Research Progresses and Applications of Fluorescent Protein Antibodies: A Review Focusing on Nanobodies
Abstract
1. Introduction
2. Overview of Fluorescent Proteins
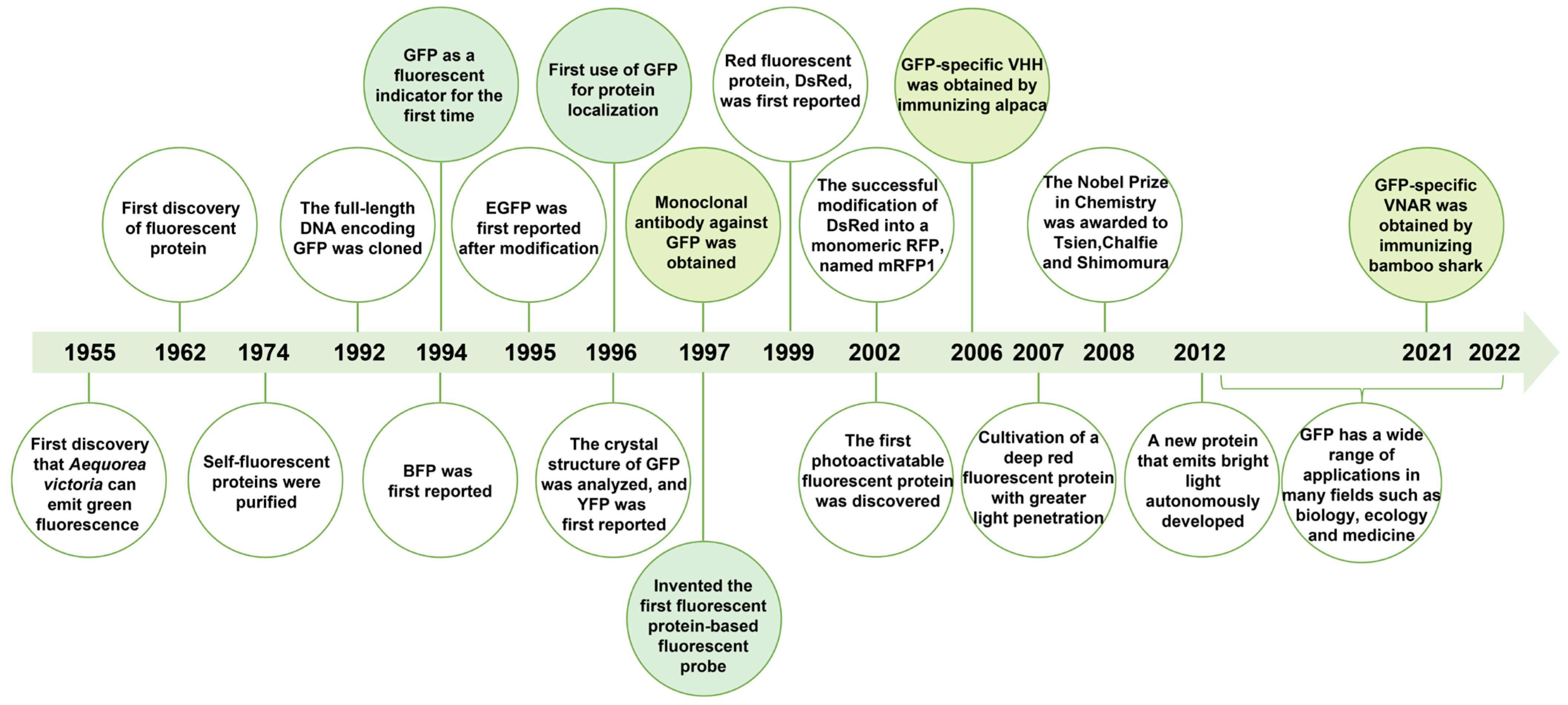
2.1. The Discovery of Fluorescent Proteins
2.2. Structure and Functional Properties of GFP
2.3. GFP Derivatives
2.4. RFP Derivatives
2.5. Near-Infrared FPs
3. Research Progresses on Fluorescent Protein Antibody
3.1. Monoclonal Antibody Targeting Fluorescent Proteins
3.2. Nanobody Targeting Fluorescent Proteins
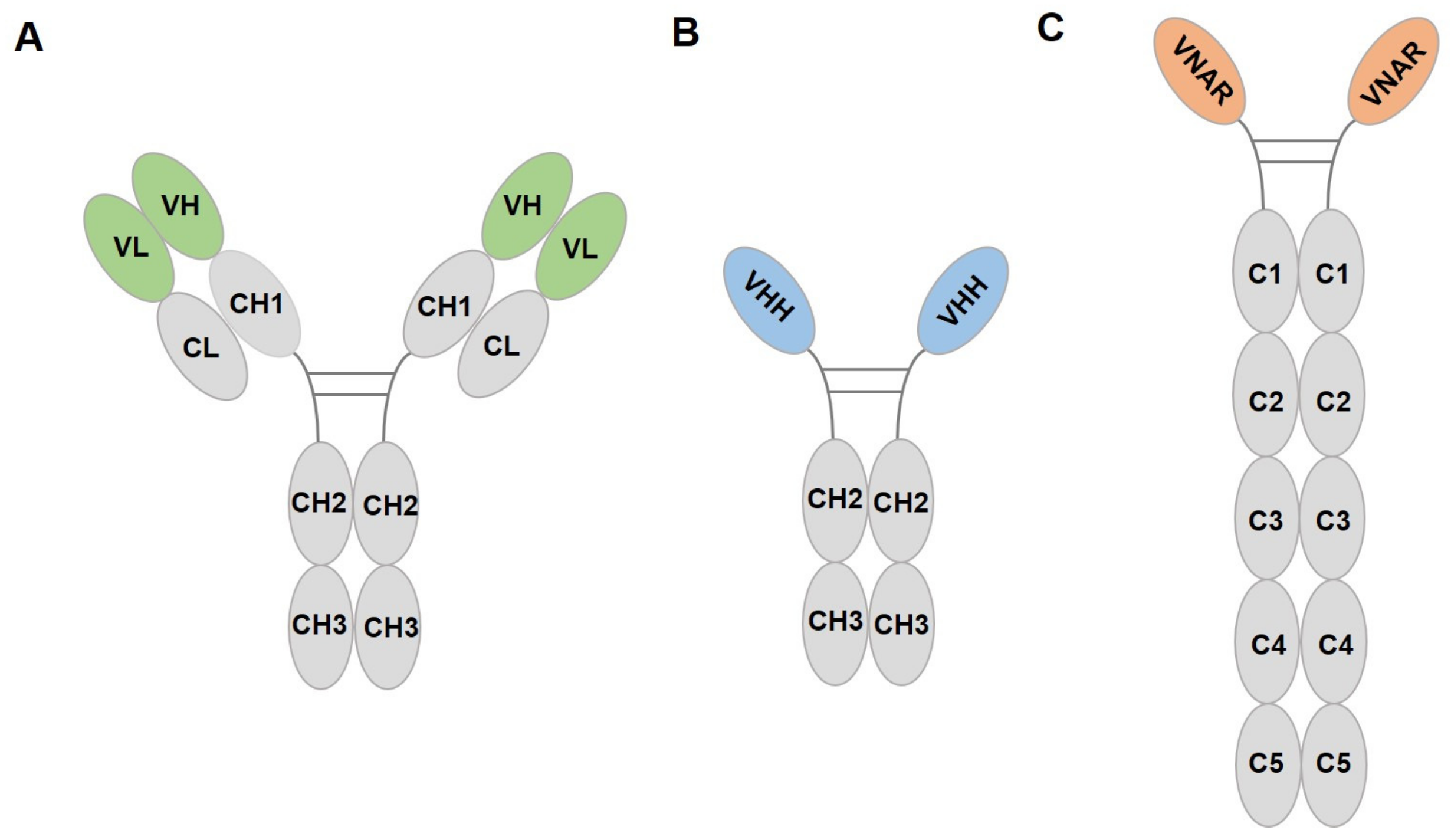
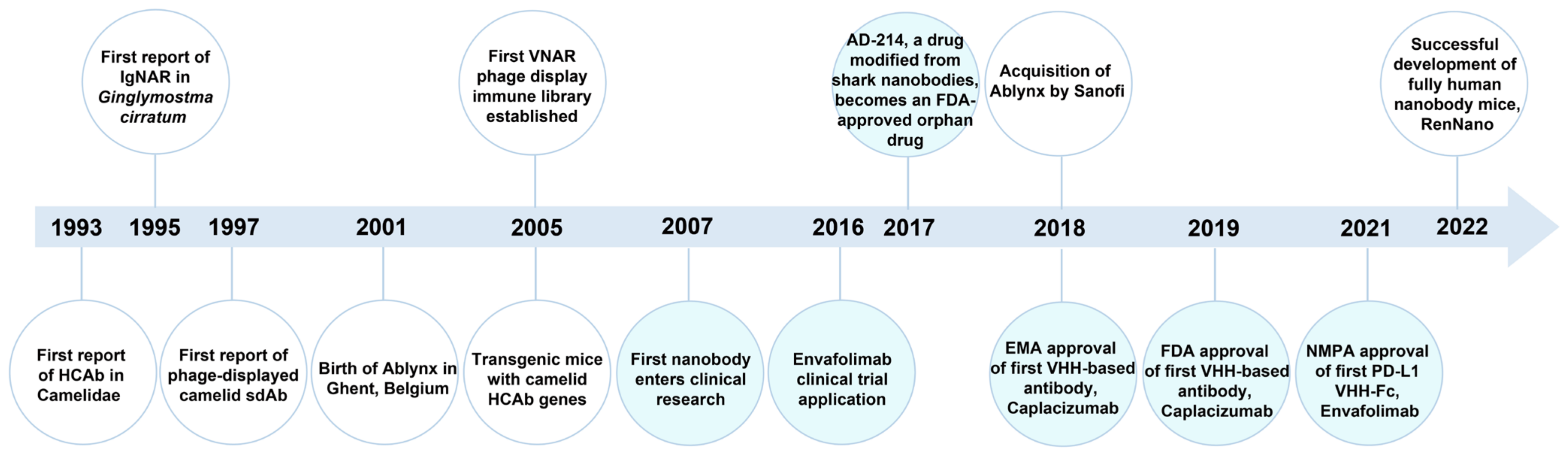
3.2.1. Introduction of Nanobody
3.2.2. Nanobody Targeting Fluorescent Proteins
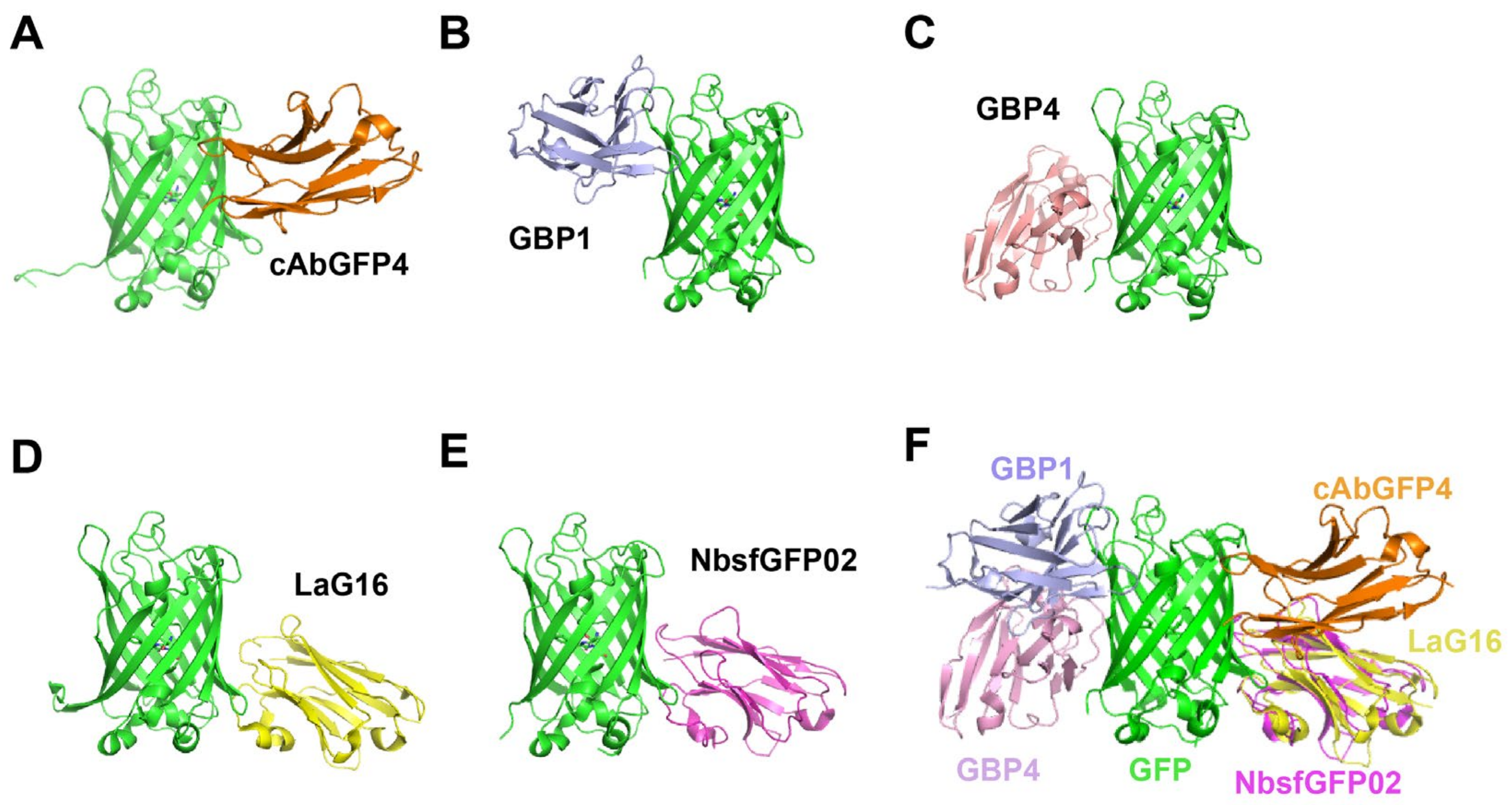
- Camelidae-derived nanobodies targeting GFP
- Shark-derived nanobodies targeting GFP.
- Nanobodies targeting other fluorescent proteins.
4. Applications of Fluorescent Protein Nanobody
4.1. Applications in Protein Detection
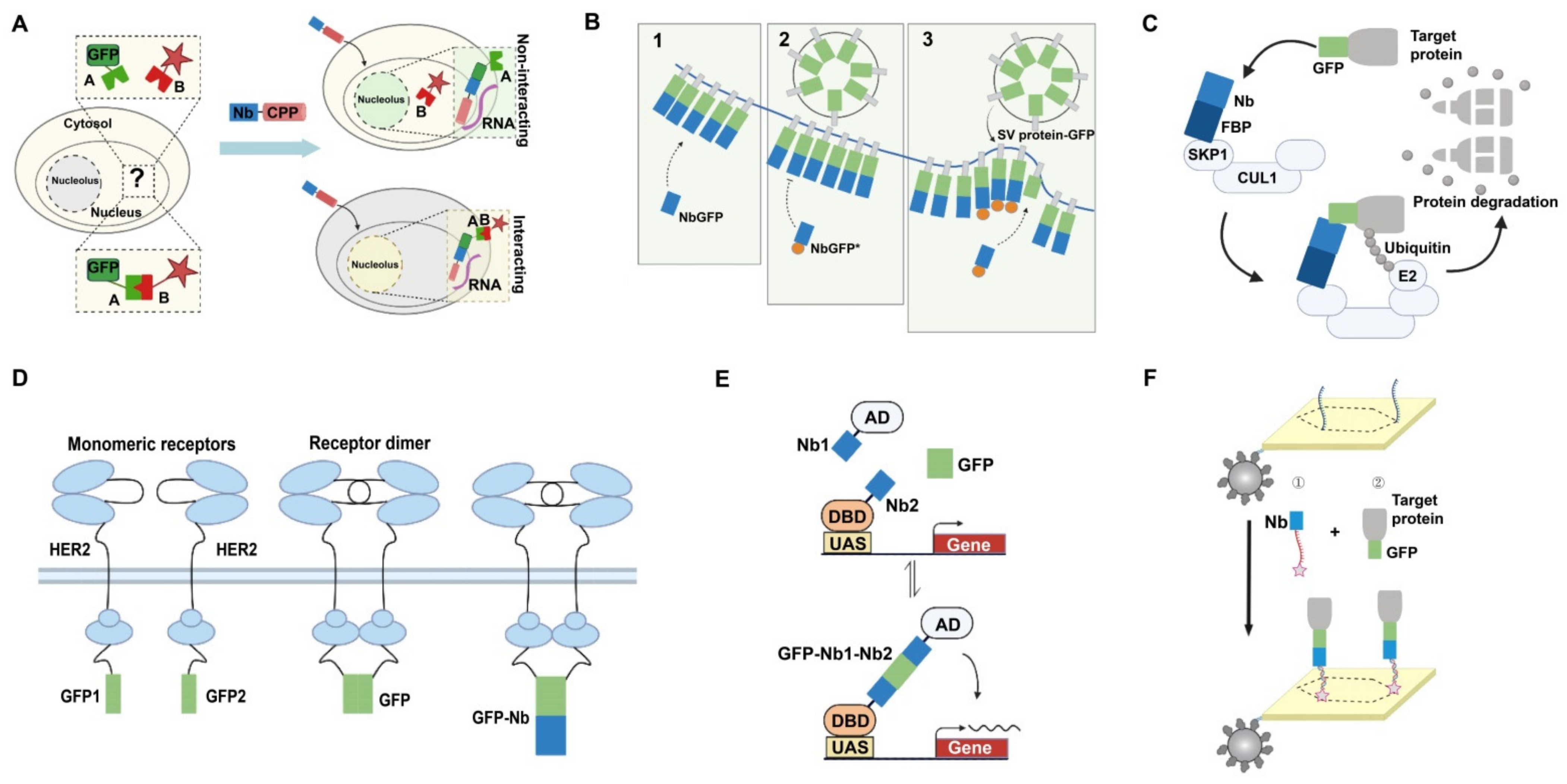
4.1.1. Detection of Intracellular Proteins
4.1.2. Detection of Proteins on the Cell Surface
4.2. Applications in Targeted Degradation of Protein Molecules
4.3. Applications in Bimolecular Complementation Affinity Purification System
4.4. Applications in Other Areas
5. Discussion on Future Outlook
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Muyldermans, S. Nanobodies: Natural Single-Domain Antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. [Google Scholar] [CrossRef] [PubMed]
- Helma, J.; Cardoso, M.C.; Muyldermans, S.; Leonhardt, H. Nanobodies and recombinant binders in cell biology. J. Cell Biol. 2015, 209, 633–644. [Google Scholar] [CrossRef] [PubMed]
- Ashour, J.; Schmidt, F.I.; Hanke, L.; Cragnolini, J.; Cavallari, M.; Altenburg, A.; Brewer, R.; Ingram, J.; Shoemaker, C.; Ploegh, H.L. Intracellular Expression of Camelid Single-Domain Antibodies Specific for Influenza Virus Nucleoprotein Uncovers Distinct Features of Its Nuclear Localization. J. Virol. 2015, 89, 2792–2800. [Google Scholar] [CrossRef] [PubMed]
- Manglik, A.; Kobilka, B.K.; Steyaert, J. Nanobodies to Study G Protein–Coupled Receptor Structure and Function. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 19–37. [Google Scholar] [CrossRef]
- Harmansa, S.; Hamaratoglu, F.; Affolter, M.; Caussinus, E. Dpp spreading is required for medial but not for lateral wing disc growth. Nature 2015, 527, 317–322. [Google Scholar] [CrossRef]
- Yamagata, M.; Sanes, J.R. Reporter–nanobody fusions (RANbodies) as versatile, small, sensitive immunohistochemical reagents. Proc. Natl. Acad. Sci. USA 2018, 115, 2126–2131. [Google Scholar] [CrossRef]
- Ariotti, N.; Hall, T.E.; Rae, J.; Ferguson, C.; McMahon, K.-A.; Martel, N.; Webb, R.E.; Webb, R.I.; Teasdale, R.D.; Parton, R.G. Modular Detection of GFP-Labeled Proteins for Rapid Screening by Electron Microscopy in Cells and Organisms. Dev. Cell 2015, 35, 513–525. [Google Scholar] [CrossRef]
