; Munzert, P.; Schulz, U.; Sonntag, F.; Dominici, L.; Michelotti, F. Direct comparison of the performance of bloch surface wave and surface plasmon polariton sensors. Sens. Actuators B 2012, 174, 292–298. [Google Scholar] [CrossRef]
- Khan, S.; Burciu, B.; Filipe, C.D.M.; Li, Y.; Dellinger, K.; Didar, T.F. DNAzyme-based biosensors: Immobilization strategies, applications, and future prospective. ACS Nano 2021, 15, 13943–13969. [Google Scholar] [CrossRef]
- Alshanski, I.; Shitrit, A.; Sukhran, Y.; Unverzagt, C.; Hurevich, M.; Yitzchaik, S. Effect of interfacial properties on impedimetric biosensing of the sialylation process with a biantennary N-glycan-based monolayer. Langmuir 2022, 38, 849–855. [Google Scholar] [CrossRef]
- Rizzo, R.; Alvaro, M.; Danz, N.; Napione, L.; Descrovi, E.; Schmieder, S.; Sinibaldi, A.; Chandrawati, R.; Rana, S.; Munzert, P.; et al. Bloch surface wave label-free and fluorescence platform for the detection of VEGF biomarker in biological matrices. Sens. Actuators B 2018, 255, 2143–2150. [Google Scholar] [CrossRef]
- Fiorilli, S.; Rivolo, P.; Descrovi, E.; Ricciardi, C.; Pasquardini, L.; Lunelli, L.; Vanzetti, L.; Pederzolli, C.; Onida, B.; Garrone, E. Vapor-phase self-assembled monolayers of aminosilane on plasma-activated silicon substrates. J. Colloid Interface Sci. 2008, 321, 235–241. [Google Scholar] [CrossRef]
- Mu, Z.; Zhao, X.; Huang, Y.; Lu, M.; Gu, Z. Photonic crystal hydrogel enhanced plasmonic staining for multiplexed protein analysis. Small 2015, 11, 6036–6043. [Google Scholar] [CrossRef]
- Slupianek, A.; Dasgupta, Y.; Ren, S.; Gurdek, E.; Donlin, M.; Nieborowska-Skorska, M.; Fleury, F.; Skorski, T. Targeting RAD51 phosphotyrosine-315 to prevent unfaithful recombination repair in BCR-ABL1 leukemia. Blood 2011, 118, 1062–1068. [Google Scholar] [CrossRef]
- Popova, M.; Shimizu, H.; Yamamoto, K.; Lebechec, M.; Takahashi, M.; Fleury, F. Detection of C-Abl kinase-promoted phosphorylation of Rad51 by specific antibodies reveals that Y54 phosphorylation is dependent on that of Y315. FEBS Lett. 2009, 583, 1867–1872. [Google Scholar] [CrossRef]
- Choe, W.; Durgannavar, T.A.; Chung, S.J. Fc-binding ligands of immunoglobulin G: An overview of high affinity proteins and peptides. Materials 2016, 9, 994. [Google Scholar] [CrossRef]
- Fishman, J.B.; Berg, E.A. Protein A and protein G purification of antibodies. Cold Spring Harb. Protoc. 2019, 1, 331–344. [Google Scholar] [CrossRef]
- Guner, H.; Ozgur, E.; Kokturk, G.; Celik, M.; Esen, E.; Topal, A.E.; Ayas, S.; Uludag, Y.; Elbuken, C.; Dana, A. A smartphone-based surface plasmon resonance imaging (SPRi) platform for on-site biodetection. Sens. Actuators B 2017, 239, 571–577. [Google Scholar] [CrossRef]
- Malinick, A.S.; Lambert, A.S.; Stuart, D.D.; Li, B.; Puente, E.; Cheng, Q. Detection of multiple sclerosis biomarkers in serum by ganglioside microarrays and surface plasmon resonance imaging. ACS Sens. 2020, 5, 3617–3626. [Google Scholar] [CrossRef]
- Yano, T.; Kajisa, T.; Ono, M.; Miyasaka, Y.; Hasegawa, Y.; Saito, A.; Otsuka, K.; Sakane, A.; Sasaki, T.; Yasutomo, K.; et al. Ultrasensitive detection of SARS-CoV-2 nucleocapsid protein using large gold nanoparticle-enhanced surface plasmon resonance. Sci. Rep. 2022, 12, 1060. [Google Scholar] [CrossRef]
- Hendriks, J.; Stojanovic, I.; Schasfoort, R.B.M.; Saris, D.B.F.; Karperien, M. Nanoparticle enhancement cascade for sensitive multiplex measurements of biomarkers in complex fluids with surface plasmon resonance imaging. Anal. Chem. 2018, 90, 6563–6571. [Google Scholar] [CrossRef]
- Lopes, L.C.; Santos, A.; Bueno, P.R. An outlook on electrochemical approaches for molecular diagnostics assays and discussions on the limitations of miniaturized technologies for point-of-care devices. Sens. Actuators Rep. 2022, 4, 100087. [Google Scholar] [CrossRef]
