Author Contributions
Conceptualization, E.D.S. and I.M.T.; methodology, E.D.S. and I.M.T.; validation, E.D.S. and I.M.T.; formal analysis, E.D.S. and I.M.T.; investigation, E.D.S., R.S.T., F.N.R. and I.M.T.; resources, F.N.R. and D.E.A.; data curation, E.D.S. and I.M.T.; writing—original draft preparation, I.M.T.; writing—review and editing, E.D.S. and I.M.T.; visualization, E.D.S.; supervision, I.M.T.; project administration, E.D.S. and I.M.T.; funding acquisition, I.N.S. All authors have read and agreed to the published version of the manuscript.
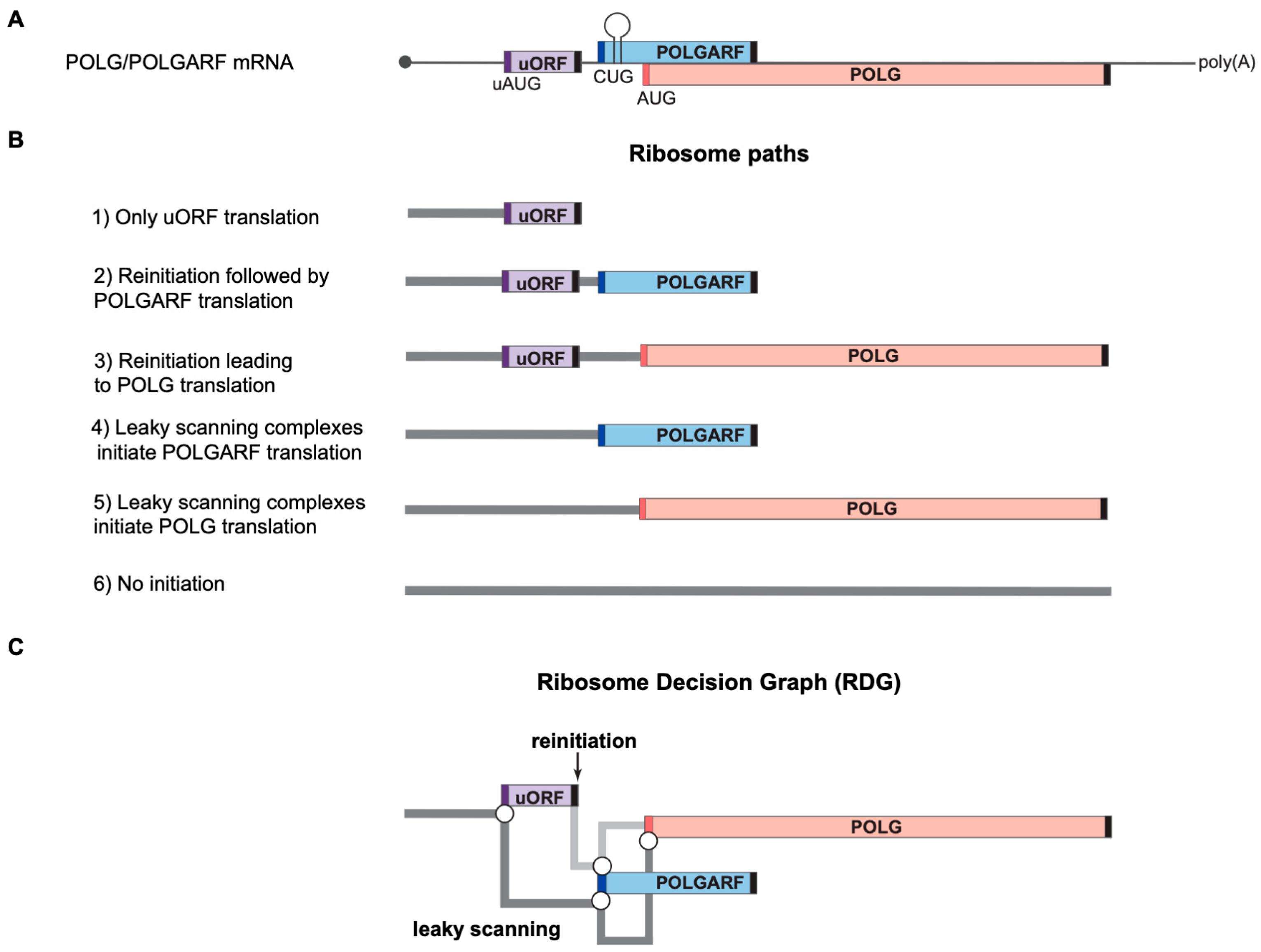
Figure 1.
Ribosome paths to initiate POLG and POLGARF translation. Please note that the scheme has been drawn not to scale for readability. (
A) A schematic representation of the natural dual-coding human POLG/POLGARF mRNA. The mRNA bears a ~300 nt long rather GC-rich (65% GC) 5′ UTR with a regulatory uORF located about 150 nt from the 5′ end and encoding 24 amino acids (the uORF is shown as a purple box, and dark purple bars depict the uAUG). There are only 11 nucleotides separating the uORF stop codon from the POLGARF start codon (the dark blue bar). The POLGARF ORF starts with the CUG codon in an unusually strong context, overlaps the main POLG ORF by more than 20%, and encodes a highly conserved ~25 kDa protein. The close-by stem-loop (referred to as SL in the other figures) ensures the efficient translation initiation on this CUG. The distance between the POLGARF start codon and the main POLG start codon (red bar) is only 53 nucleotides. Black bars depict stop codons. (
B) Ribosome paths through the POLG/POLGARF mRNA. Reinitiation and leaky scanning are specified with respect to the uORF. All ribosomes that initiate translation on the main POLG start codon have leaked through the POLGARF start codon, regardless of their path through the uORF (leaky scanning or reinitiation). (
C) Ribosome Decision Graph (RDG) representing translation as multiple ribosome paths through the POLG/POLGARF 5′ UTR. Boxes depict the ORFs and circles depict branching points where the ribosome makes a “decision” about whether it initiates or not. The path of leaky scanning complexes towards the downstream start codons is shown in dark grey. The light grey path represents the post-terminating small ribosome subunit that resumes scanning and can initiate on the downstream POLGARF or POLG start codons (the reinitiation path). The concept of visualization was adopted from [
20].
Figure 1.
Ribosome paths to initiate POLG and POLGARF translation. Please note that the scheme has been drawn not to scale for readability. (
A) A schematic representation of the natural dual-coding human POLG/POLGARF mRNA. The mRNA bears a ~300 nt long rather GC-rich (65% GC) 5′ UTR with a regulatory uORF located about 150 nt from the 5′ end and encoding 24 amino acids (the uORF is shown as a purple box, and dark purple bars depict the uAUG). There are only 11 nucleotides separating the uORF stop codon from the POLGARF start codon (the dark blue bar). The POLGARF ORF starts with the CUG codon in an unusually strong context, overlaps the main POLG ORF by more than 20%, and encodes a highly conserved ~25 kDa protein. The close-by stem-loop (referred to as SL in the other figures) ensures the efficient translation initiation on this CUG. The distance between the POLGARF start codon and the main POLG start codon (red bar) is only 53 nucleotides. Black bars depict stop codons. (
B) Ribosome paths through the POLG/POLGARF mRNA. Reinitiation and leaky scanning are specified with respect to the uORF. All ribosomes that initiate translation on the main POLG start codon have leaked through the POLGARF start codon, regardless of their path through the uORF (leaky scanning or reinitiation). (
C) Ribosome Decision Graph (RDG) representing translation as multiple ribosome paths through the POLG/POLGARF 5′ UTR. Boxes depict the ORFs and circles depict branching points where the ribosome makes a “decision” about whether it initiates or not. The path of leaky scanning complexes towards the downstream start codons is shown in dark grey. The light grey path represents the post-terminating small ribosome subunit that resumes scanning and can initiate on the downstream POLGARF or POLG start codons (the reinitiation path). The concept of visualization was adopted from [
20].