- Früholz, S.; Fäßler, F.; Kolukisaoglu, Ü.; Pimpl, P. Nanobody-triggered lockdown of VSRs reveals ligand reloading in the Golgi. Nat. Commun. 2018, 9, 643. [Google Scholar] [CrossRef]
- Prole, D.L.; Taylor, C.W. A genetically encoded toolkit of functionalized nanobodies against fluorescent proteins for visualizing and manipulating intracellular signalling. BMC Biol. 2019, 17, 41. [Google Scholar] [CrossRef]
- Shimomura, O.; Johnson, F.H.; Saiga, Y. Extraction, Purification and Properties of Aequorin, a Bioluminescent Protein from the Luminous Hydromedusan, Aequorea. J. Cell. Comp. Physiol. 1962, 59, 223–239. [Google Scholar] [CrossRef]
- Prasher, D.; McCann, R.O.; Cormier, M.J. Cloning and expression of the cDNA coding for aequorin, a bioluminescent calcium-binding protein. Biochem. Biophys. Res. Commun. 1985, 126, 1259–1268. [Google Scholar] [CrossRef]
- Heim, R.; Prasher, D.C.; Tsien, R.Y. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl. Acad. Sci. USA 1994, 91, 12501–12504. [Google Scholar] [CrossRef]
- Matz, M.; Fradkov, A.F.; Labas, Y.A.; Savitsky, A.; Zaraisky, A.G.; Markelov, M.L.; Lukyanov, S.A. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 1999, 17, 969–973. [Google Scholar] [CrossRef]
- Park, N.; Song, J.; Jeong, S.; Tran, T.T.; Ko, H.W.; Kim, E.Y. Vaccinia-related kinase 3 (VRK3) sets the circadian period and amplitude by affecting the subcellular localization of clock proteins in mammalian cells. Biochem. Biophys. Res. Commun. 2017, 487, 320–326. [Google Scholar] [CrossRef]
- Zhao, D.; Xue, C.; Lin, S.; Shi, S.; Li, Q.; Liu, M.; Cai, X.; Lin, Y. Notch Signaling Pathway Regulates Angiogenesis via Endothelial Cell in 3D Co-Culture Model. J. Cell. Physiol. 2016, 232, 1548–1558. [Google Scholar] [CrossRef]
- Rodriguez, E.A.; Campbell, R.E.; Lin, J.Y.; Lin, M.Z.; Miyawaki, A.; Palmer, A.E.; Shu, X.; Zhang, J.; Tsien, R.Y. The Growing and Glowing Toolbox of Fluorescent and Photoactive Proteins. Trends Biochem. Sci. 2017, 42, 111–129. [Google Scholar] [CrossRef]
- Yang, F.; Moss, L.G.; Phillips, G.N., Jr. The molecular structure of green fluorescent protein. Nat. Biotechnol. 1996, 14, 1246–1251. [Google Scholar] [CrossRef]
- Ormö, M.; Cubitt, A.B.; Kallio, K.; Gross, L.A.; Tsien, R.Y.; Remington, S.J. Crystal Structure of the Aequorea victoria Green Fluorescent Protein. Science 1996, 273, 1392–1395. [Google Scholar] [CrossRef]
- Reid, B.G.; Flynn, G.C. Chromophore Formation in Green Fluorescent Protein. Biochemistry 1997, 36, 6786–6791. [Google Scholar] [CrossRef]
- Zhang, G.; Gurtu, V.; Kain, S.R. An Enhanced Green Fluorescent Protein Allows Sensitive Detection of Gene Transfer in Mammalian Cells. Biochem. Biophys. Res. Commun. 1996, 227, 707–711. [Google Scholar] [CrossRef]
