Thermostable Lactonases Inhibit Pseudomonas aeruginosa Biofilm: Effect In Vitro and in Drosophila melanogaster Model of Chronic Infection
Abstract
1. Introduction
2. Results and Discussion
2.1. Expression and Purification of Phosphotriesterase-like Lactonases SacPox and SsoPox
2.2. Substrate Specificity on Lactones (TBBL, 3-Oxo-C12-HSL, C4-HSL)
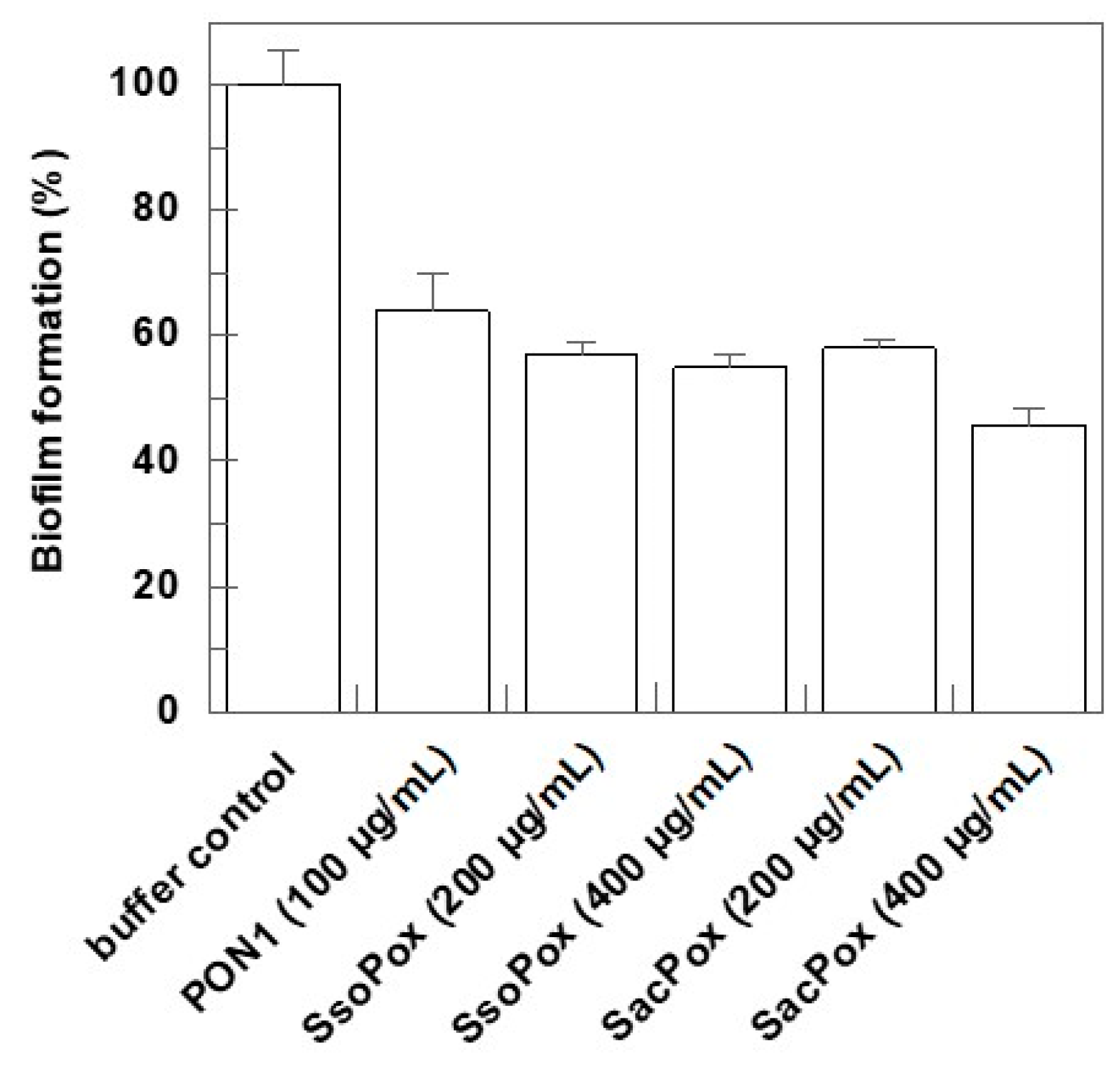
2.3. Effect on Biofilm Formation
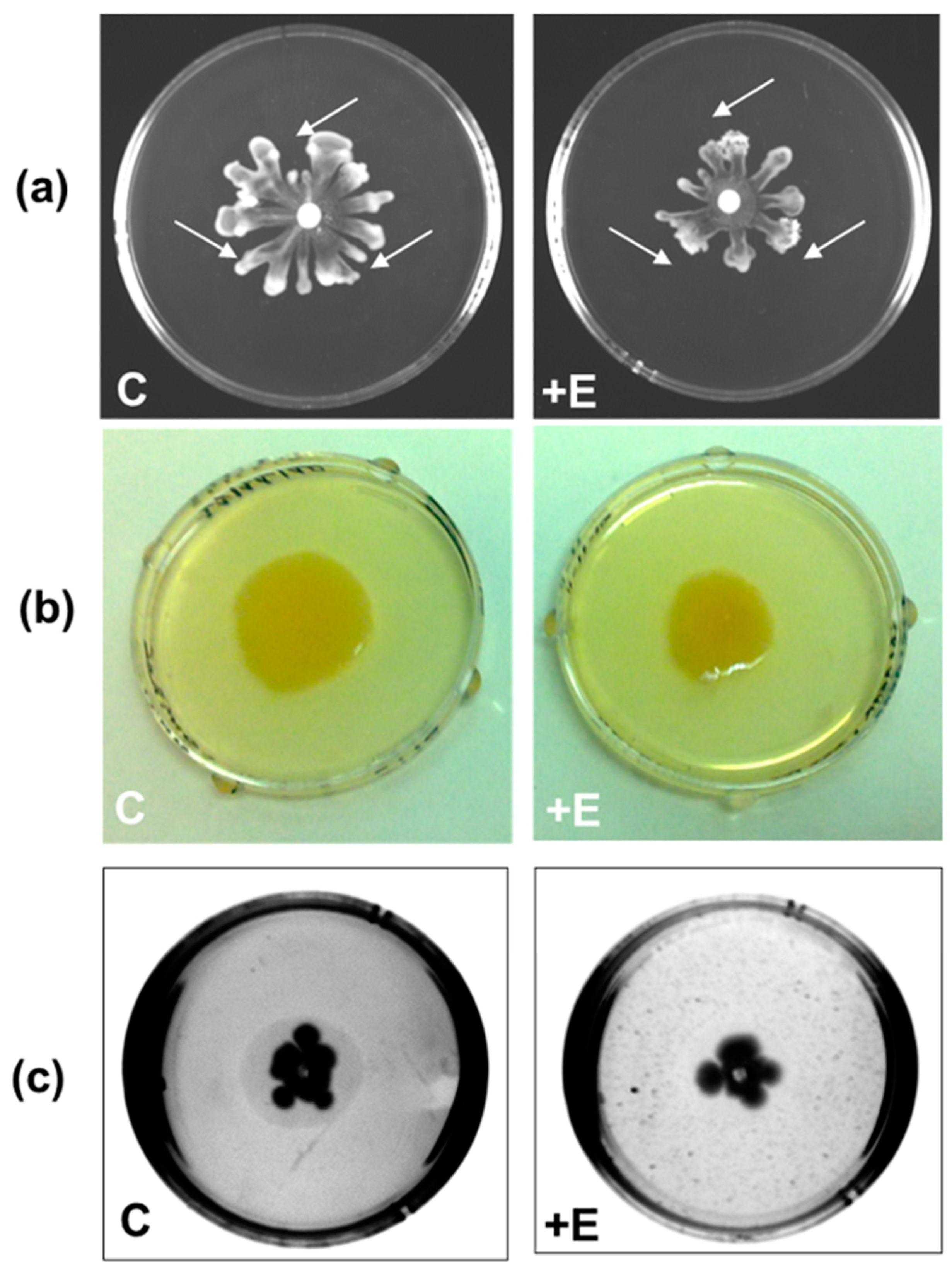
2.4. Effect of SacPox on Liquid PAO1 Culture: Growth and Pathogenicity Marker Analysis
Evaluation of the Effect of SacPox on Swarming, Swimming, and Twitching Motilities
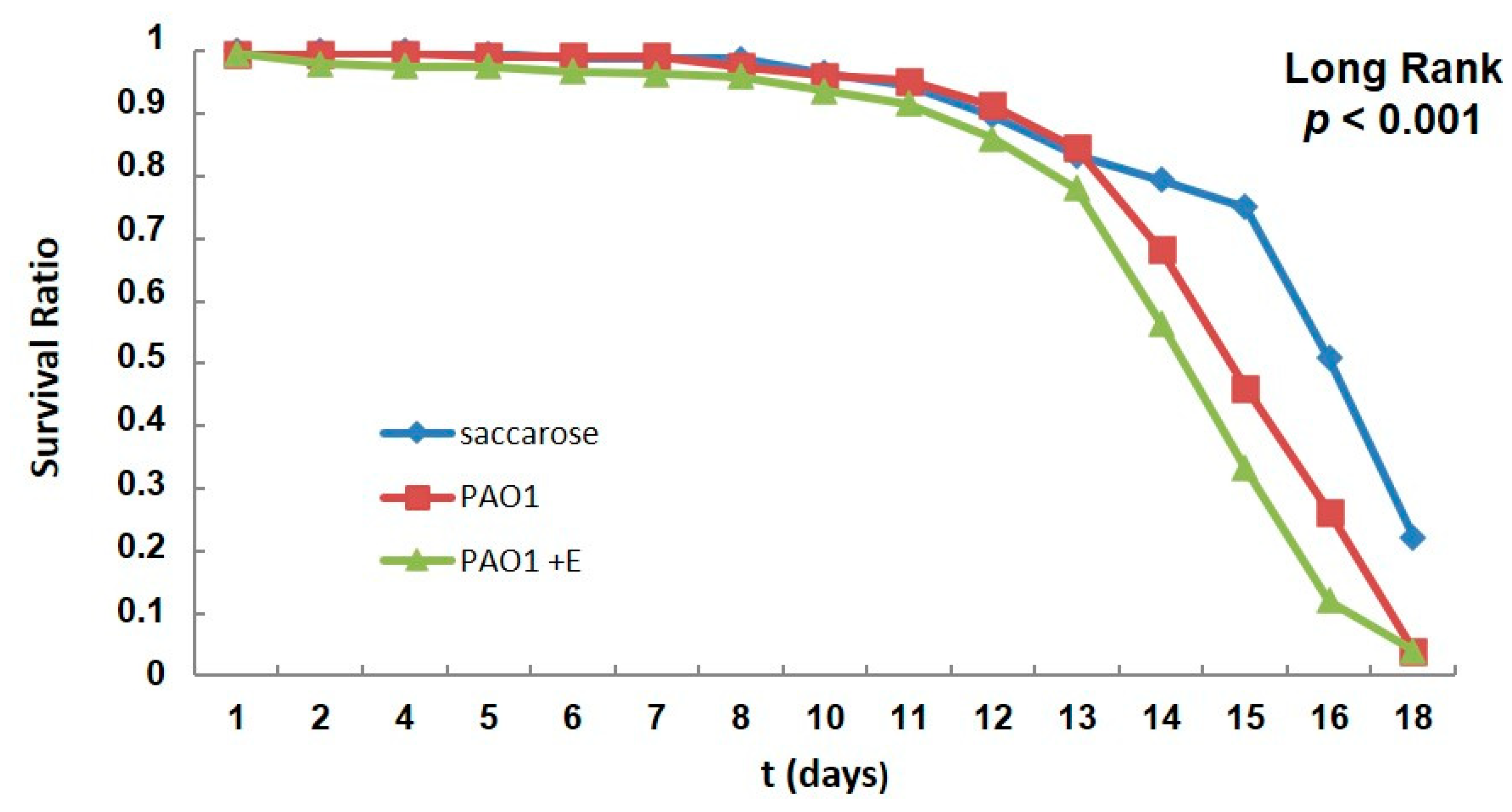
2.5. PAO1-Induced Lethality in Wild-Type Flies Treated with Enzyme: Chronic Infection Model “by Feeding”
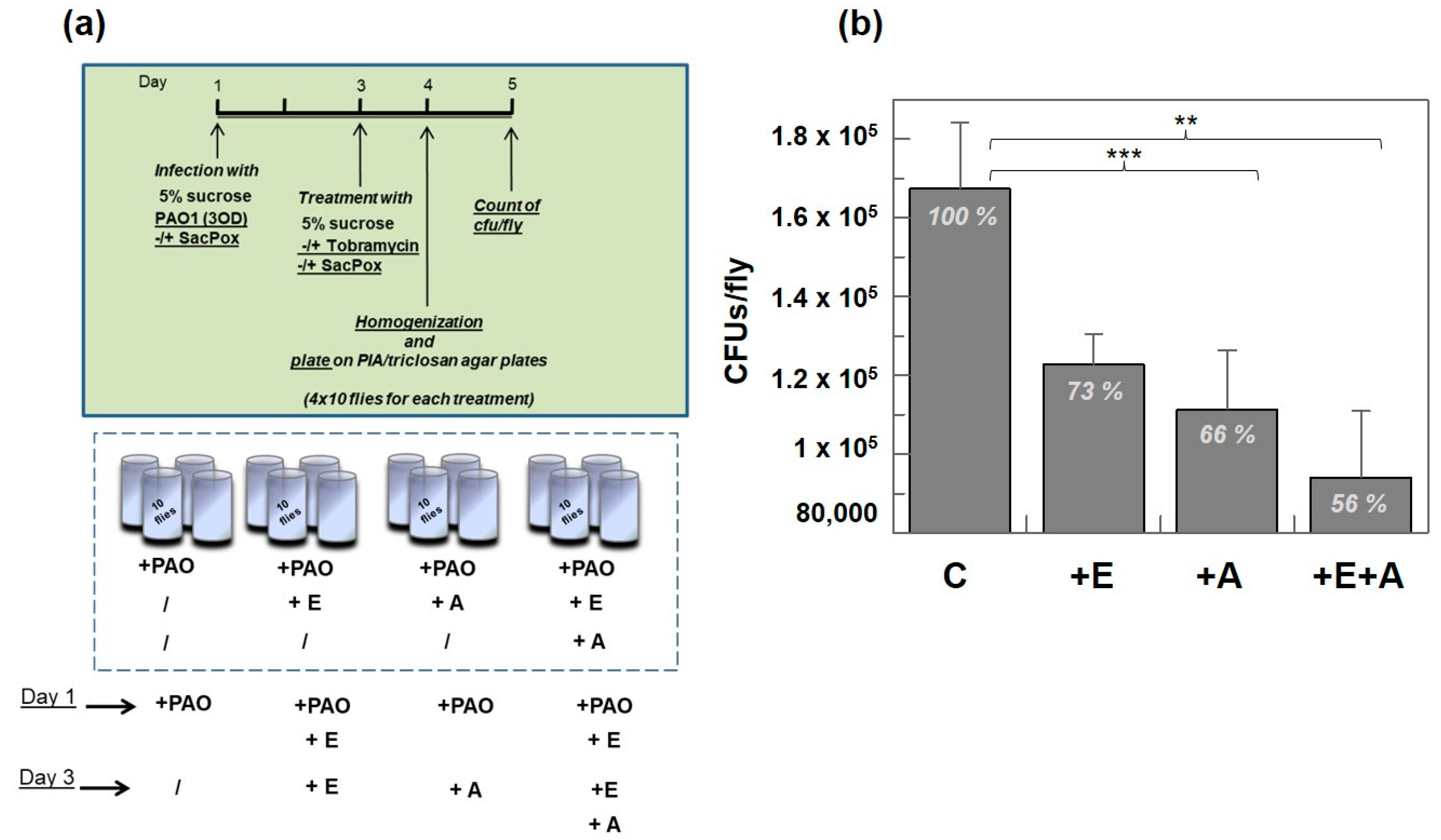
Treatment with Enzyme SacPox and Antibiotic
3. Materials and Methods
3.1. Protein Expression and Purification
3.2. P. aeruginosa Growth and Biofilm Inhibition
3.3. Lactonase Activity
3.4. PAO1 Liquid Culture: Elastase and Pyocyanin Extraction
3.5. Motility Assays on Agar Plates
