Distribution and Location of BEVs in Different Genotypes of Bananas Reveal the Coevolution of BSVs and Bananas
Abstract
:1. Introduction
2. Results
2.1. Detection of BEVs
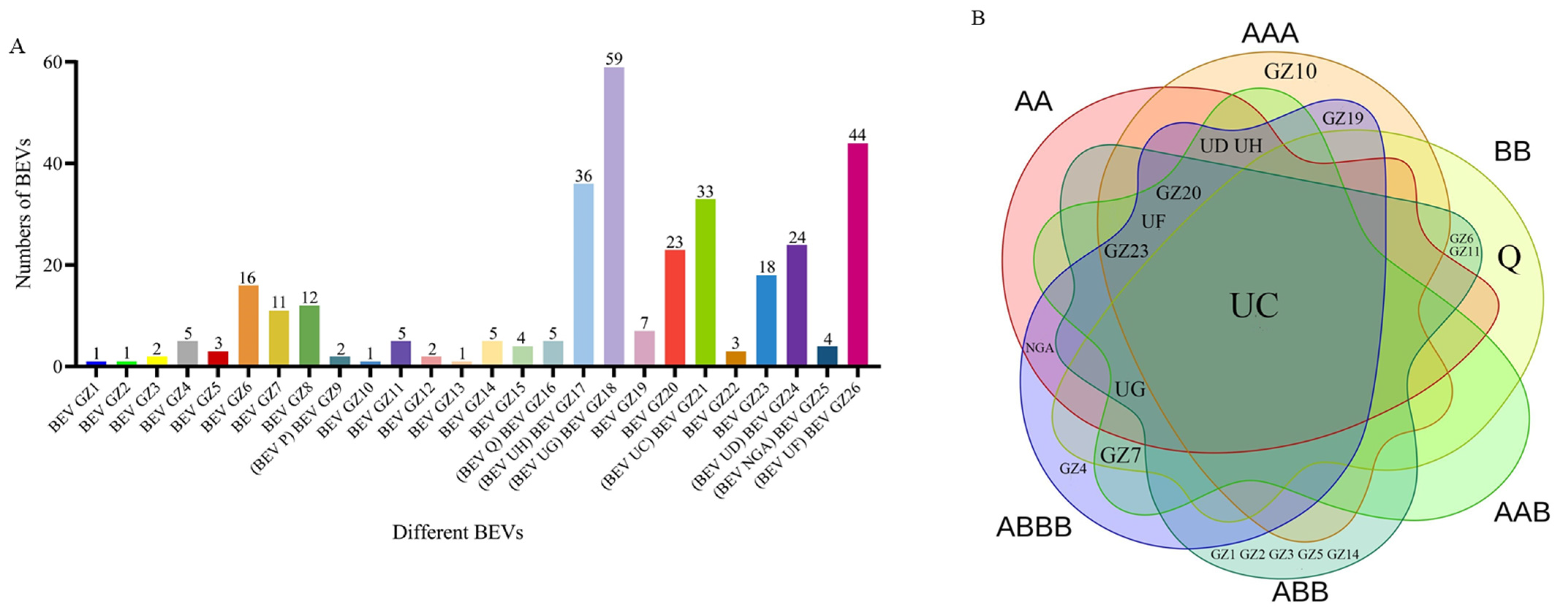
2.2. BEVs in Different Genotypes of Bananas
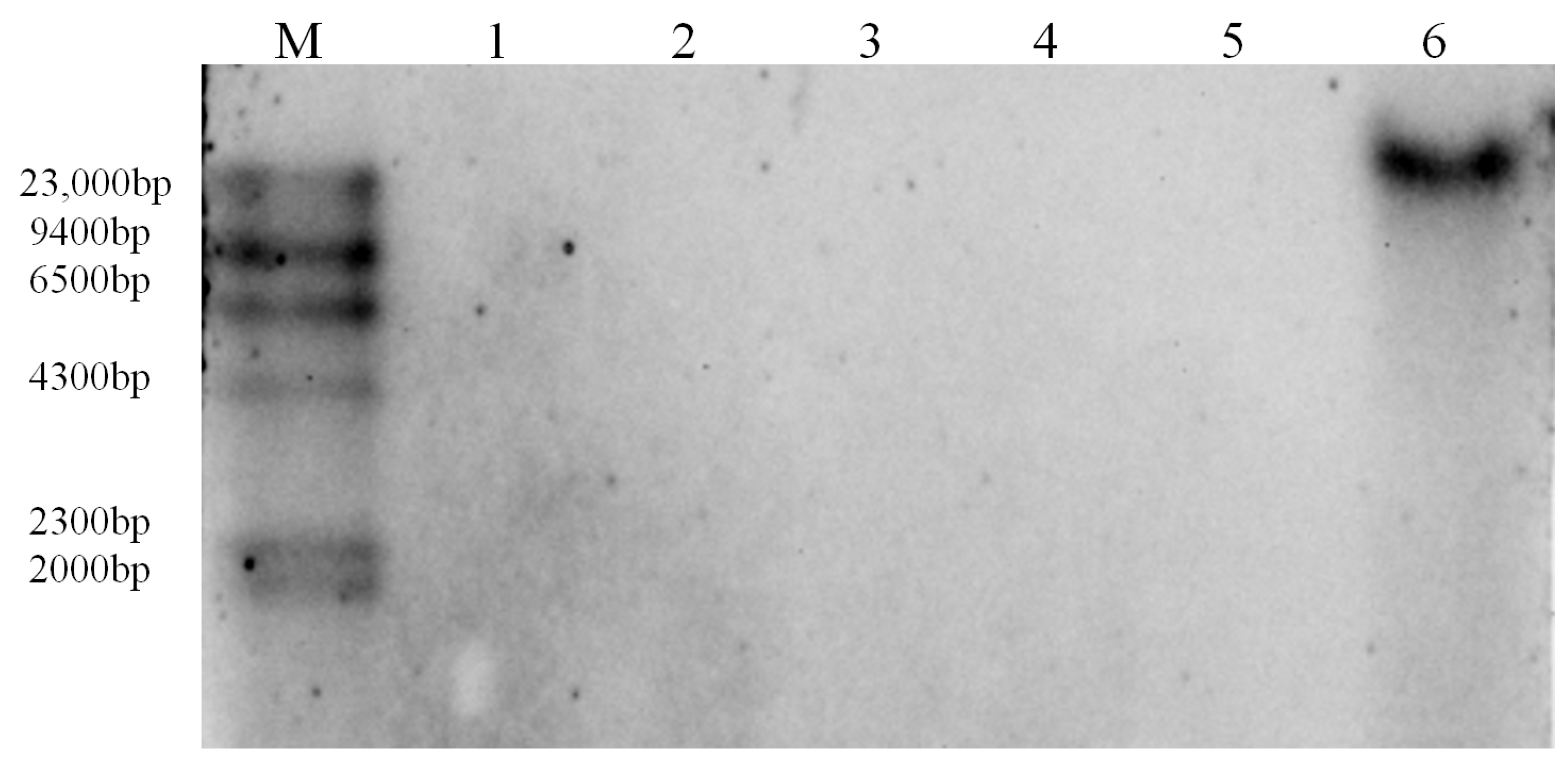
2.3. Southern Blot Analyses of BEVs
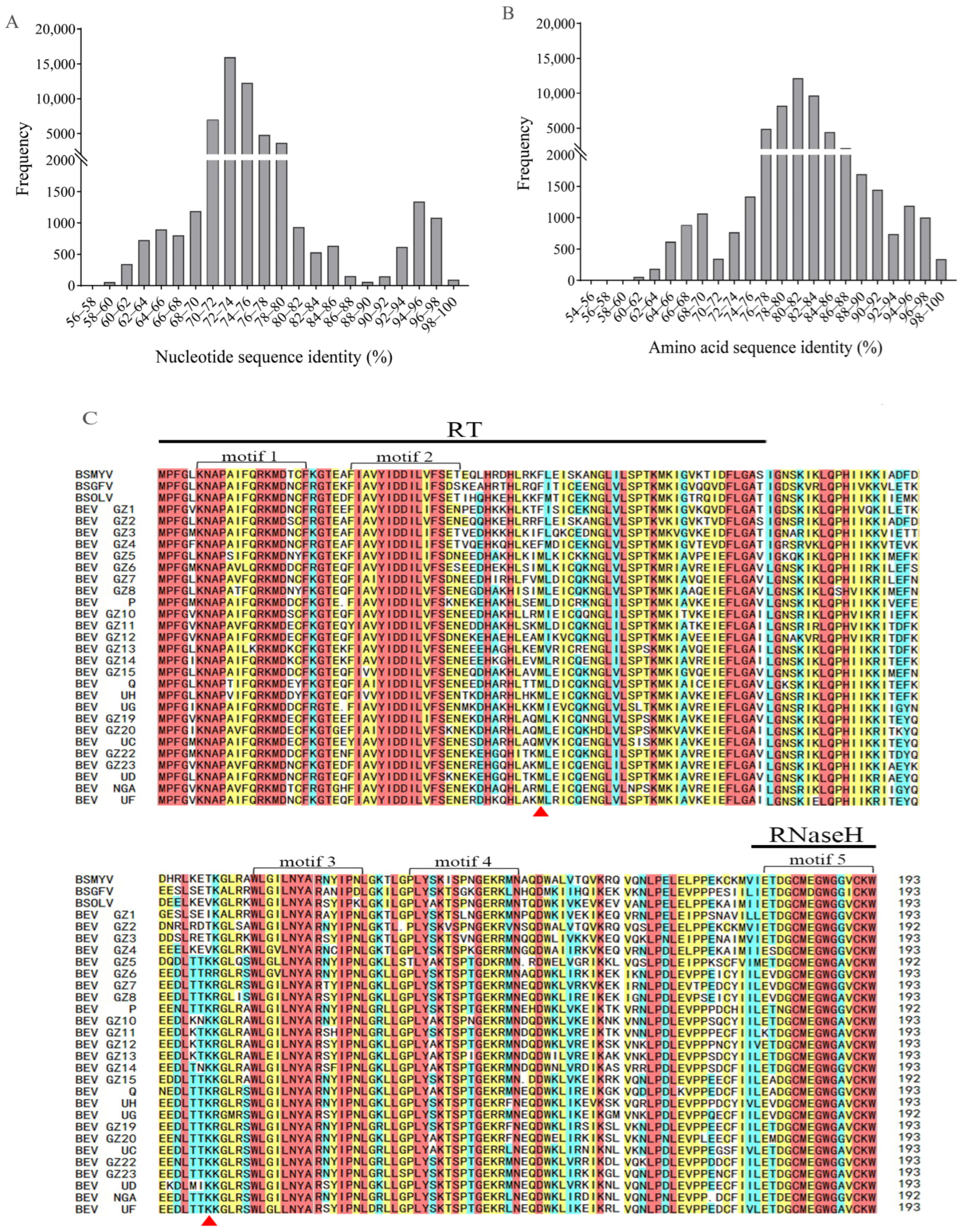
2.4. Sequence Analyses between BEVs and BSVs
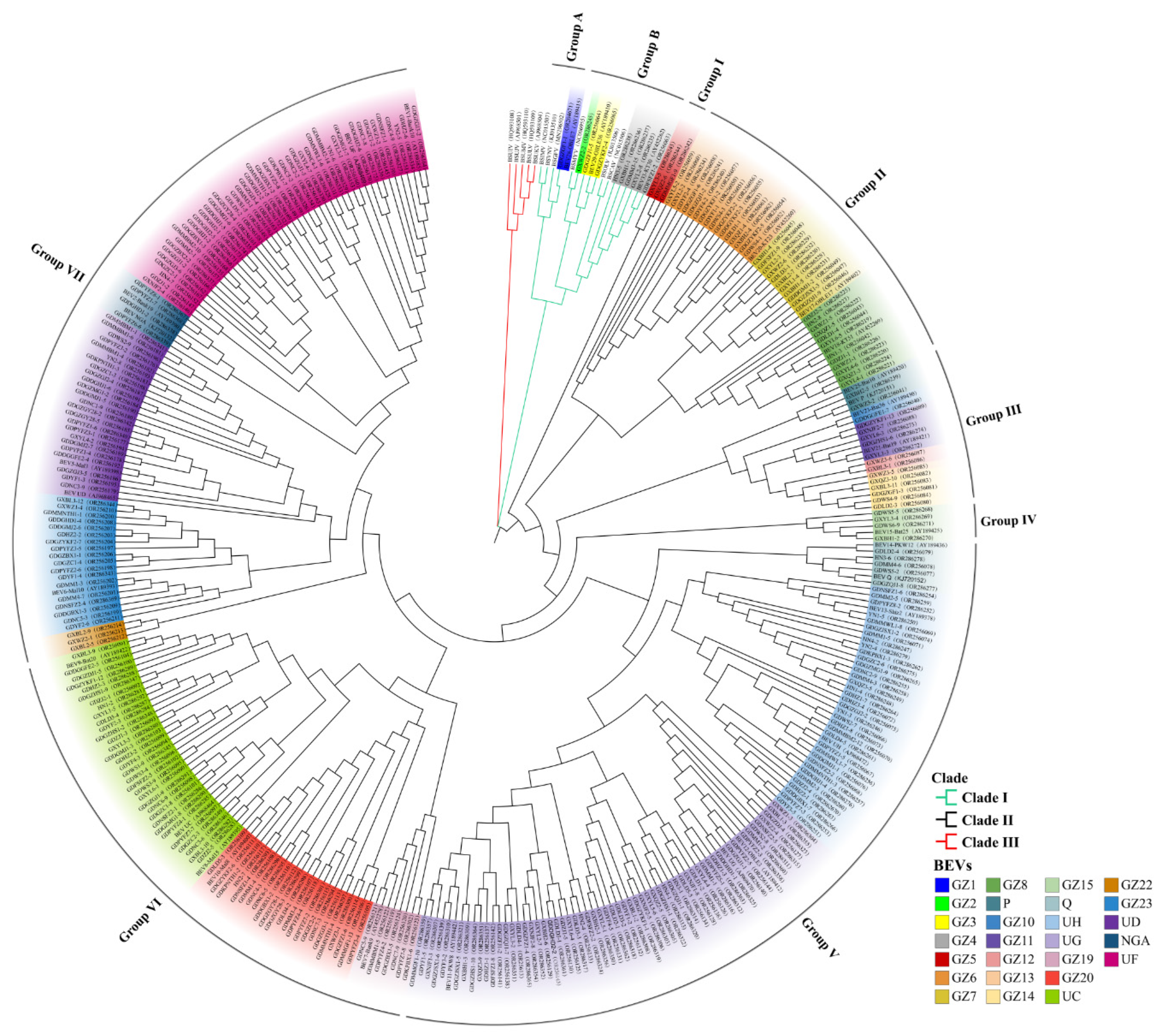
2.5. Phylogenetic Analyses of BEVs
2.6. Relationship between BEVs in Different Clades and the Genotypes of Bananas
2.7. Distribution and Locations of BEVs on Chromosomes of Different Genotypes of Bananas
2.7.1. Distribution and Location of BEVs on the Chromosomes of M. acuminata subsp. DH-Pahang (AA)
2.7.2. Distribution and Location of BEVs on the Chromosomes of M. balbisiana subsp. PKW (BB)
2.7.3. Distribution and Location of BEVs on the Chromosomes of M. schizocarpa subsp. HN8 (SS)
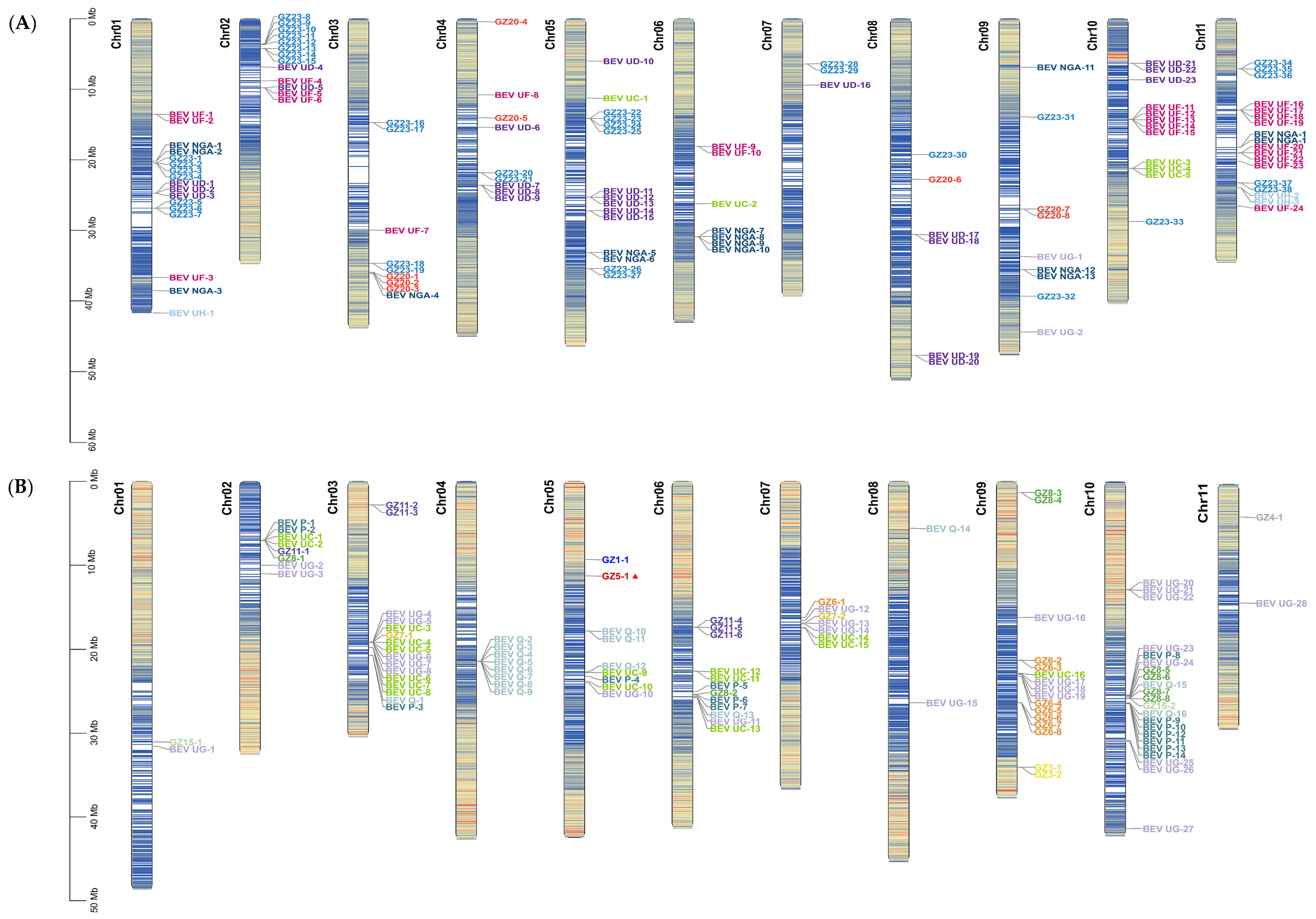
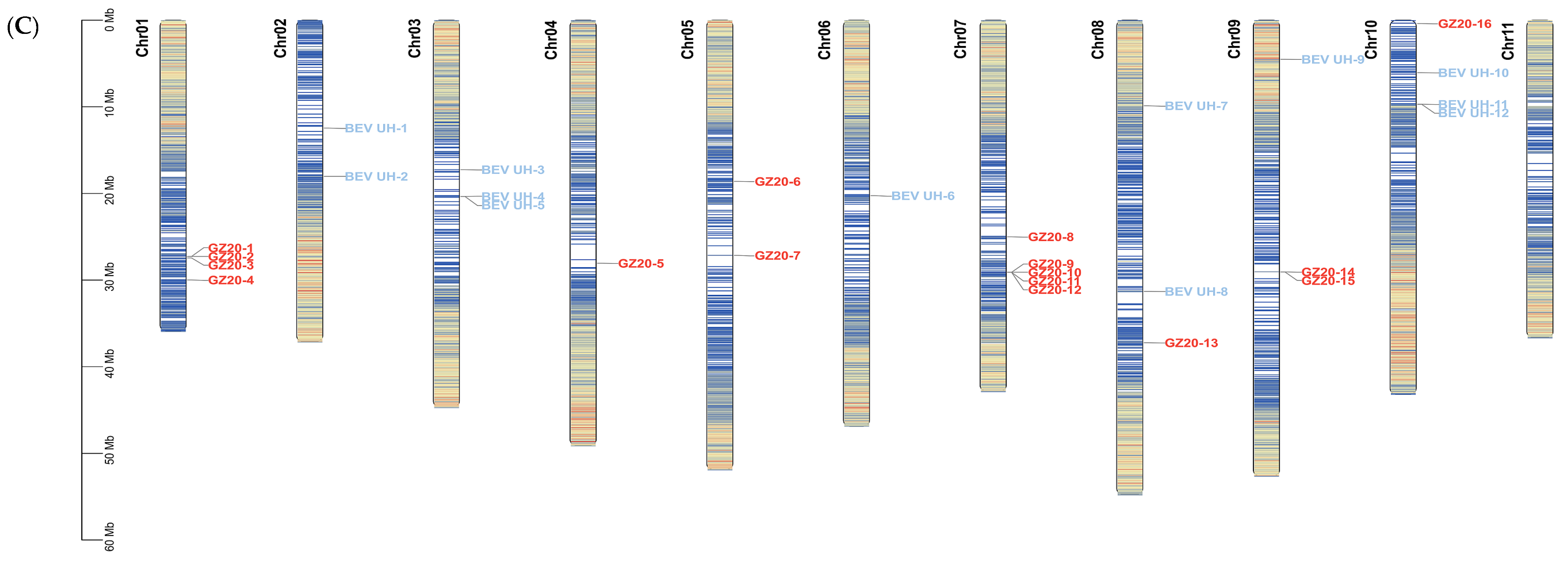
2.7.4. Analyses of BEVs in Different Clades on the Different Chromosomes
2.8. Synteny Analyses of BEVs on the Chromosomes of Different Bananas
3. Discussion
3.1. BEVs in Different Genotypes of Bananas
3.2. Genetic Diversity of BEVs
3.3. Location of BEVs on Different Banana Chromosomes
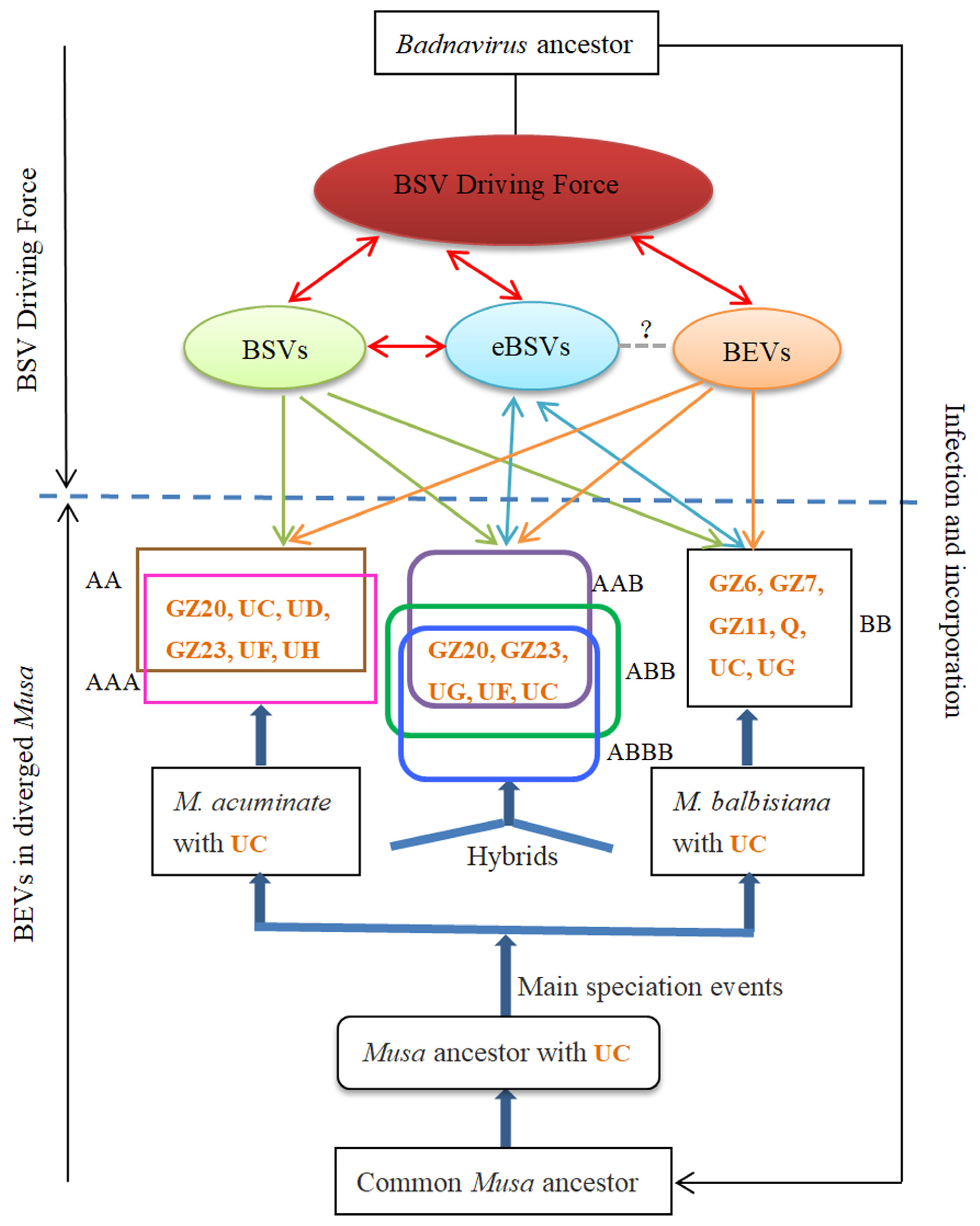
3.4. Coevolution of BEVs with BSVs and Bananas
4. Materials and Methods
4.1. Plant Materials
4.2. DNA Extractions of Banana Samples and Cloning and Sequencing
4.3. Southern Blot
4.4. BEVs Sequence Analyses and Phylogenetic Analyses
4.5. Analyses of BEVs Distribution on Banana Chromosomes
4.6. Collinear Analyses of BEVs in Banana Genome
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Staginnus, C.; Iskra-Caruana, M.L.; Lockhart, B.; Hohn, T.; Richert-Pöggeler, K.R. Suggestions for a nomenclature of endogenous pararetroviral sequences in plants. Arch. Virol. 2009, 154, 1189–1193. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.P.; Lockhart, B.E.; Olszewski, N.E. The ORF I and II proteins of commelina yellow mottle virus are virion-associated. Virology 1996, 223, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Jaufeerally-Fakim, Y.; Khorugdharry, A.; Harper, G. Genetic variants of banana streak virus in Mauritius. Virus Res. 2006, 115, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Kunii, M.; Kanda, M.; Nagano, H.; Uyeda, I.; Kishima, Y.; Sano, Y. Reconstruction of putative DNA virus from endogenous rice tungro bacilliform virus-like sequences in the rice genome: Implications for integration and evolution. BMC Genom. 2004, 5, 80. [Google Scholar] [CrossRef] [PubMed]