- Pédelacq, J.-D.; Cabantous, S.; Tran, T.; Terwilliger, T.; Waldo, G.S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006, 24, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Crameri, A.; Whitehorn, E.A.; Tate, E.; Stemmer, W.P. Improved Green Fluorescent Protein by Molecular Evolution Using DNA Shuffling. Nat. Biotechnol. 1996, 14, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Miura, H.; Inoko, H.; Inoue, I.; Tanaka, M.; Sato, M.; Ohtsuka, M. Simple cloning strategy using GFPuv gene as positive/negative indicator. Anal. Biochem. 2011, 416, 237–239. [Google Scholar] [CrossRef] [PubMed]
- Mena, M.A.; Treynor, T.P.; Mayo, S.L.; Daugherty, P.S. Blue fluorescent proteins with enhanced brightness and photostability from a structurally targeted library. Nat. Biotechnol. 2006, 24, 1569–1571. [Google Scholar] [CrossRef] [PubMed]
- Ai, H.; Shaner, N.; Cheng, Z.; Tsien, R.Y.; Campbell, R.E. Exploration of New Chromophore Structures Leads to the Identification of Improved Blue Fluorescent Proteins. Biochemistry 2007, 46, 5904–5910. [Google Scholar] [CrossRef]
- Subach, O.M.; Gundorov, I.S.; Yoshimura, M.; Subach, F.V.; Zhang, J.; Grüenwald, D.; Souslova, E.A.; Chudakov, D.M.; Verkhusha, V.V. Conversion of Red Fluorescent Protein into a Bright Blue Probe. Chem. Biol. 2008, 15, 1116–1124. [Google Scholar] [CrossRef]
- Merzlyak, E.; Goedhart, J.; Shcherbo, D.; Bulina, M.E.; Shcheglov, A.S.; Fradkov, A.F.; Gaintzeva, A.; Lukyanov, K.; Lukyanov, S.; Gadella, T.; et al. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat. Methods 2007, 4, 555–557. [Google Scholar] [CrossRef]
- Cubitt, A.B.; Heim, R.; Adams, S.R.; Boyd, A.E.; Gross, L.A.; Tsien, R.Y. Understanding, improving and using green fluorescent proteins. Trends Biochem. Sci. 1995, 20, 448–455. [Google Scholar] [CrossRef]
- Tomosugi, W.; Matsuda, T.; Tani, T.; Nemoto, T.; Kotera, I.; Saito, K.; Horikawa, K.; Nagai, T. An ultramarine fluorescent protein with increased photostability and pH insensitivity. Nat. Methods 2009, 6, 351–353. [Google Scholar] [CrossRef]
- Goedhart, J.; van Weeren, L.; Hink, M.A.; Vischer, N.O.E.; Jalink, K.; Gadella, T.W.J. Bright cyan fluorescent protein variants identified by fluorescence lifetime screening. Nat. Methods 2010, 7, 137–139. [Google Scholar] [CrossRef]
- Day, R.N.; Davidson, M.W. The fluorescent protein palette: Tools for cellular imaging. Chem. Soc. Rev. 2009, 38, 2887–2921. [Google Scholar] [CrossRef]
- Griesbeck, O.; Baird, G.S.; Campbell, R.E.; Zacharias, D.A.; Tsien, R.Y. Reducing the Environmental Sensitivity of Yellow Fluorescent Protein. J. Biol. Chem. 2001, 276, 29188–29194. [Google Scholar] [CrossRef]
- Nagai, T.; Ibata, K.; Park, E.S.; Kubota, M.; Mikoshiba, K.; Miyawaki, A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002, 20, 87–90. [Google Scholar] [CrossRef]
- Shaner, N.C.; Campbell, R.E.; Steinbach, P.A.; Giepmans, B.N.; Palmer, A.E.; Tsien, R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004, 22, 1567–1572. [Google Scholar] [CrossRef]