![Ijms 24 17149 g001]()
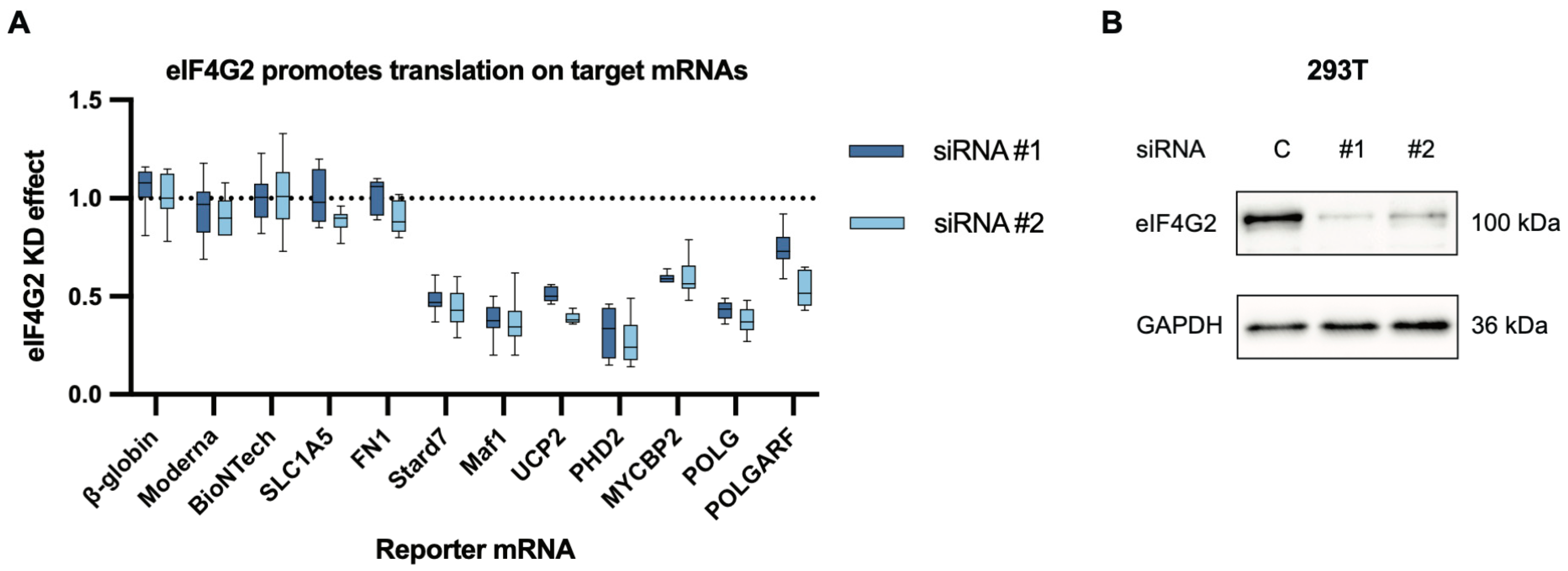
Figure 2.
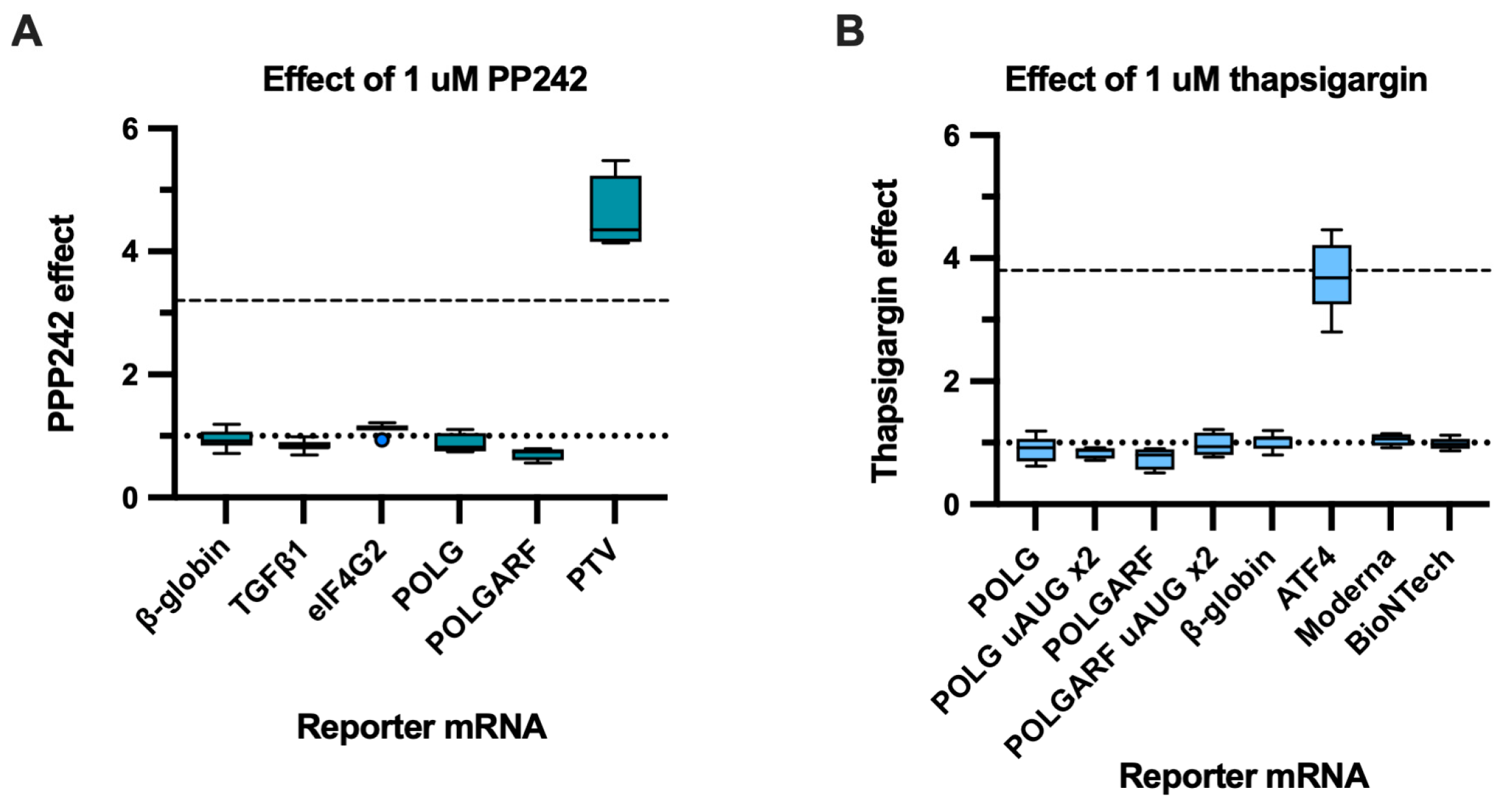
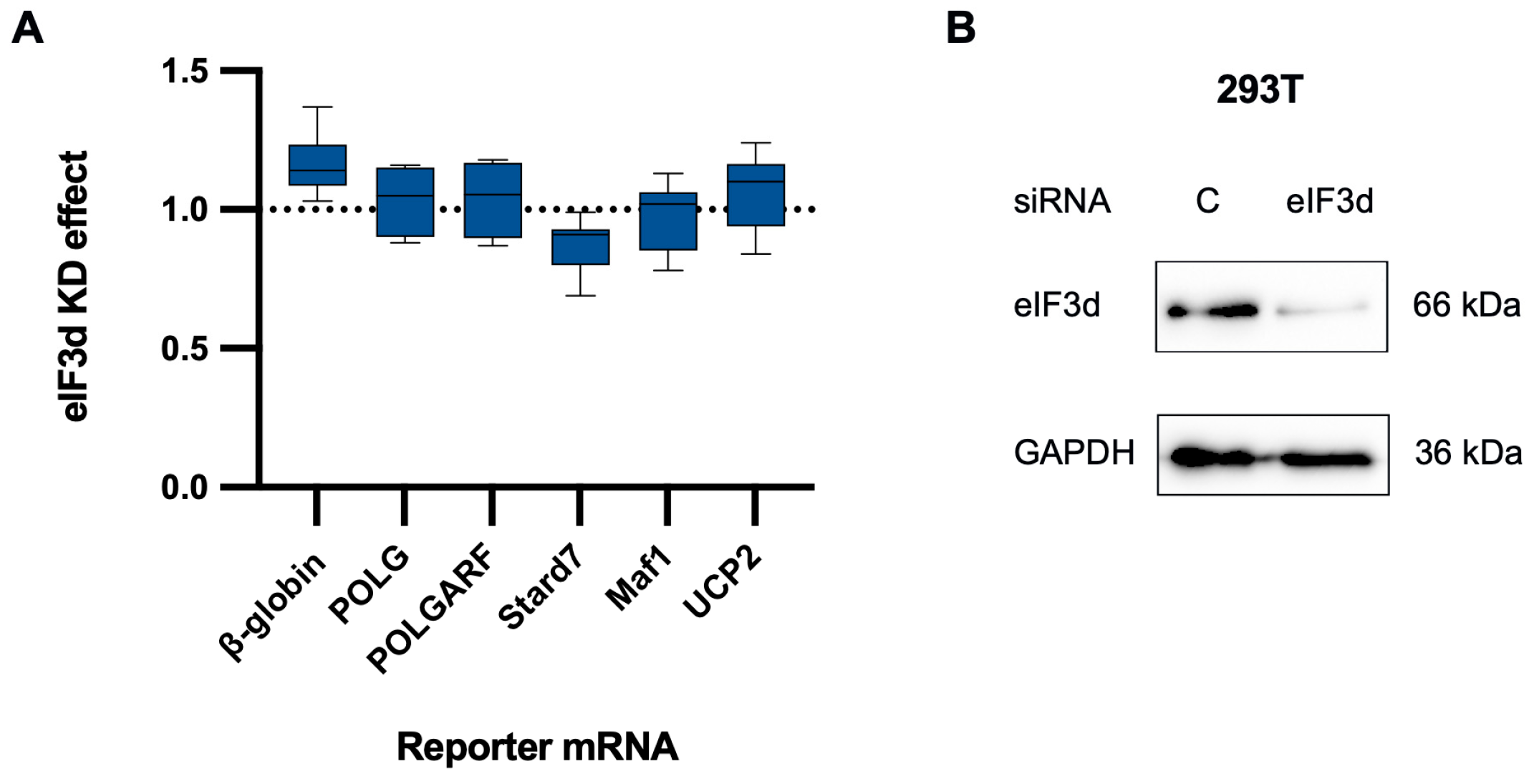
eIF4G2 participates in the translation of both POLGARF and POLG. (A) The contribution of eIF4G2 to translation was assessed using reporter mRNA transfection in 293T cells depleted of eIF4G2. Cells pretreated with either control or anti-eIF4G2 siRNAs for 72 h were transfected with in vitro transcribed m7G-capped and polyadenylated reporters. The Nluc-coding reference β-globin reporter mRNA was co-transfected with all of the reporters. The data are presented as ratios of normalized reporter expression in the eIF4G2-depleted to the control cells (the eIF4G2 KD effect). The knockdown effect < 1 corresponds to translation inhibition in the absence of eIF4G2. The translation of Stard7, Maf1, and UCP2 mRNAs has previously been shown to require eIF4G2. The reporter mRNAs with β-globin, SLC1A5, FN1, Moderna, and BioNTech 5′ UTR do not need eIF4G2 and serve as a negative control. For all eIF4G2 targets and β-globin reporters, the number of replicates exceeds 20. For the non-targets n ≥ 5. (B) Western blot analysis of the eIF4G2 knockdown in 293T cells, GAPDH as a loading control.