3.6. D. melanogaster Infection Assays by Feeding
Treatment with Antibiotic Tobramycin
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Streeter, K.; Katouli, M. Pseudomonas aeruginosa: A review of their pathogenesis and prevalence in clinical settings and the environment. Infect. Epidemiol. Med. 2016, 2, 25–32. [Google Scholar] [CrossRef]
- Green, S.K.; Schroth, M.N.; Cho, J.J.; Kominos, S.K.; Vitan-za-jack, V.B. Agricultural plants and soil as a reservoir for Pseudomonas aeruginosa. Appl. Microbiol. 1974, 28, 987–991. [Google Scholar] [CrossRef]
- Kerr, K.G.; Snelling, A.M. Pseudomonas aeruginosa: Aformidable and ever-present adversary. J. Hosp. Infect. 2009, 73, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Stover, C.K.; Pham, X.Q.; Erwin, A.L.; Mizoguchi, S.D.; Warrener, P.; Hickey, M.J.; Brinkman, F.S.; Hufnagle, W.O.; Kowalik, D.J.; Lagrou, M.; et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef]
- Crone, S.; Vives-Flórez, M.; Kvich, L.; Saunders, A.M.; Malone, M.; Nicolaisen, M.H.; Martínez-García, E.; Rojas-Acosta, C.; Catalina Gomez-Puerto, M.; Calum, H.; et al. The environmental occurrence of Pseudomonas aeruginosa. APMIS 2020, 128, 220–231. [Google Scholar] [CrossRef] [PubMed]