- Shcherbo, D.; Merzlyak, E.M.; Chepurnykh, T.V.; Fradkov, A.F.; Ermakova, G.V.; Solovieva, E.A.; Lukyanov, K.A.; Bogdanova, E.A.; Zaraisky, A.G.; Lukyanov, S.; et al. Bright far-red fluorescent protein for whole-body imaging. Nat. Methods 2007, 4, 741–746. [Google Scholar] [CrossRef]
- Shcherbo, D.; Murphy, C.S.; Ermakova, G.V.; Solovieva, E.A.; Chepurnykh, T.V.; Shcheglov, A.S.; Verkhusha, V.; Pletnev, V.Z.; Hazelwood, K.L.; Roche, P.M.; et al. Far-red fluorescent tags for protein imaging in living tissues. Biochem. J. 2009, 418, 567–574. [Google Scholar] [CrossRef]
- Oliinyk, O.S.; Chernov, K.G.; Verkhusha, V.V. Bacterial Phytochromes, Cyanobacteriochromes and Allophycocyanins as a Source of Near-Infrared Fluorescent Probes. Int. J. Mol. Sci. 2017, 18, 1691. [Google Scholar] [CrossRef]
- Shu, X.; Royant, A.; Lin, M.Z.; Aguilera, T.A.; Lev-Ram, V.; Steinbach, P.A.; Tsien, R.Y. Mammalian Expression of Infrared Fluorescent Proteins Engineered from a Bacterial Phytochrome. Science 2009, 324, 804–807. [Google Scholar] [CrossRef]
- Yu, D.; Gustafson, W.C.; Han, C.; Lafaye, C.; Noirclerc-Savoye, M.; Ge, W.-P.; Thayer, D.A.; Huang, H.; Kornberg, T.B.; Royant, A.; et al. An improved monomeric infrared fluorescent protein for neuronal and tumour brain imaging. Nat. Commun. 2014, 5, 3626. [Google Scholar] [CrossRef]
- Yu, D.; Baird, M.A.; Allen, J.R.; Howe, E.S.; Klassen, M.P.; Reade, A.; Makhijani, K.; Song, Y.; Liu, S.; Murthy, Z.; et al. A naturally monomeric infrared fluorescent protein for protein labeling in vivo. Nat. Methods 2015, 12, 763–765. [Google Scholar] [CrossRef]
- Shcherbakova, D.M.; Baloban, M.; Emelyanov, A.V.; Brenowitz, M.; Guo, P.; Verkhusha, V.V. Bright monomeric near-infrared fluorescent proteins as tags and biosensors for multiscale imaging. Nat. Commun. 2016, 7, 12405. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, E.A.; Tran, G.N.; Gross, L.A.; Crisp, J.L.; Shu, X.; Lin, J.Y.; Tsien, R.Y. A far-red fluorescent protein evolved from a cyanobacterial phycobiliprotein. Nat. Methods 2016, 13, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Oliinyk, O.S.; Shemetov, A.A.; Pletnev, S.; Shcherbakova, D.M.; Verkhusha, V.V. Smallest near-infrared fluorescent protein evolved from cyanobacteriochrome as versatile tag for spectral multiplexing. Nat. Commun. 2019, 10, 279. [Google Scholar] [CrossRef]
- Oliinyk, O.S.; Baloban, M.; Clark, C.L.; Carey, E.; Pletnev, S.; Nimmerjahn, A.; Verkhusha, V.V. Single-domain near-infrared protein provides a scaffold for antigen-dependent fluorescent nanobodies. Nat. Methods 2022, 19, 740–750. [Google Scholar] [CrossRef] [PubMed]
- Tiller, K.E.; Tessier, P.M. Advances in Antibody Design. Annu. Rev. Biomed. Eng. 2015, 17, 191–216. [Google Scholar] [CrossRef]