Figure 2.
eIF4G2 participates in the translation of both POLGARF and POLG. (A) The contribution of eIF4G2 to translation was assessed using reporter mRNA transfection in 293T cells depleted of eIF4G2. Cells pretreated with either control or anti-eIF4G2 siRNAs for 72 h were transfected with in vitro transcribed m7G-capped and polyadenylated reporters. The Nluc-coding reference β-globin reporter mRNA was co-transfected with all of the reporters. The data are presented as ratios of normalized reporter expression in the eIF4G2-depleted to the control cells (the eIF4G2 KD effect). The knockdown effect < 1 corresponds to translation inhibition in the absence of eIF4G2. The translation of Stard7, Maf1, and UCP2 mRNAs has previously been shown to require eIF4G2. The reporter mRNAs with β-globin, SLC1A5, FN1, Moderna, and BioNTech 5′ UTR do not need eIF4G2 and serve as a negative control. For all eIF4G2 targets and β-globin reporters, the number of replicates exceeds 20. For the non-targets n ≥ 5. (B) Western blot analysis of the eIF4G2 knockdown in 293T cells, GAPDH as a loading control.
![Ijms 24 17149 g002]()
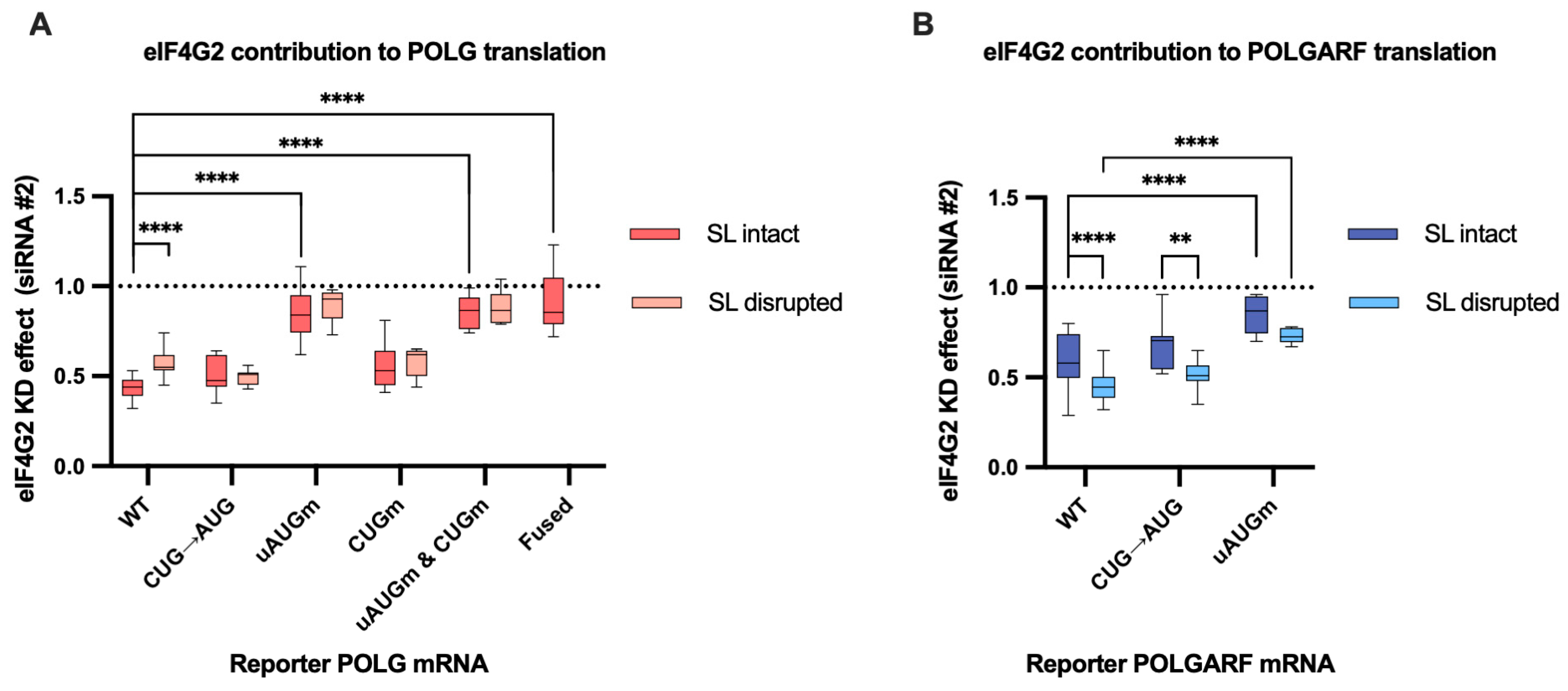
Figure 3.