- Diggle, S.P.; Whiteley, M. Microbe Profile: Pseudomonas aeruginosa: Opportunistic pathogen and lab rat. Microbiology 2020, 166, 30–33, Corrigendum in Microbiology 2021, 167, 001073. [Google Scholar] [CrossRef] [PubMed]
- Ito, C.A.S.; Bail, L.; Arend, L.; Nogueira, K.D.S.; Tuon, F.F. The activity of ceftazidime/avibactam against carbapenem-resistant Pseudomonas aeruginosa. Infect. Dis. 2021, 53, 386–389. [Google Scholar] [CrossRef]
- Kalluf, K.O.; Arend, L.N.; Wuicik, T.E.; Pilonetto, M.; Tuon, F.F. Molecular epidemiology of SPM-1-producing Pseudomonas aeruginosa by rep-PCR in hospitals in Parana, Brazil. Infect. Genet. Evol. 2017, 49, 130–133. [Google Scholar] [CrossRef] [PubMed]
- Ramos, G.P.; Rocha, J.L.; Tuon, F.F. Seasonal humidity may influence Pseudomonas aeruginosa hospital-acquired infection rates. Int. J. Infect. Dis. 2013, 17, e757–e761. [Google Scholar] [CrossRef]
- Tuon, F.F.; Gortz, L.W.; Rocha, J.L. Risk factors for pan-resistant Pseudomonas aeruginosa bacteremia and the adequacy antibiotic therapy. Braz. J. Infect. Dis. 2012, 16, 351–356. [Google Scholar] [CrossRef][Green Version]
- Tuon, F.F.; Dantas, L.R.; Suss, P.H.; Tasca Ribeiro, V.S. Pathogenesis of the Pseudomonas aeruginosa Biofilm: A Review. Pathogens 2022, 11, 300. [Google Scholar] [CrossRef]
- Rossi, E.; La Rosa, R.; Bartell, J.A.; Marvig, R.L.; Haagensen, J.A.; Sommer, L.M.; Molin, S.; Johansen, H.K. Pseudomonas aeruginosa adaptation and evolution in patients with cystic fibrosis. Nat. Rev. Microbiol. 2021, 19, 331–342. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial Resistance in ESKAPE Pathogens. Clin. Microbiol. Rev. 2020, 13, e00181-19. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015, 6, 26–41. [Google Scholar] [CrossRef]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Turkina, M.V.; Vikström, E. Bacteria-Host Crosstalk: Sensing of the Quorum in the Context of Pseudomonas aeruginosa Infections. J. Innate Immun. 2019, 11, 263–279. [Google Scholar] [CrossRef]
- Mann, E.E.; Wozniak, D.J. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol. Rev. 2012, 36, 893–916. [Google Scholar] [CrossRef]