- Gayral, P.; Noa-Carrazana, J.C.; Lescot, M.; Lheureux, F.; Lockhart, B.E.; Matsumoto, T.; Piffanelli, P.; Iskra-Caruana, M.L. A single banana streak virus integration event in the banana genome as the origin of infectious endogenous pararetrovirus. J. Virol. 2008, 82, 6697–6710. [Google Scholar] [CrossRef] [PubMed]
- Harper, G.; Osuji, J.O.; Heslop-Harrison, J.S.; Hull, R. Integration of banana streak badnavirus into the Musa genome: Molecular and cytogenetic evidence. Virology 1999, 255, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Ndowora, T.U.O.M.; Dehal, G.; LaFleur, D.; Harper, G.; Hull, R.; Olszewski, N.E.; Lockhart, B. Evidence that badnavirus infection in Musa can originate from integrated pararetroviral sequences. Virology 1999, 255, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Chabannes, M.; Baurens, F.; Duroy, P.; Bocs, S.; Vernerey, M.; Rodier-Goud, M.; Barbe, V.; Gayral, P.; Iskra-Caruana, M. Three infectious viral species lying in wait in the banana genome. J. Virol. 2013, 87, 8624–8637. [Google Scholar] [CrossRef]
- Duroy, P.O.; Perrier, X.; Laboureau, N.; Jacquemoud-Collet, J.P.; Iskra-Caruana, M.L. How endogenous plant pararetroviruses shed light on Musa evolution. Ann. Bot. 2016, 117, 625–641. [Google Scholar] [CrossRef]
- Geering, A.D.W.; Olszewski, N.E.; Harper, G.; Lockhart, B.E.L.; Hull, R.; Thomas, J.E. Banana contains a diverse array of endogenous badnaviruses. J. Gen. Virol. 2005, 86, 511–520. [Google Scholar] [CrossRef]
- Chen, S.; Zheng, H.; Kishima, Y. Genomic fossils reveal adaptation of non-autonomous pararetroviruses driven by concerted evolution of noncoding regulatory sequences. PLoS Pathog. 2017, 13, e1006413. [Google Scholar] [CrossRef] [PubMed]
- Gayral, P.; Iskra-Caruana, M.L. Phylogeny of banana streak virus reveals recent and repetitive endogenization in the genome of its banana host (Musa sp.). J. Mol. Evol. 2009, 69, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Feschotte, C.; Gilbert, C. Endogenous viruses: Insights into viral evolution and impact on host biology. Nat. Rev. Genet. 2012, 13, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Staginnus, C.; Richert-Poggeler, K.R. Endogenous pararetroviruses: Two-faced travelers in the plant genome. Trends Plant Sci. 2006, 11, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Iskra-Caruana, M.L.; Duroy, P.O.; Chabannes, M.; Muller, E. The common evolutionary history of badnaviruses and banana. Infect. Genet. Evol. 2014, 21, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Iskra-Caruana, M.L.; Chabannes, M.; Duroy, P.O.; Muller, E. A possible scenario for the evolution of banana streak virus in banana. Virus Res. 2014, 186, 155–162. [Google Scholar] [CrossRef] [PubMed]