eIF4G2 promotes POLG and POLGARF translation. (A) In vitro transcribed m7G-capped and polyadenylated POLG reporters with the indicated wild-type (WT) or mutated 5′ UTRs were transfected into mock- and eIF4G2-depleted (siRNA#1) 293T cells along with the reference β-globin reporter mRNA (n ≥ 10). All assayed reporters were tested with either the wild-type or the disrupted stem-loop (referred to as SL intact or SL disrupted, respectively). The effect of the knockdown (eIF4G2 KD effect) is calculated by dividing the normalized reporter expression in eIF4G2-depleted cells by that in control cells. The knockdown effect < 1 reflects translation inhibition by the eIF4G2 depletion. The statistical significance is determined using the Mann–Whitney U test. Three and four asterisks stand for p < 0.001 and p < 0.0001, respectively. (B) Results of POLGARF reporter mRNA transfections (similar to panel A). (C) Schematic representation of the POLG reporters examined in panel A (not to scale). The POLG start codon (red bar) drives translation of the chimeric reporter protein, which consists of 15 N-terminal POLG amino acids (shown in pink) fused to the firefly luciferase (Fluc). The POLGARF CUG codon (the dark blue bar) drives the translation of an ORF that overlaps out of frame with the Fluc and encodes the 60-aa-long peptide with 33 N-terminal POLGARF amino acids (CUG-driven uORF is displayed in blue). The purple box depicts the regulatory uORF. The start codons are shown in corresponding color bars, and black bars depict the stop codons. The stem-loop is omitted for the sake of readability. Crosses display the positions of the upstream (with respect to the POLG ORF) start codons that were substituted for the stop codons in the corresponding reporters. In the reporter mRNA called “fused”, the uORF is fused in frame to the chimeric POLG/Fluc sequence. This resulting chimaera evaluates the role of eIF4G2 in the POLG/POLGARF mRNA translation initiation from the 5′-end to the uAUG. (D) Panel D is similar to panel C, with the exception that POLGARF reporter mRNAs are displayed. The POLGARF CUG start codon drives translation of a chimeric reporter protein, consisting of 33 N-terminal POLGARF amino acids (shown in blue) fused to the Fluc. The POLG start codon is out of frame with Fluc and provides translation of a 20-amino-acid-long stub (shown in pink).
Figure 3.
eIF4G2 promotes POLG and POLGARF translation. (A) In vitro transcribed m7G-capped and polyadenylated POLG reporters with the indicated wild-type (WT) or mutated 5′ UTRs were transfected into mock- and eIF4G2-depleted (siRNA#1) 293T cells along with the reference β-globin reporter mRNA (n ≥ 10). All assayed reporters were tested with either the wild-type or the disrupted stem-loop (referred to as SL intact or SL disrupted, respectively). The effect of the knockdown (eIF4G2 KD effect) is calculated by dividing the normalized reporter expression in eIF4G2-depleted cells by that in control cells. The knockdown effect < 1 reflects translation inhibition by the eIF4G2 depletion. The statistical significance is determined using the Mann–Whitney U test. Three and four asterisks stand for p < 0.001 and p < 0.0001, respectively. (B) Results of POLGARF reporter mRNA transfections (similar to panel A). (C) Schematic representation of the POLG reporters examined in panel A (not to scale). The POLG start codon (red bar) drives translation of the chimeric reporter protein, which consists of 15 N-terminal POLG amino acids (shown in pink) fused to the firefly luciferase (Fluc). The POLGARF CUG codon (the dark blue bar) drives the translation of an ORF that overlaps out of frame with the Fluc and encodes the 60-aa-long peptide with 33 N-terminal POLGARF amino acids (CUG-driven uORF is displayed in blue). The purple box depicts the regulatory uORF. The start codons are shown in corresponding color bars, and black bars depict the stop codons. The stem-loop is omitted for the sake of readability. Crosses display the positions of the upstream (with respect to the POLG ORF) start codons that were substituted for the stop codons in the corresponding reporters. In the reporter mRNA called “fused”, the uORF is fused in frame to the chimeric POLG/Fluc sequence. This resulting chimaera evaluates the role of eIF4G2 in the POLG/POLGARF mRNA translation initiation from the 5′-end to the uAUG. (D) Panel D is similar to panel C, with the exception that POLGARF reporter mRNAs are displayed. The POLGARF CUG start codon drives translation of a chimeric reporter protein, consisting of 33 N-terminal POLGARF amino acids (shown in blue) fused to the Fluc. The POLG start codon is out of frame with Fluc and provides translation of a 20-amino-acid-long stub (shown in pink).
![Ijms 24 17149 g003]()
Figure 4.
Mutating the start codons affects POLG and POLGARF translation efficiencies. m7G-capped and polyadenylated reporters with the indicated wild-type (WT) or mutated 5′ UTR were transfected into 293T cells along with the reference β-globin reporter mRNA (n ≥ 15). Nluc activity was used to normalize Fluc reporter expression. All assayed reporters were tested with either the wild-type or the disrupted stem-loop sequence (referred to as SL intact and SL disrupted, respectively). Relative translation efficiency is calculated by dividing the normalized expression of a mutant reporter (with the start codon altered) by the normalized expression of the wild-type reporter. When reporters with disrupted stem-loops are used, relative translation efficiency is shown with respect to the corresponding constructs with wild-type start codons and disrupted stem-loops. The dotted line at 1 corresponds to translation efficiency identical to that of the wild-type reporter mRNA. (A) Results of POLG reporter mRNA transfections. The statistical significance is determined using the Mann–Whitney U test. Four and two asterisks indicate p < 0.0001 and p < 0.01, respectively. (B) Results of POLGARF reporter mRNA transfections (similar to panel A). Four asterisks stand for p < 0.0001.
Figure 4.
Mutating the start codons affects POLG and POLGARF translation efficiencies. m7G-capped and polyadenylated reporters with the indicated wild-type (WT) or mutated 5′ UTR were transfected into 293T cells along with the reference β-globin reporter mRNA (n ≥ 15). Nluc activity was used to normalize Fluc reporter expression. All assayed reporters were tested with either the wild-type or the disrupted stem-loop sequence (referred to as SL intact and SL disrupted, respectively). Relative translation efficiency is calculated by dividing the normalized expression of a mutant reporter (with the start codon altered) by the normalized expression of the wild-type reporter. When reporters with disrupted stem-loops are used, relative translation efficiency is shown with respect to the corresponding constructs with wild-type start codons and disrupted stem-loops. The dotted line at 1 corresponds to translation efficiency identical to that of the wild-type reporter mRNA. (A) Results of POLG reporter mRNA transfections. The statistical significance is determined using the Mann–Whitney U test. Four and two asterisks indicate p < 0.0001 and p < 0.01, respectively. (B) Results of POLGARF reporter mRNA transfections (similar to panel A). Four asterisks stand for p < 0.0001.