- Gengyo-Ando, K.; Mitani, S. 1020 Production and characterization of a monoclonal antibody against the green fluorescent protein (GFP). Neurosci. Res. 1997, 28, S122. [Google Scholar] [CrossRef]
- Zhuang, R.; Zhang, Y.; Zhang, R.; Song, C.; Yang, K.; Yang, A.; Jin, B. Purification of GFP fusion proteins with high purity and yield by monoclonal antibody-coupled affinity column chromatography. Protein Expr. Purif. 2008, 59, 138–143. [Google Scholar] [CrossRef]
- Ward, E.S.; Güssow, D.; Griffiths, A.D.; Jones, P.T.; Winter, G. Binding activities of a repertoire of single immunoglobulin variable domains secreted from Escherichia coli. Nature 1989, 341, 544–546. [Google Scholar] [CrossRef]
- Hamers-Casterman, C.; Atarhouch, T.; Muyldermans, S.; Robinson, G.; Hammers, C.; Songa, E.B.; Bendahman, N.; Hammers, R. Naturally occurring antibodies devoid of light chains. Nature 1993, 363, 446–448. [Google Scholar] [CrossRef]
- Greenberg, A.S.; Avila, D.; Hughes, M.; Hughes, A.; McKinney, E.C.; Flajnik, M.F. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nat. Cell Biol. 1995, 374, 168–173. [Google Scholar] [CrossRef]
- De Meyer, T.; Muyldermans, S.; Depicker, A. Nanobody-based products as research and diagnostic tools. Trends Biotechnol. 2014, 32, 263–270. [Google Scholar] [CrossRef]
- Skerra, A. Alternative non-antibody scaffolds for molecular recognition. Curr. Opin. Biotechnol. 2007, 18, 295–304. [Google Scholar] [CrossRef]
- Muyldermans, S.; Baral, T.; Retamozzo, V.C.; De Baetselier, P.; De Genst, E.; Kinne, J.; Leonhardt, H.; Magez, S.; Nguyen, V.; Revets, H.; et al. Camelid immunoglobulins and nanobody technology. Veter. Immunol. Immunopathol. 2009, 128, 178–183. [Google Scholar] [CrossRef]
- Rothbauer, U.; Zolghadr, K.; Tillib, S.; Nowak, D.; Schermelleh, L.; Gahl, A.; Backmann, N.; Conrath, K.; Muyldermans, S.; Cardoso, M.C.; et al. Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods 2006, 3, 887–889. [Google Scholar] [CrossRef]
- Kubala, M.H.; Kovtun, O.; Alexandrov, K.; Collins, B.M. Structural and thermodynamic analysis of the GFP:GFP-nanobody complex. Protein Sci. 2010, 19, 2389–2401. [Google Scholar] [CrossRef]
- Fang, Z.; Cao, D.; Qiu, J. Development and production of nanobodies specifically against green fluorescence protein. Appl. Microbiol. Biotechnol. 2020, 104, 4837–4848. [Google Scholar] [CrossRef]
- Kirchhofer, A.; Helma, J.; Schmidthals, K.; Frauer, C.; Cui, S.; Karcher, A.; Pellis, M.; Muyldermans, S.; Casas-Delucchi, C.S.; Cardoso, M.C.; et al. Modulation of protein properties in living cells using nanobodies. Nat. Struct. Mol. Biol. 2009, 17, 133–138. [Google Scholar] [CrossRef]
- Rothbauer, U.; Zolghadr, K.; Muyldermans, S.; Schepers, A.; Cardoso, M.C.; Leonhardt, H. A Versatile Nanotrap for Biochemical and Functional Studies with Fluorescent Fusion Proteins. Mol. Cell. Proteom. 2008, 7, 282–289. [Google Scholar] [CrossRef]