- King, A.M.Q.; Adams, M.J.; Lefkowitz, E.J.; Carstens, E.B. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses; Academic Press: San Diego, CA, USA, 2012; p. 1463. [Google Scholar]
- Harper, G.; Hart, D.; Moult, S.; Hull, R.; Geering, A.; Thomas, J. The diversity of banana streak virus isolates in Uganda. Arch. Virol. 2005, 150, 2407–2420. [Google Scholar] [CrossRef]
- James, A.P.; Geijskes, R.J.; Dale, J.L.; Harding, R.M. Development of a novel rolling-circle amplification technique to detect banana streak virus that also discriminates between integrated and episomal virus sequences. Plant Dis. 2011, 95, 57–62. [Google Scholar] [CrossRef]
- Chabannes, M.; Gabriel, M.; Aksa, A.; Galzi, S.; Dufayard, J.F.; Iskra-Caruana, M.L.; Muller, E. Badnaviruses and banana genomes: A long association sheds light on Musa phylogeny and origin. Mol. Plant Pathol. 2021, 22, 216–230. [Google Scholar] [CrossRef]
- D’Hont, A.; Denoeud, F.; Aury, J.M.; Baurens, F.C.; Carreel, F.; Garsmeur, O.; Noel, B.; Bocs, S.; Droc, G.; Rouard, M.; et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 2012, 488, 213–217. [Google Scholar] [CrossRef]
- Simmonds, N.W.; Weatherup, S.T.C. Numerical taxonomy of the wild bananas (Musa). New Phytol. 1990, 115, 567–571. [Google Scholar] [CrossRef] [PubMed]
- Geering, A.D.; Olszewski, N.E.; Dahal, G.; Thomas, J.E.; Lockhart, B.E. Analysis of the distribution and structure of integrated banana streak virus DNA in a range of Musa cultivars. Mol. Plant Pathol. 2001, 2, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Harper, G.; Hull, R.; Lockhart, B.; Olszewski, N. Viral sequences integrated into plant genomes. Annu. Rev. Phytopathol. 2002, 40, 119–136. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Wang, X.; Lu, Z.; Xu, Y.; Deng, X.; Xu, Q. Endogenous pararetrovirus sequences are widely present in citrinae genomes. Virus Res. 2019, 262, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Kuriyama, K.; Tabara, M.; Moriyama, H.; Kanazawa, A.; Koiwa, H.; Takahashi, H.; Fukuhara, T. Disturbance of floral colour pattern by activation of an endogenous pararetrovirus, petunia vein clearing virus, in aged petunia plants. Plant. J. 2020, 103, 497–511. [Google Scholar] [CrossRef] [PubMed]