![Ijms 24 17149 g004]()
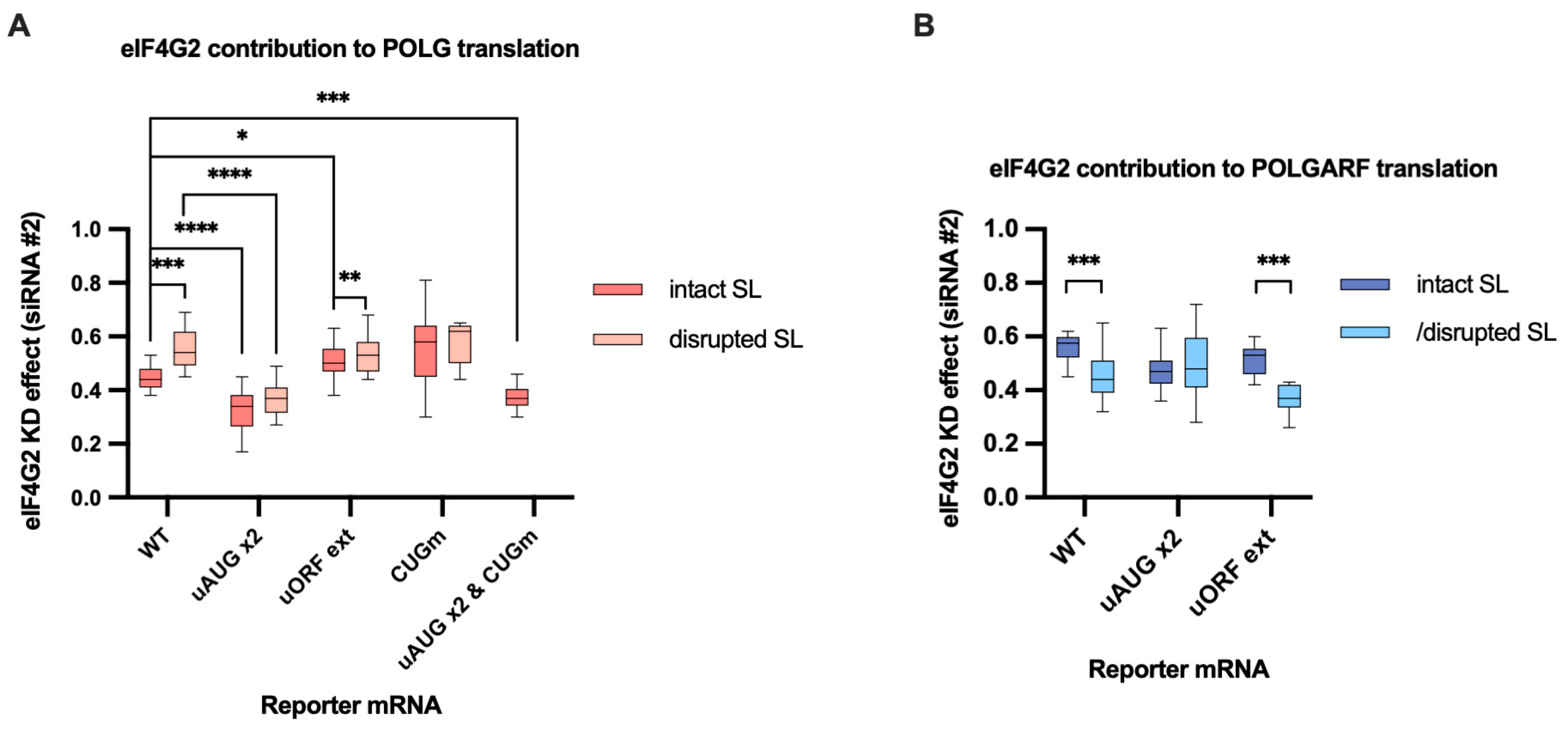
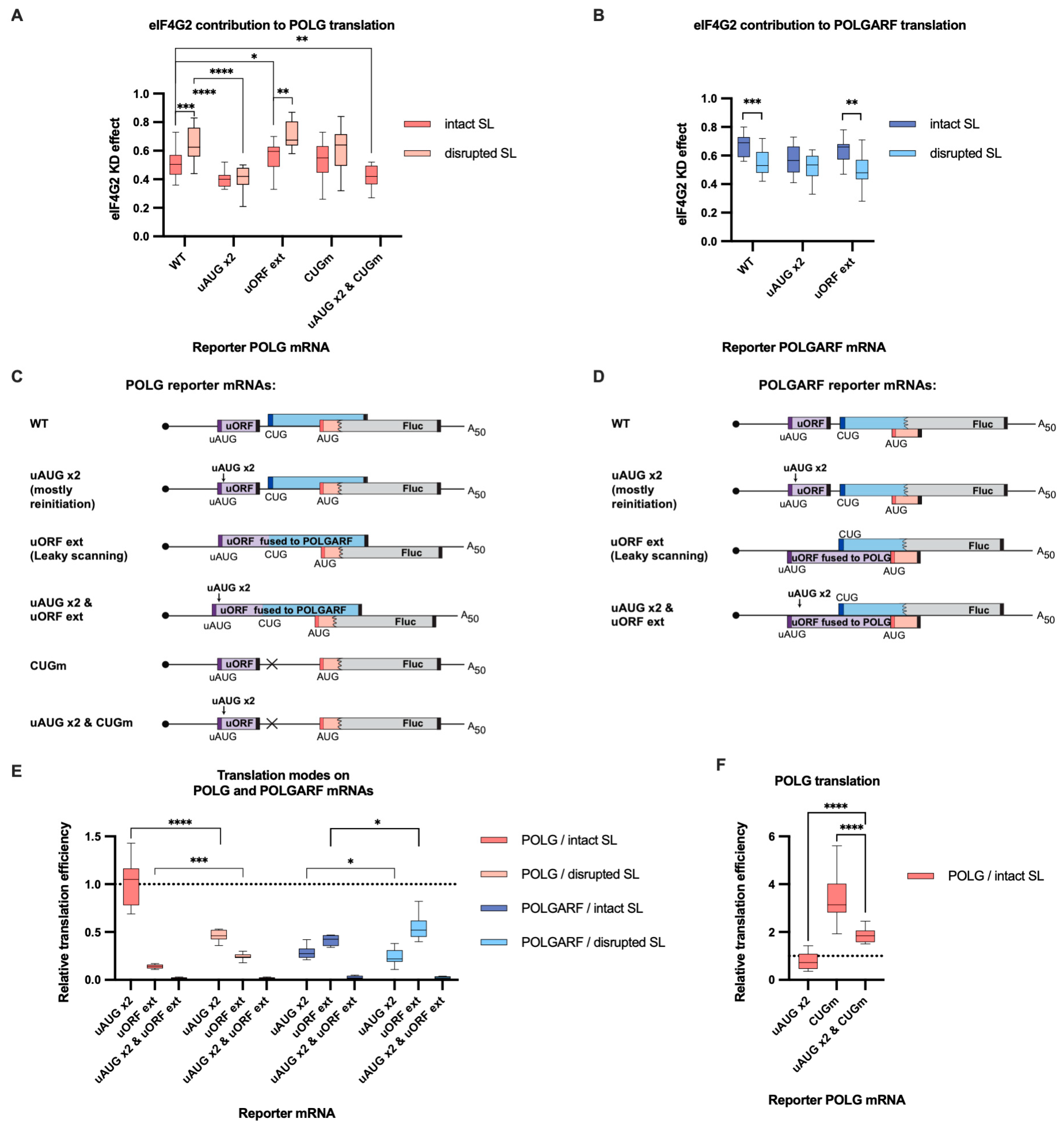
Figure 5.