- Fridy, P.; Li, Y.; Keegan, S.; Thompson, M.K.; Nudelman, I.; Scheid, J.F.; Oeffinger, M.; Nussenzweig, M.C.; Fenyö, D.; Chait, B.T.; et al. A robust pipeline for rapid production of versatile nanobody repertoires. Nat. Methods 2014, 11, 1253–1260. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, Y.; Ding, Y.; Hattori, M. Structure-based engineering of anti-GFP nanobody tandems as ultra-high-affinity reagents for purification. Sci. Rep. 2020, 10, 6239. [Google Scholar] [CrossRef]
- Twair, A.; Al-Okla, S.; Zarkawi, M.; Abbady, A.Q. Characterization of camel nanobodies specific for superfolder GFP fusion proteins. Mol. Biol. Rep. 2014, 41, 6887–6898. [Google Scholar] [CrossRef] [PubMed]
- Zhong, P.; Wang, Z.; Cheng, S.; Zhang, Y.; Jiang, H.; Liu, R.; Ding, Y. Structural insights into two distinct nanobodies recognizing the same epitope of green fluorescent protein. Biochem. Biophys. Res. Commun. 2021, 565, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Hao, R.; Chen, C.; Su, Z.; Zhao, L.; Luo, Z.; Xie, W. Rapid Delivery of Nanobodies/VHHs into Living Cells via Expressing In Vitro-Transcribed mRNA. Mol. Ther.-Methods Clin. Dev. 2020, 17, 401–408. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Wang, M.; Xiang, H.; Jiang, Y.; Gong, J.; Su, D.; Al Azad, M.A.R.; Dong, H.; Feng, L.; Wu, J.; et al. Bamboo Shark as a Small Animal Model for Single Domain Antibody Production. Front. Bioeng. Biotechnol. 2021, 9, 792111. [Google Scholar] [CrossRef]
- Li, S.; Shan, H.; Wang, T.; Zheng, X.; Shi, M.; Chen, B.; Lu, H.; Zhang, Y.; Zhao, S.; Hua, Z. Generation of mWasabi fluorescent protein-binding nanobodies. Anal. Biochem. 2020, 608, 113875. [Google Scholar] [CrossRef]
- Ries, J.; Kaplan, C.; Platonova, E.; Eghlidi, H.M.; Ewers, H. A simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nat. Methods 2012, 9, 582–584. [Google Scholar] [CrossRef]
- Beghein, E.; Gettemans, J. Nanobody Technology: A Versatile Toolkit for Microscopic Imaging, Protein–Protein Interaction Analysis, and Protein Function Exploration. Front. Immunol. 2017, 8, 771. [Google Scholar] [CrossRef]
- Platonova, E.; Winterflood, C.M.; Junemann, A.; Albrecht, D.; Faix, J.; Ewers, H. Single-molecule microscopy of molecules tagged with GFP or RFP derivatives in mammalian cells using nanobody binders. Methods 2015, 88, 89–97. [Google Scholar] [CrossRef]
- Herce, H.D.; Schumacher, D.; Schneider, A.F.; Ludwig, A.K.; Mann, F.A.; Fillies, M.; Kasper, M.-A.; Reinke, S.; Krause, E.; Leonhardt, H.; et al. Cell-permeable nanobodies for targeted immunolabelling and antigen manipulation in living cells. Nat. Chem. 2017, 9, 762–771. [Google Scholar] [CrossRef]
- Payne, N.C.; Mazitschek, R. Tiny Titans: Nanobodies as Powerful Tools for TR-FRET Assay Development. Anal. Sens. 2022, 2, e202200020. [Google Scholar] [CrossRef]
- Wendel, S.; Fischer, E.C.; Martínez, V.; Seppälä, S.; Nørholm, M.H.H. A nanobody:GFP bacterial platform that enables functional enzyme display and easy quantification of display capacity. Microb. Cell Factories 2016, 15, 71. [Google Scholar] [CrossRef]