- Geering, A.D.; Maumus, F.; Copetti, D.; Choisne, N.; Zwickl, D.J.; Zytnicki, M.; McTaggart, A.R.; Scalabrin, S.; Vezzulli, S.; Wing, R.A.; et al. Endogenous florendoviruses are major components of plant genomes and hallmarks of virus evolution. Nat. Commun. 2014, 5, 5269. [Google Scholar] [CrossRef]
- Ge, X.J.; Liu, M.H.; Wang, W.K.; Schaal, B.A.; Chiang, T.Y. Population structure of wild bananas, Musa balbisiana, in China determined by SSR fingerprinting and cpDNA PCR-RFLP. Mol. Ecol. 2005, 14, 933–944. [Google Scholar] [CrossRef]
- Perrier, X.; De Langhe, E.; Donohue, M.; Lentfer, C.; Vrydaghs, L.; Bakry, F.; Carreel, F.; Hippolyte, I.; Horry, J.P.; Jenny, C.; et al. Multidisciplinary perspectives on banana (Musa spp.) domestication. Proc. Natl. Acad. Sci. USA 2011, 108, 11311–11318. [Google Scholar] [CrossRef]
- Xiong, Y.; Eickbush, T.H. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990, 9, 3353–3362. [Google Scholar] [CrossRef]
- Richert-Poggeler, K.R.; Vijverberg, K.; Alisawi, O.; Chofong, G.N.; Heslop-Harrison, J.; Schwarzacher, T. Participation of multifunctional RNA in replication, recombination and regulation of endogenous plant pararetroviruses (EPRVs). Front. Plant Sci. 2021, 12, 689307. [Google Scholar] [CrossRef]
- Cui, J.; Holmes, E.C. Endogenous RNA viruses of plants in insect genomes. Virology 2012, 427, 77–79. [Google Scholar] [CrossRef] [PubMed]
- Hull, R.; Harper, G.; Lockhart, B. Viral sequences integrated into plant genomes. Trends Plant Sci. 2000, 5, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Mette, M.F.; Kanno, T.; Aufsatz, W.; Jakowitsch, J.; van der Winden, J.; Matzke, M.A.; Matzke, A.J. Endogenous viral sequences and their potential contribution to heritable virus resistance in plants. EMBO J. 2002, 21, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Jakowitsch, J.; Mette, M.F.; van Der Winden, J.; Matzke, M.A.; Matzke, A.J. Integrated pararetroviral sequences define a unique class of dispersed repetitive DNA in plants. Proc. Natl. Acad. Sci. USA 1999, 96, 13241–13246. [Google Scholar] [CrossRef] [PubMed]
- Iskra-Caruana, M.; Baurens, F.; Gayral, P.; Chabannes, M. A four-partner plant–virus interaction: Enemies can also come from within. Mol. Plant. Microbe Interact. 2010, 23, 1394–1402. [Google Scholar] [CrossRef]
- Geering, A.D.; McMichael, L.A.; Dietzgen, R.G.; Thomas, J.E. Genetic diversity among banana streak virus isolates from Australia. Phytopathology 2000, 90, 921–927. [Google Scholar] [CrossRef]