eIF4G2 promotes both leaky scanning and reinitiation on POLG and POLGARF mRNAs. (A) In vitro transcribed m7G-capped and polyadenylated POLG reporters with the indicated wild-type (WT) or mutated 5′ UTRs were transfected into mock- and eIF4G2-depleted 293T cells alongside reference β-globin reporter mRNA (n ≥ 10). The knockdown was accomplished using siRNA #1 against eIF4G2. Almost all assayed reporters were tested with either a wild-type or a disrupted stem-loop sequence (referred to as “SL intact” and “SL disrupted”, respectively). The knockdown effect (eIF4G2 KD effect) is calculated by dividing normalized reporter expression in eIF4G2-depleted cells by normalized expression in control cells. The knockdown effect < 1 signifies a translation inhibition in the absence of eIF4G2. The statistical significance is determined using the Mann–Whitney U test. The asterisks indicate p-value (one, two, three, and four asterisks stand for p < 0.05, p < 0.01, p < 0.001, and p < 0.001, respectively). (B) Results of POLGARF reporter mRNA transfections (similar to panel A). (C) Schematic representation of the POLG reporters examined in panel A (not to scale). Ribosomes can reach the main POLG start codon via leaky scanning through the uAUG or by reinitiation after completing the translation of the uORF. To estimate the eIF4G2 contribution to the leaky scanning, the uORF stop codon was mutated so that the extended uORF became fused to the POLGARF ORF and overlapped out of frame with the firefly luciferase ORF (referred to as “uORF ext”). To significantly reduce the leaky scanning through the uORF, two extra start AUG codons (referred to as “uAUG x2”) were inserted in frame and near the uAUG codon. On such a 5′ UTR, ribosomes mostly reach the main POLG start codon via reinitiation. Both mutations were introduced into the POLG 5′ UTR (referred to as “uAUG x2 & uORF ext”) to estimate the scanning complexes’ retention at the uORF in-frame start codons. The reinitiated scanning complexes on their way to the POLG start codon might interfere with the 80S ribosomes on the POLGARF ORF, thus increasing the need for eIF4G2. To estimate if this is the case, we compare the eIF4G2 contribution to reinitiation on the “POLG uAUG x2” reporter with that on the corresponding reporter mRNA with a mutated POLGARF start codon (referred to as “uAUG x2 & CUGm”). The start codons are shown in corresponding color bars, and black bars depict stop codons. (D) Panel D is similar to panel C, with the exception that POLGARF reporter mRNAs are displayed. The uORF stop codon was mutated so that the extended uORF became fused to the POLG ORF stub and overlapped out of frame with the firefly luciferase ORF (referred to as “uORF ext”). The POLGARF “uAUG x2” and “uAUG x2 & uORF ext” reporters are constructed similarly to corresponding POLG reporters. (E) Contribution of leaky scanning and reinitiation to translation of POLG and POLGARF mRNAs. Relative translation efficiency is calculated by dividing the normalized expression of a mutant reporter (designated as “uAUG x2” or “uORF ext” to address leaky scanning and reinitiation, respectively) by the normalized expression of a wild-type reporter. When reporters with disrupted stem-loops were used, the relative translation efficiency is shown with respect to corresponding reporters having wild-type start codons and a disrupted stem-loop. The dotted line at 1 corresponds to translation efficiency identical to that of the wild-type reporter mRNA. Note that the contribution of the reinitiation mode can be estimated through the subtraction of leaky scanning mode from 1. The “uAUG x2” reporter constructs arguably reflect the upper estimate for the contribution of reinitiation mode on the wild-type mRNA. The statistical significance was determined using the Mann–Whitney U test. The asterisks indicate p-value (four, three, and one asterisks stand for p < 0.0001, p < 0.001, and p < 0.01, respectively). (F) Reinitiating ribosomes mainly bypass the POLGARF CUG start codon. Relative translation efficiency is calculated in the same way as in Panel E, and statistical significance is determined similarly. Asterisks denote p < 0.0001.
Figure 5.
eIF4G2 promotes both leaky scanning and reinitiation on POLG and POLGARF mRNAs. (A) In vitro transcribed m7G-capped and polyadenylated POLG reporters with the indicated wild-type (WT) or mutated 5′ UTRs were transfected into mock- and eIF4G2-depleted 293T cells alongside reference β-globin reporter mRNA (n ≥ 10). The knockdown was accomplished using siRNA #1 against eIF4G2. Almost all assayed reporters were tested with either a wild-type or a disrupted stem-loop sequence (referred to as “SL intact” and “SL disrupted”, respectively). The knockdown effect (eIF4G2 KD effect) is calculated by dividing normalized reporter expression in eIF4G2-depleted cells by normalized expression in control cells. The knockdown effect < 1 signifies a translation inhibition in the absence of eIF4G2. The statistical significance is determined using the Mann–Whitney U test. The asterisks indicate p-value (one, two, three, and four asterisks stand for p < 0.05, p < 0.01, p < 0.001, and p < 0.001, respectively). (B) Results of POLGARF reporter mRNA transfections (similar to panel A). (C) Schematic representation of the POLG reporters examined in panel A (not to scale). Ribosomes can reach the main POLG start codon via leaky scanning through the uAUG or by reinitiation after completing the translation of the uORF. To estimate the eIF4G2 contribution to the leaky scanning, the uORF stop codon was mutated so that the extended uORF became fused to the POLGARF ORF and overlapped out of frame with the firefly luciferase ORF (referred to as “uORF ext”). To significantly reduce the leaky scanning through the uORF, two extra start AUG codons (referred to as “uAUG x2”) were inserted in frame and near the uAUG codon. On such a 5′ UTR, ribosomes mostly reach the main POLG start codon via reinitiation. Both mutations were introduced into the POLG 5′ UTR (referred to as “uAUG x2 & uORF ext”) to estimate the scanning complexes’ retention at the uORF in-frame start codons. The reinitiated scanning complexes on their way to the POLG start codon might interfere with the 80S ribosomes on the POLGARF ORF, thus increasing the need for eIF4G2. To estimate if this is the case, we compare the eIF4G2 contribution to reinitiation on the “POLG uAUG x2” reporter with that on the corresponding reporter mRNA with a mutated POLGARF start codon (referred to as “uAUG x2 & CUGm”). The start codons are shown in corresponding color bars, and black bars depict stop codons. (D) Panel D is similar to panel C, with the exception that POLGARF reporter mRNAs are displayed. The uORF stop codon was mutated so that the extended uORF became fused to the POLG ORF stub and overlapped out of frame with the firefly luciferase ORF (referred to as “uORF ext”). The POLGARF “uAUG x2” and “uAUG x2 & uORF ext” reporters are constructed similarly to corresponding POLG reporters. (E) Contribution of leaky scanning and reinitiation to translation of POLG and POLGARF mRNAs. Relative translation efficiency is calculated by dividing the normalized expression of a mutant reporter (designated as “uAUG x2” or “uORF ext” to address leaky scanning and reinitiation, respectively) by the normalized expression of a wild-type reporter. When reporters with disrupted stem-loops were used, the relative translation efficiency is shown with respect to corresponding reporters having wild-type start codons and a disrupted stem-loop. The dotted line at 1 corresponds to translation efficiency identical to that of the wild-type reporter mRNA. Note that the contribution of the reinitiation mode can be estimated through the subtraction of leaky scanning mode from 1. The “uAUG x2” reporter constructs arguably reflect the upper estimate for the contribution of reinitiation mode on the wild-type mRNA. The statistical significance was determined using the Mann–Whitney U test. The asterisks indicate p-value (four, three, and one asterisks stand for p < 0.0001, p < 0.001, and p < 0.01, respectively). (F) Reinitiating ribosomes mainly bypass the POLGARF CUG start codon. Relative translation efficiency is calculated in the same way as in Panel E, and statistical significance is determined similarly. Asterisks denote p < 0.0001.