- Seitz, K.J.; Rizzoli, S.O. GFP nanobodies reveal recently-exocytosed pHluorin molecules. Sci. Rep. 2019, 9, 7773. [Google Scholar] [CrossRef]
- Caussinus, E.; Kanca, O.; Affolter, M. Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat. Struct. Mol. Biol. 2011, 19, 117–121. [Google Scholar] [CrossRef]
- Shin, Y.J.; Park, S.K.; Jung, Y.J.; Na Kim, Y.; Kim, K.S.; Park, O.K.; Kwon, S.-H.; Jeon, S.H.; Trinh, L.A.; Fraser, S.; et al. Nanobody-targeted E3-ubiquitin ligase complex degrades nuclear proteins. Sci. Rep. 2015, 5, 14269. [Google Scholar] [CrossRef]
- Baudisch, B.; Pfort, I.; Sorge, E.; Conrad, U. Nanobody-Directed Specific Degradation of Proteins by the 26S-Proteasome in Plants. Front. Plant Sci. 2018, 9, 130. [Google Scholar] [CrossRef]
- Croucher, D.R.; Iconomou, M.; Hastings, J.F.; Kennedy, S.P.; Han, J.Z.R.; Shearer, R.F.; McKenna, J.; Wan, A.; Lau, J.; Aparicio, S.; et al. Bimolecular complementation affinity purification (BiCAP) reveals dimer-specific protein interactions for ERBB2 dimers. Sci. Signal. 2016, 9, 436. [Google Scholar] [CrossRef]
- Tang, J.C.; Szikra, T.; Kozorovitskiy, Y.; Teixiera, M.; Sabatini, B.L.; Roska, B.; Cepko, C.L. A Nanobody-Based System Using Fluorescent Proteins as Scaffolds for Cell-Specific Gene Manipulation. Cell 2013, 154, 928–939. [Google Scholar] [CrossRef]
- Sommese, R.F.; Hariadi, R.F.; Kim, K.; Liu, M.; Tyska, M.J.; Sivaramakrishnan, S. Patterning protein complexes on DNA nanostructures using a GFP nanobody. Protein Sci. 2016, 25, 2089–2094. [Google Scholar] [CrossRef]
| Classification | Protein | Excitation Peak (nm) | Emission Peak (nm) | Molar Extinction Coefficient (×103 M−1 cm−1) | Quantum Yield | Brightness (Relative to EGFP, %) | Structure |
|---|---|---|---|---|---|---|---|
| Green | wtGFP | 395 | 509 | 25 | 0.79 | 48 | monomer |
| EGFP | 488 | 507 | 56 | 0.60 | 100 | monomer | |
| sfGFP | 485 | 510 | 83.3 | 0.65 | 160 | monomer | |
| GFPuv | 395 | 509 | N.A. | N.A. | N.A. | monomer | |
| Blue | EBFP | 380 | 440 | 29 | 0.30 | 27 | monomer |
| Azurite | 383 | 447 | 26.2 | 0.55 | 43 | monomer | |
| EBFP2 | 383 | 448 | 32 | 0.56 | 53 | monomer | |
| mTagBFP | 402 | 457 | 52 | 0.63 | 98 | monomer | |
| Ultramarine | Sirius | 355 | 424 | 15 | 0.24 | 11 | monomer |
| Cyan | ECFP | 433 | 475 | 26 | 0.40 | 39 | monomer |
| Cerulean | 433 | 475 | 32.5 | 0.6 | 79 | monomer | |
| mTFP | 462 | 492 | 64 | 0.85 | 162 | monomer | |
| mTurquoise | 434 | 474 | 30 | 0.84 | 75 | monomer | |
| Yellow | EYFP | 514 | 527 | 83.4 | 0.60 | 150 | monomer |
| mCitrine | 516 | 529 | 77 | 0.8 | 174 | monomer | |
| mVenus | 515 | 528 | 92.2 | 0.60 | 156 | monomer | |
| Red | DsRed | 558 | 583 | 75 | 0.79 | 165 | tetramer |
| TagRFP | 555 | 584 | 100 | 0.48 | 143 | monomer | |