- Rothnie, H.M.; Chapdelaine, Y.; Hohn, T. Pararetroviruses and retroviruses: A comparative review of viral structure and gene expression strategies. Adv. Virus. Res. 1994, 44, 1–67. [Google Scholar] [CrossRef]
- Matzke, M.; Gregor, W.; Mette, M.F.; Ufsatz, W.; Kanno, T.; Jakowitsch, J.; Matzke, A.J.M. Endogenous pararetroviruses of allotetraploid Nicotiana tabacum and its diploid progenitors, N. sylvestris and N. tomentosiformis. Biol. J. Linn. Soc. 2004, 82, 627–638. [Google Scholar] [CrossRef]
- Carreel, F.; de Leon, D.G.; Lagoda, P.; Lanaud, C.; Jenny, C.; Horry, J.P.; du Montcel, H.T. Ascertaining maternal and paternal lineage within Musa by chloroplast and mitochondrial DNA RFLP analyses. Genome 2002, 45, 679–692. [Google Scholar] [CrossRef]
- White, S.E.; Habera, L.F.; Wessler, S.R. Retrotransposons in the flanking regions of normal plant genes: A role for copia-like elements in the evolution of gene structure and expression. Proc. Natl. Acad. Sci. USA 1994, 91, 11792–11796. [Google Scholar] [CrossRef]
- Hughes, J.F.; Coffin, J.M. Evidence for genomic rearrangements mediated by human endogenous retroviruses during primate evolution. Nat. Genet. 2001, 29, 487–489. [Google Scholar] [CrossRef] [PubMed]
- Staginnus, C.; Gregor, W.; Mette, M.F.; Teo, C.H.; Borroto-Fernandez, E.G.; Machado, M.L.; Matzke, M.; Schwarzacher, T. Endogenous pararetroviral sequences in tomato (Solanum lycopersicum) and related species. BMC Plant Biol. 2007, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Serfraz, S.; Sharma, V.; Maumus, F.; Aubriot, X.; Geering, A.; Teycheney, P.Y. Insertion of badnaviral DNA in the late blight resistance gene (R1a) of brinjal eggplant (Solanum melongena). Front. Plant Sci. 2021, 12, 683681. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.I.; Hohn, T.; Selvarajan, R. Badnaviruses: The current global scenario. Viruses 2016, 8, 177. [Google Scholar] [CrossRef] [PubMed]
- Cote, F.X.; Galzi, S.; Folliot, M.; Lamagnere, Y.; Teycheney, P.Y.; Iskra-Caruana, M.L. Micropropagation by tissue culture triggers differential expression of infectious endogenous banana streak virus sequences (eBSV) present in the B genome of natural and synthetic interspecific banana plantains. Mol. Plant Pathol. 2010, 11, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.B.; Kasdorf, G.; Nel, L.H.; Pietersen, G. Transmission of activated-episomal banana streak OL (badna) virus (BSOLV) to cv. Williams banana (Musa sp.) by three mealybug species. Plant Dis. 2008, 92, 1158–1163. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, J.N.; Ntui, V.O.; Ron, M.; Muiruri, S.K.; Britt, A.; Tripathi, L. CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Commun. Biol. 2019, 2, 46. [Google Scholar] [CrossRef]