![Ijms 24 17149 g005]()
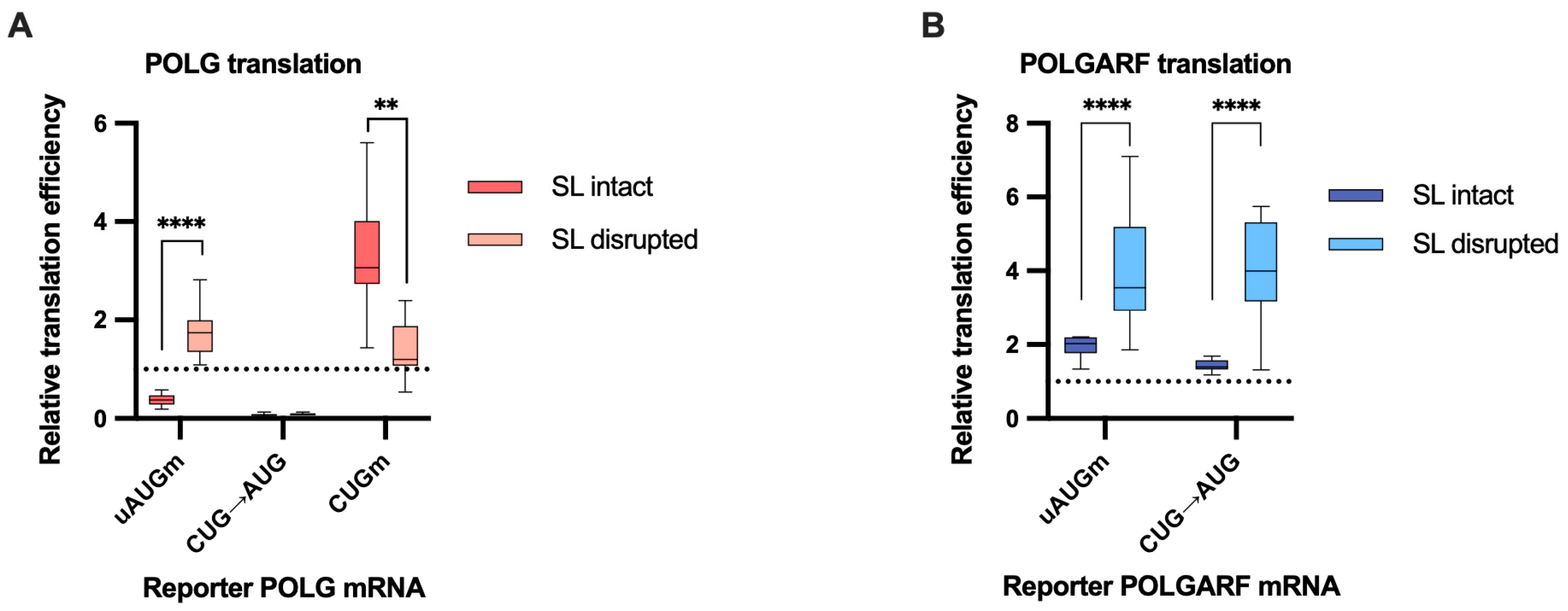
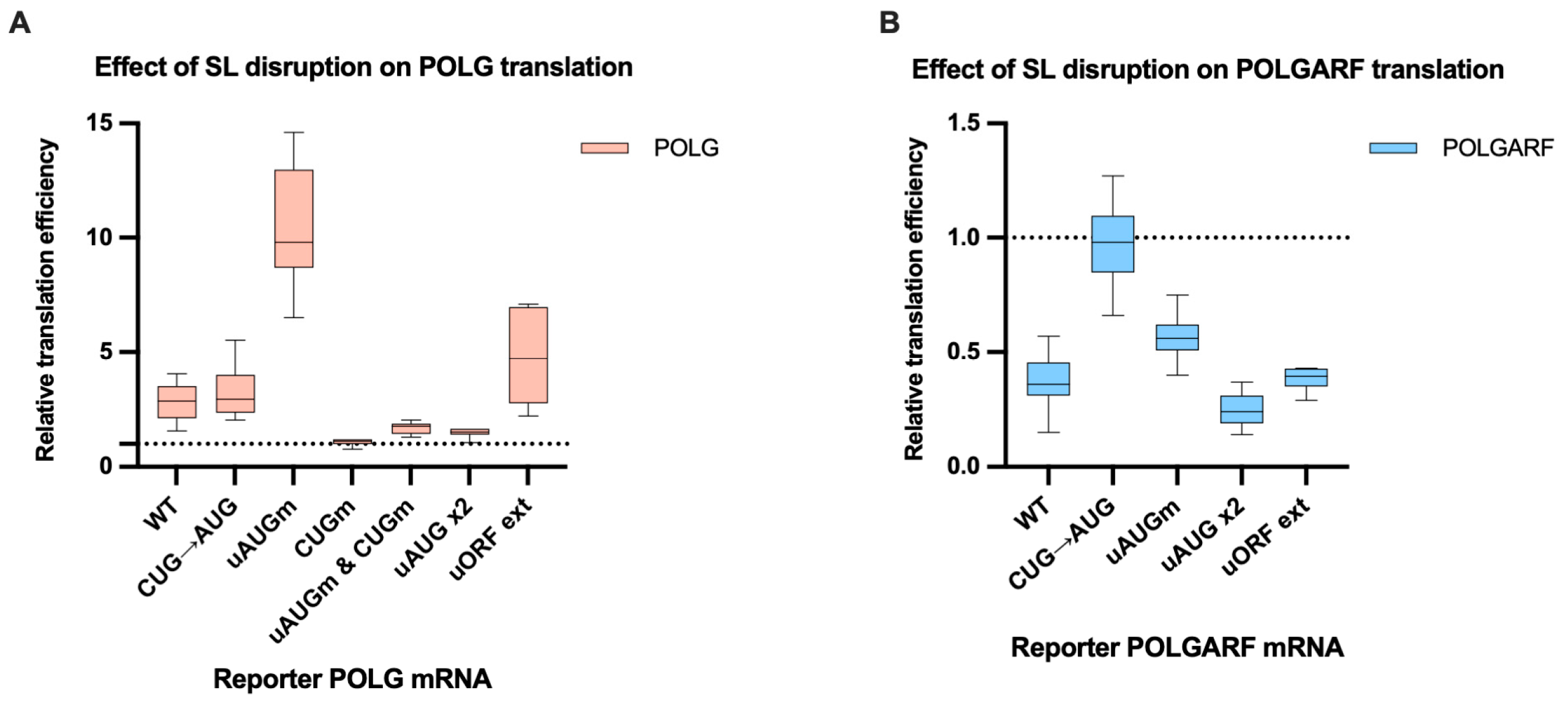
Figure 6.
The disruption of the stem-loop affects the translation efficiency of POLG and POLGARF mRNAs. m7G-capped and polyadenylated reporters with the indicated wild-type (WT) or mutated 5′ UTR were transfected into 293T cells along with the reference β-globin reporter mRNA (n ≥ 15). Nluc activity was used to normalize Fluc reporter expression. All assayed reporters were tested with either a wild-type or a disrupted stem-loop sequence (SL disruption). The effect of the stem-loop disruption is calculated by dividing the normalized expression of a mutant reporter with the disrupted stem-loop by the normalized expression of a corresponding reporter with the intact stem-loop. The dotted line at 1 corresponds to translation efficiency identical to that of the wild-type reporter mRNA. (A) Results of reporter POLG mRNA transfections. (B) Results of reporter POLGARF mRNA transfections (similar to panel (A)).
Figure 6.
The disruption of the stem-loop affects the translation efficiency of POLG and POLGARF mRNAs. m7G-capped and polyadenylated reporters with the indicated wild-type (WT) or mutated 5′ UTR were transfected into 293T cells along with the reference β-globin reporter mRNA (n ≥ 15). Nluc activity was used to normalize Fluc reporter expression. All assayed reporters were tested with either a wild-type or a disrupted stem-loop sequence (SL disruption). The effect of the stem-loop disruption is calculated by dividing the normalized expression of a mutant reporter with the disrupted stem-loop by the normalized expression of a corresponding reporter with the intact stem-loop. The dotted line at 1 corresponds to translation efficiency identical to that of the wild-type reporter mRNA. (A) Results of reporter POLG mRNA transfections. (B) Results of reporter POLGARF mRNA transfections (similar to panel (A)).
Figure 7.