- Mulcahy, H.; Sibley, C.D.; Surette, M.G.; Lewenza, S. Drosophila melanogaster as an animal model for the study of Pseudomonas aeruginosa biofilm infections in vivo. PLoS Pathog. 2011, 7, e1002299. [Google Scholar] [CrossRef]
- Bjarnsholt, T.; Jensen, P.Ø.; Burmølle, M.; Hentzer, M.; Haagensen, J.A.J.; Hougen, H.P.; Calum, H.; Madsen, K.G.; Moser, C.; Molin, S.; et al. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology 2005, 151 Pt 2, 373–383. [Google Scholar] [CrossRef]
- Langendonk, R.F.; Neill, D.R.; Fothergill, J.L. The Building Blocks of Antimicrobial Resistance in Pseudomonas aeruginosa: Implications for Current Resistance-Breaking Therapies. Front. Cell. Infect. Microbiol. 2021, 11, 665759. [Google Scholar] [CrossRef]
- Kiran, S.; Sharma, P.; Harjai, K.; Capalash, N. Enzymatic quorum quenching increases antibiotic susceptibility of multidrug resistant Pseudomonas aeruginosa. Iran. J. Microbiol. 2011, 3, 1–12. [Google Scholar]
- Mion, S.; Rémy, B.; Plener, L.; Brégeon, F.; Chabrière, E.; Daudé, D. Quorum Quenching Lactonase Strengthens Bacteriophage and Antibiotic Arsenal Against Pseudomonas aeruginosa Clinical Isolates. Front. Microbiol. 2019, 10, 2049. [Google Scholar] [CrossRef]