- Mujahid, A.; Mustafa, G.; Dickert, F.L. Label-free bioanalyte detection from nanometer to micrometer dimensions—Molecular imprinting and QCMs. Biosensors 2018, 8, 52. [Google Scholar] [CrossRef]
- Biswas, S.; Lan, Q.; Xie, Y.; Sun, X.; Wang, Y. Label-free electrochemical immunosensor for ultrasensitive detection of carbohydrate antigen 125 based on antibody-immobilized biocompatible MOF-808/CNT. ACS Appl. Mater. Interfaces 2021, 13, 3295–3302. [Google Scholar] [CrossRef]
- Jozghorbani, M.; Fathi, M.; Kazemi, S.H.; Alinejadian, N. Determination of carcinoembryonic antigen as a tumor marker using a novel graphene-based label-free electrochemical immunosensor. Anal. Biochem. 2021, 613, 114017. [Google Scholar] [CrossRef]
- Magliulo, M.; De Tullio, D.; Vikholm-Lundin, I.; Albers, W.M.; Munter, T.; Manoli, K.; Palazzo, G.; Torsi, L. Label-free C-reactive protein electronic detection with an electrolyte-gated organic field-effect transistor-based immunosensor. Anal. Bioanal. Chem. 2016, 408, 3943–3952. [Google Scholar] [CrossRef]
- Filipiak, M.S.; Rother, M.; Andoy, N.M.; Knudsen, A.C.; Grimm, S.; Bachran, C.; Swee, L.K.; Zaumseil, J.; Tarasov, A. Highly sensitive, selective and label-free protein detection in physiological solutions using carbon nanotube transistors with nanobody receptors. Sens. Actuators B 2018, 255, 1507–1516. [Google Scholar] [CrossRef]
- Genco, E.; Fattori, M.; Harpe, P.J.A.; Modena, F.; Viola, F.A.; Caironi, M.; Wheeler, M.; Fichet, G.; Torricelli, F.; Sarcina, L.; et al. A 4 × 4 biosensor array with a 42-μw/channel multiplexed current sensitive front-end featuring 137-DB DR and zeptomolar sensitivity. IEEE Open J. Solid-State Circuits Soc. 2022, 2, 193–207. [Google Scholar] [CrossRef]
- Velic, D.; Demeyer, A.; Peterlini, T.; Benhelli-Mokrani, H.; Mathé-Allainmat, M.; Masson, J.-Y.; Fleury, F. Molecular determinant of DIDS analogs targeting RAD51 activity. Molecules 2021, 26, 5460. [Google Scholar] [CrossRef]
- Konopsky, V.N.; Alieva, E.V. Imaging biosensor based on planar optical waveguide. Opt. Laser Technol. 2019, 115, 171–175. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nifontova, G.; Petrova, I.; Gerasimovich, E.; Konopsky, V.N.; Ayadi, N.; Charlier, C.; Fleury, F.; Karaulov, A.; Sukhanova, A.; Nabiev, I. Label-Free Multiplexed Microfluidic Analysis of Protein Interactions Based on Photonic Crystal Surface Mode Imaging. Int. J. Mol. Sci. 2023, 24, 4347. https://doi.org/10.3390/ijms24054347
Nifontova G, Petrova I, Gerasimovich E, Konopsky VN, Ayadi N, Charlier C, Fleury F, Karaulov A, Sukhanova A, Nabiev I. Label-Free Multiplexed Microfluidic Analysis of Protein Interactions Based on Photonic Crystal Surface Mode Imaging. International Journal of Molecular Sciences. 2023; 24(5):4347. https://doi.org/10.3390/ijms24054347
Chicago/Turabian StyleNifontova, Galina, Irina Petrova, Evgeniia Gerasimovich, Valery N. Konopsky, Nizar Ayadi, Cathy Charlier, Fabrice Fleury, Alexander Karaulov, Alyona Sukhanova, and Igor Nabiev. 2023. "Label-Free Multiplexed Microfluidic Analysis of Protein Interactions Based on Photonic Crystal Surface Mode Imaging" International Journal of Molecular Sciences 24, no. 5: 4347. https://doi.org/10.3390/ijms24054347
APA StyleNifontova, G., Petrova, I., Gerasimovich, E., Konopsky, V. N., Ayadi, N., Charlier, C., Fleury, F., Karaulov, A., Sukhanova, A., & Nabiev, I. (2023). Label-Free Multiplexed Microfluidic Analysis of Protein Interactions Based on Photonic Crystal Surface Mode Imaging. International Journal of Molecular Sciences, 24(5), 4347. https://doi.org/10.3390/ijms24054347