eIF4G2 promotes both leaky scanning and reinitiation on Stard7, Maf1, and UCP2 mRNAs. (A) In vitro transcribed m7G-capped and polyadenylated Maf1, Stard7, and UCP2 reporters with the indicated wild-type (WT) or mutated 5′ UTRs were transfected into mock- and eIF4G2-depleted (siRNA#1) 293T cells alongside reference β-globin reporter mRNA (n ≥ 10). The effect of the knockdown (eIF4G2 KD effect) is calculated by dividing normalized reporter expression in eIF4G2-depleted cells by normalized expression in control cells. The knockdown effect <1 shows translation inhibition in the absence of eIF4G2. (B) Panel B is similar to panel A, except siRNA #2 was used for the eIF4G2 depletion in 293T cells (n ≥ 10). (C) Schematic representation of the reporters examined in panels A, B, and E (not in scale). Stard7, Maf1, and UCP2 5′UTRs contain uORFs (shown in blue box). Dark blue and red bars depict uAUG and main start codons, respectively, and black bars depict stop codons. All analyzed 5′ UTRs were mutated to address leaky scanning and reinitiation separately. Leaky scanning is addressed using an mRNA reporter with an extended uORF that overlaps significantly with the Fluc coding sequence (referred to as “uORF ext”). The insertion of two additional uAUGs into uORF (designated as “uAUG x2”) makes the reinitiation nearly the only way for ribosomes to reach the main start codon. To estimate the scanning complexes retention at the uORF in-frame start codons, both mutations were introduced into these 5′ UTRs (referred to as “uAUG x2 & uORF ext”). (D) Ribosome Decision Graphs representing translation as multiple ribosome paths through the wild-type or mutated 5′ UTRs. Boxes demonstrate ORFs. Circles depict branching points where the ribosome makes a “decision” of whether to initiate or not. The path of leaky scanning complexes towards the downstream start codon is shown in dark grey. Light grey paths represent the post-terminating small ribosome subunit that resumes scanning and can initiate on the main start codon (reinitiation path). The dotted paths represent the significant decrease in the corresponding mode of initiation. (E) The contributions of leaky scanning and reinitiation to the translation of Stard7, Maf1, and UCP2 mRNAs. Relative translation efficiency is calculated by dividing the normalized expression of a mutant reporter (designated as “uAUG x2” or “uORF ext” to address leaky scanning and reinitiation, respectively) by the normalized expression of a wild-type reporter (n ≥ 10). The dotted line at 1 corresponds to translation efficiency identical to that of the wild-type reporter mRNA. Note that the contribution of the reinitiation mode can be estimated through subtraction of the leaky scanning mode from 1. The “uAUG x2” reporter constructs reflect the upper estimate for the contribution of the reinitiation mode on the wild-type mRNA.
Figure 7.
eIF4G2 promotes both leaky scanning and reinitiation on Stard7, Maf1, and UCP2 mRNAs. (A) In vitro transcribed m7G-capped and polyadenylated Maf1, Stard7, and UCP2 reporters with the indicated wild-type (WT) or mutated 5′ UTRs were transfected into mock- and eIF4G2-depleted (siRNA#1) 293T cells alongside reference β-globin reporter mRNA (n ≥ 10). The effect of the knockdown (eIF4G2 KD effect) is calculated by dividing normalized reporter expression in eIF4G2-depleted cells by normalized expression in control cells. The knockdown effect <1 shows translation inhibition in the absence of eIF4G2. (B) Panel B is similar to panel A, except siRNA #2 was used for the eIF4G2 depletion in 293T cells (n ≥ 10). (C) Schematic representation of the reporters examined in panels A, B, and E (not in scale). Stard7, Maf1, and UCP2 5′UTRs contain uORFs (shown in blue box). Dark blue and red bars depict uAUG and main start codons, respectively, and black bars depict stop codons. All analyzed 5′ UTRs were mutated to address leaky scanning and reinitiation separately. Leaky scanning is addressed using an mRNA reporter with an extended uORF that overlaps significantly with the Fluc coding sequence (referred to as “uORF ext”). The insertion of two additional uAUGs into uORF (designated as “uAUG x2”) makes the reinitiation nearly the only way for ribosomes to reach the main start codon. To estimate the scanning complexes retention at the uORF in-frame start codons, both mutations were introduced into these 5′ UTRs (referred to as “uAUG x2 & uORF ext”). (D) Ribosome Decision Graphs representing translation as multiple ribosome paths through the wild-type or mutated 5′ UTRs. Boxes demonstrate ORFs. Circles depict branching points where the ribosome makes a “decision” of whether to initiate or not. The path of leaky scanning complexes towards the downstream start codon is shown in dark grey. Light grey paths represent the post-terminating small ribosome subunit that resumes scanning and can initiate on the main start codon (reinitiation path). The dotted paths represent the significant decrease in the corresponding mode of initiation. (E) The contributions of leaky scanning and reinitiation to the translation of Stard7, Maf1, and UCP2 mRNAs. Relative translation efficiency is calculated by dividing the normalized expression of a mutant reporter (designated as “uAUG x2” or “uORF ext” to address leaky scanning and reinitiation, respectively) by the normalized expression of a wild-type reporter (n ≥ 10). The dotted line at 1 corresponds to translation efficiency identical to that of the wild-type reporter mRNA. Note that the contribution of the reinitiation mode can be estimated through subtraction of the leaky scanning mode from 1. The “uAUG x2” reporter constructs reflect the upper estimate for the contribution of the reinitiation mode on the wild-type mRNA.
![Ijms 24 17149 g007]()
Figure 8.
In vitro translation with eIF1. Reporter mRNAs were translated in S20 cell extract from Expi293F cells in the presence of eIF1 or a storage buffer (n ≥ 5). The Fluc activity (or Nluc in the case of the eIF1 reporter) was measured. Translation inhibition by eIF1 is demonstrated relative to reporter activity in the presence of a storage buffer. The dotted line at 1 represents translation that is completely unresponsive to eIF1. (A) eIF1 inhibits translation of reporter POLG mRNAs. (B) The effect of eIF1 on the translation of POLGARF reporter mRNAs. (C) The effect of eIF1 on the translation of β-globin, eIF5, eIF1, and eIF4G2 reporter mRNAs. The reporter mRNA for eIF1 and eIF4G2 has wild-type GUG start codons.
Figure 8.
In vitro translation with eIF1. Reporter mRNAs were translated in S20 cell extract from Expi293F cells in the presence of eIF1 or a storage buffer (n ≥ 5). The Fluc activity (or Nluc in the case of the eIF1 reporter) was measured. Translation inhibition by eIF1 is demonstrated relative to reporter activity in the presence of a storage buffer. The dotted line at 1 represents translation that is completely unresponsive to eIF1. (A) eIF1 inhibits translation of reporter POLG mRNAs. (B) The effect of eIF1 on the translation of POLGARF reporter mRNAs. (C) The effect of eIF1 on the translation of β-globin, eIF5, eIF1, and eIF4G2 reporter mRNAs. The reporter mRNA for eIF1 and eIF4G2 has wild-type GUG start codons.
Figure 9.
The proposed eIF4G2′s roles in cap-dependent translation of mRNA with uORFs. (
A) The initial steps of translation initiation occur in accordance with the conventional Kozak’s mechanism. The small ribosome subunit is recruited to the 5′ end of mRNA via interaction between m
7G-cap and eIF4F. Then eIF4G1 facilitates scanning from the very 5′ end. The 80S ribosome is depicted translating the uORF. uORF translation leads to the need for eIF4G2 that can mediate both leaky scanning and reinitiation on a single mRNA at the same time. (
B) The role of eIF4G2 in leaky scanning has been linked to a partial loss of eIF4G1 caused by collisions of the scanning complex and the 80S ribosome within the uORF. Through interaction with eIF4E, the eIF4G1 gets back to the m
7G-cap, and the role of helicase is then assumed by the eIF4G2–eIF4A complex, thereby mediating leaky scanning. (
C) eIF4G2 can also facilitate reinitiation on the same mRNA. eIF4G2 assists the post-terminating 80S ribosome to resume scanning and thus contributes to reinitiation. The scheme from [
5] was elaborated.
Figure 9.
The proposed eIF4G2′s roles in cap-dependent translation of mRNA with uORFs. (
A) The initial steps of translation initiation occur in accordance with the conventional Kozak’s mechanism. The small ribosome subunit is recruited to the 5′ end of mRNA via interaction between m
7G-cap and eIF4F. Then eIF4G1 facilitates scanning from the very 5′ end. The 80S ribosome is depicted translating the uORF. uORF translation leads to the need for eIF4G2 that can mediate both leaky scanning and reinitiation on a single mRNA at the same time. (
B) The role of eIF4G2 in leaky scanning has been linked to a partial loss of eIF4G1 caused by collisions of the scanning complex and the 80S ribosome within the uORF. Through interaction with eIF4E, the eIF4G1 gets back to the m
7G-cap, and the role of helicase is then assumed by the eIF4G2–eIF4A complex, thereby mediating leaky scanning. (
C) eIF4G2 can also facilitate reinitiation on the same mRNA. eIF4G2 assists the post-terminating 80S ribosome to resume scanning and thus contributes to reinitiation. The scheme from [
5] was elaborated.
![Ijms 24 17149 g009]()