- Umber, M.; Pressat, G.; Fort, G.; Plaisir, P.K.; Guiougiou, C.; Lambert, F.; Farinas, B.; Pichaut, J.P.; Janzac, B.; Delos, J.M.; et al. Risk assessment of infectious endogenous banana streak viruses in Guadeloupe. Front. Plant Sci. 2022, 13, 951285. [Google Scholar] [CrossRef]
- Abraham, A.; Winter, S.; Richert-Pöggeler, K.R.; Menzel, W. Molecular characterization of a new badnavirus associated with streak symptoms on enset (Ensete ventricosum, Musaceae). J. Phytopathol. 2018, 166, 565–571. [Google Scholar] [CrossRef]
- Yang, I.C.; Hafner, G.J.; Revill, P.A.; Dale, J.L.; Harding, R.M. Sequence diversity of South Pacific isolates of taro bacilliform virus and the development of a PCR-based diagnostic test. Arch. Virol. 2003, 148, 1957–1968. [Google Scholar] [CrossRef]
- Le Provost, G.; Iskra-Caruana, M.L.; Acina, I.; Teycheney, P.Y. Improved detection of episomal banana streak viruses by multiplex immunocapture PCR. J. Virol. Methods 2006, 137, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Belser, C.; Baurens, F.C.; Noel, B.; Martin, G.; Cruaud, C.; Istace, B.; Yahiaoui, N.; Labadie, K.; Hribova, E.; Dolezel, J.; et al. Telomere-to-telomere gapless chromosomes of banana using nanopore sequencing. Commun. Biol. 2021, 4, 1047. [Google Scholar] [CrossRef] [PubMed]
- Belser, C.; Istace, B.; Denis, E.; Dubarry, M.; Baurens, F.C.; Falentin, C.; Genete, M.; Berrabah, W.; Chevre, A.M.; Delourme, R.; et al. Chromosome-scale assemblies of plant genomes using nanopore long reads and optical maps. Nat. Plants 2018, 4, 879–887. [Google Scholar] [CrossRef]
- Davey, M.W.; Gudimella, R.; Harikrishna, J.A.; Sin, L.W.; Khalid, N.; Keulemans, J. A draft Musa balbisiana genome sequence for molecular genetics in polyploid, inter-and intra-specific Musa hybrids. BMC Genom. 2013, 14, 683. [Google Scholar] [CrossRef]

| BEVs | Loci | Chromosomes |
|---|---|---|
| GZ20 | 8 | Chr03, Chr04, Chr08, Chr09 |
| GZ23 | 38 | Chr01, Chr02, Chr03, Chr04, Chr05, Chr07, Chr08Chr09, Chr10, Chr11 |
| NGA | 15 | Chr01, Chr03, Chr05, Chr06, Chr09 |
| UC | 5 | Chr05, Chr06, Chr10 |
| UD | 23 | Chr01, Chr02, Chr04, Chr05, Chr07, Chr08, Chr10 |
| UF | 25 | Chr01, Chr02, Chr03, Chr04, Chr06, Chr10, Chr11 |
| UH | 3 | Chr01, Chr11 |