| mCherry | 587 | 610 | 72 | 0.2 | 43 | monomer | |
| mStrawberry | 574 | 596 | 90 | 0.29 | 78 | monomer | |
| dTomato | 554 | 581 | 69 | 0.69 | 142 | dimer | |
| mTangerine | 568 | 585 | 38 | 0.30 | 34 | monomer | |
| mKate | 588 | 635 | 45 | 0.33 | 45 | monomer | |
| mKate2 | 588 | 635 | 62.5 | 0.4 | 74 | monomer | |
| Katushka | 588 | 635 | 65 | 0.34 | 67 | dimer | |
| Orange | mOrange | 548 | 562 | 71 | 0.70 | 148 | monomer |
| mBanana | 540 | 553 | 6 | 0.70 | 12.5 | monomer |
| Photoreceptor | Protein | Excitation Peak (nm) | Emission Peak (nm) | Molar Extinction Coefficient (×103 M−1 cm−1) | Quantum Yield | Brightness (Relative to miRFP670nano, %) |
|---|---|---|---|---|---|---|
| BphP | IFP1.4 | 684 | 708 | 88 | 7 | N.A. |
| IFP2.0 | 690 | 711 | 86 | 8 | N.A. | |
| mIFP | 683 | 704 | 82 | 8.4 | 67 | |
| miRFP670 | 642 | 670 | 87.4 | 14 | 119 | |
| miRFP703 | 674 | 703 | 90.9 | 8.6 | 76 | |
| miRFP709 | 683 | 709 | 78.4 | 5.4 | 41 | |
| ApcF | BDFP1.5 | 688 | 711 | 74 | 5.0 | 36 |
| CBCR | miRFP670nano | 645 | 670 | 95 | 10.8 | 100 |
| miRFP670nano3 | 645 | 670 | 129 | 18.5 | 233 |
| Nanobodies | Targets | Source | Reference |
|---|---|---|---|
| cAbGFP4 | GFP | alpaca | [54] |
| GBP1-7 | GFP | camel | [57] |
| LaGs | GFP | llama | [59] |
| NbsfGFP01, NbsfGFP02, NbsfGFP03, NbsfGFP04, NbsfGFP06, NbsfGFP07, NbsfGFP08 | sfGFP | camel | [61] |
| D5, E6, A12, B9 | GFP | alpaca | [56] |
| BsG3, 73, 80, 89, 93, 98, 105 | GFP | bamboo shark | [64] |
| Nb4, Nb6, Nb27 | mWasabi | camel | [65] |
| LaMs | mCherry | llama | [59] |
| BSR1, BSR3, BSR4 | iRFP713 | bamboo shark | [64] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.-L.; Xie, X.-X.; Zhong, N.; Sun, L.-C.; Lin, D.; Zhang, L.-J.; Weng, L.; Jin, T.; Cao, M.-J. Research Progresses and Applications of Fluorescent Protein Antibodies: A Review Focusing on Nanobodies. Int. J. Mol. Sci. 2023, 24, 4307. https://doi.org/10.3390/ijms24054307
Chen Y-L, Xie X-X, Zhong N, Sun L-C, Lin D, Zhang L-J, Weng L, Jin T, Cao M-J. Research Progresses and Applications of Fluorescent Protein Antibodies: A Review Focusing on Nanobodies. International Journal of Molecular Sciences. 2023; 24(5):4307. https://doi.org/10.3390/ijms24054307
Chicago/Turabian StyleChen, Yu-Lei, Xin-Xin Xie, Ning Zhong, Le-Chang Sun, Duanquan Lin, Ling-Jing Zhang, Ling Weng, Tengchuan Jin, and Min-Jie Cao. 2023. "Research Progresses and Applications of Fluorescent Protein Antibodies: A Review Focusing on Nanobodies" International Journal of Molecular Sciences 24, no. 5: 4307. https://doi.org/10.3390/ijms24054307
APA StyleChen, Y.-L., Xie, X.-X., Zhong, N., Sun, L.-C., Lin, D., Zhang, L.-J., Weng, L., Jin, T., & Cao, M.-J. (2023). Research Progresses and Applications of Fluorescent Protein Antibodies: A Review Focusing on Nanobodies. International Journal of Molecular Sciences, 24(5), 4307. https://doi.org/10.3390/ijms24054307