| UG | 2 | Chr09 |
| BEVs | Loci | Chromosomes |
|---|---|---|
| GZ1 | 1 | Chr05 |
| GZ3 | 2 | Chr09 |
| GZ4 | 1 | Chr11 |
| GZ5 | 1 | Chr05 |
| GZ6 | 8 | Chr07, Chr09 |
| GZ7 | 2 | Chr03, Chr07 |
| GZ8 | 8 | Chr02, Chr06, Chr09, Chr10 |
| GZ11 | 6 | Chr02, Chr03, Chr06 |
| GZ15 | 2 | Chr01, Chr10 |
| P | 14 | Chr02, Chr03, Chr05, Chr06, Chr10 |
| Q | 16 | Chr03, Chr04, Chr05, Chr06, Chr08, Chr10 |
| UC | 16 | Chr02, Chr03, Chr05, Chr06, Chr07, Chr09 |
| UF | 1 | Chr01 |
| UG | 28 | Chr01, Chr02, Chr03, Chr05, Chr06, Chr07, Chr08, Chr09, Chr10, Chr11 |
| Banana Cultivars | Genotypes | No. | Collection Time | Collecting Place | Isolates |
|---|---|---|---|---|---|
| GY28 | AA | 1 | December 2021 | College of Horticulture, SCAU | GDGZGY28 |
| Gongjiao | AA | 3 | March 2022 | Institute of Fruit Tree Research, GDAAS | GDGZGJ1–3 |
| Neiguijiao | AA | 1 | March 2022 | Institute of Fruit Tree Research, GDAAS | GDGZMG1 |
| Huangdijiao | AA | 2 | October 2022 | Dongguan, Guangdong | GDDGHD1–2 |
| Hongjiao | AAA | 2 | October 2022 | Dongguan, Guangdong | GDDGHJ1–2 |
| Guifeijiao | AAA | 2 | October 2022 | Dongguan, Guangdong | GDDGGFE1–2 |
| Nantianhuang | AAA | 1 | April 2021 | Maoming, Guangdong | GDMMNTH1 |
| Nantianhuang | AAA | 1 | April 2021 | Kaiping, Guangdong | GDKPNTH1 |
| Weiliansi | AAA | 1 | December 2021 | Maoming, Guangdong | GDMMWL1 |
| Baxijiao | AAA | 1 | December 2021 | College of Horticulture, SCAU | GDGZBX1 |
| Baxijiao | AAA | 1 | September 2021 | Guangzhou, Guangdong | GDGZBX2 |
| Baxijiao | AAA | 1 | April 2021 | Dongguan, Guangdong | GDDGBX1 |
| Baxijiao | AAA | 1 | April 2021 | Kaiping, Guangdong | GDKPBX1 |
| Baxijiao | AAA | 1 | February 2021 | Yunnan Province | YNBX1 |
| Qujiangyejiao | BB | 1 | March 2022 | Institute of Fruit Tree Research, GDAAS | GDGZQJ1 |
| Huashiyejiao | BB | 1 | March 2022 | Institute of Fruit Tree Research, GDAAS | GDGZHS1 |
| Jinshaxiang | AAB | 1 | March 2022 | Institute of Fruit Tree Research, GDAAS | GDGZJSX1 |
| C.La.Nang Tiem | AAB | 2 | March 2022 | Institute of Fruit Tree Research, GDAAS | GDGZC1–2 |
| Dajiao | ABB | 1 | December 2021 | College of Horticulture, SCAU | GDGZDJ1 |
| Fenjiao | ABB | 1 | December 2021 | College of Horticulture, SCAU | GDGZFJ1 |
| Jinfen | ABB | 2 | April 2021 | Guangxi Academy of Agricultural Sciences | GXNJF1–2 |
| Yuekangfen 1 | ABB | 2 | September 2021 | Guangzhou, Guangdong | GDGZYKF1–2 |
| Guangfen 1 | ABB | 1 | April 2021 | Maoming, Guangdong | GDMMGF1 |
| Guangfen 1 | ABB | 1 | September 2021 | Guangzhou, Guangdong | GDGZGF1 |
| Fenza 1 | ABBB | 1 | September 2021 | Guangzhou, Guangdong | GDGZFZ1 |
| Fenza 1 | ABBB | 8 | April 2021 | Guangzhou, Guangdong | GDPYFZ1–8 |
| Fenza 1 | ABBB | 2 | April 2021 | Guangzhou, Guangdong | GDNSFZ1–2 |
| Fenza 1 | ABBB | 2 | September 2021 | Fosan, Guangdong | GDFSFZ1–2 |
| Malaijiao | Unknown | 2 | April 2021 | Maoming, Guangdong | GDMMBM1–2 |
| Yunjiao | Unknown | 1 | February 2021 | Yunnan Province | YN2 |
| Majiao | Unknown | 2 | October 2022 | Dongguan, Guangdong | GDDGMJ1–2 |
| Unknown | Unknown | 6 | February 2021 | Greenhouse, SCAU | GDWS1–6 |
| Unknown | Unknown | 7 | February 2021 | Farm, SCAU | GDNC1–7 |
| Unknown | Unknown | 3 | February 2021 | Huizhou, Guangdong | GDHZ1–3 |
| Unknown | Unknown | 5 | May 2021 | Luoding, Guangdong | GDLD1–5 |
| Unknown | Unknown | 4 | May 2021 | Yunfu, Guangdong | GDYF1–4 |
| Unknown | Unknown | 2 | May 2021 | Zhangjiang, Guangdong | GDZJ1–2 |
| Unknown | Unknown | 4 | May 2021 | Maoming, Guangdong | GDMM1–4 |
| Unknown | Unknown | 2 | May 2021 | Beihai, Guangxi | GXBH1–2 |
| Unknown | Unknown | 3 | May 2021 | Beiliu, Guangxi | GXBL1–3 |
| Unknown | Unknown | 3 | May 2021 | Qinzhou, Guangxi | GXQZ1–3 |
| Unknown | Unknown | 3 | May 2021 | Wuzhou, Guangxi | GXWZ1–3 |
| Unknown | Unknown | 6 | May 2021 | Yulin, Guangxi | GXYL1–6 |
| Unknown | Unknown | 4 | February 2021 | Hainan Province | HN1–4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rao, X.; Chen, H.; Lu, Y.; Liu, R.; Li, H. Distribution and Location of BEVs in Different Genotypes of Bananas Reveal the Coevolution of BSVs and Bananas. Int. J. Mol. Sci. 2023, 24, 17064. https://doi.org/10.3390/ijms242317064
Rao X, Chen H, Lu Y, Liu R, Li H. Distribution and Location of BEVs in Different Genotypes of Bananas Reveal the Coevolution of BSVs and Bananas. International Journal of Molecular Sciences. 2023; 24(23):17064. https://doi.org/10.3390/ijms242317064
Chicago/Turabian StyleRao, Xueqin, Huazhou Chen, Yongsi Lu, Runpei Liu, and Huaping Li. 2023. "Distribution and Location of BEVs in Different Genotypes of Bananas Reveal the Coevolution of BSVs and Bananas" International Journal of Molecular Sciences 24, no. 23: 17064. https://doi.org/10.3390/ijms242317064
APA StyleRao, X., Chen, H., Lu, Y., Liu, R., & Li, H. (2023). Distribution and Location of BEVs in Different Genotypes of Bananas Reveal the Coevolution of BSVs and Bananas. International Journal of Molecular Sciences, 24(23), 17064. https://doi.org/10.3390/ijms242317064