- Chen, F.; Gao, Y.; Chen, X.; Yu, Z.; Li, X. Quorum Quenching Enzymes and Their Application in Degrading Signal Molecules to Block Quorum Sensing-Dependent Infection. Int. J. Mol. Sci. 2013, 14, 17477–17500. [Google Scholar] [CrossRef]
- Afriat, L.; Roodveldt, C.; Manco, G.; Tawfik, D.S. The latent promiscuity of newly identified microbial lactonases is linked to a recently diverged phosphotriesterase. Biochemistry 2006, 45, 13677–13686. [Google Scholar] [CrossRef]
- Elias, M.; Dupuy, J.; Merone, L.; Mandrich, L.; Porzio, E.; Moniot, S.; Rochu, D.; Lecomte, C.; Rossi, M.; Masson, P.; et al. Structural basis for natural lactonase and promiscuous phosphotriesterase activities. J. Mol. Biol. 2008, 379, 1017–1028. [Google Scholar] [CrossRef]
- Billot, R.; Plener, L.; Jacquet, P.; Elias, M.; Chabrière, E.; Daudé, D. Engineering acyl-homoserine lactone-interfering enzymes toward bacterial control. J. Biol. Chem. 2020, 295, 12993–13007. [Google Scholar] [CrossRef] [PubMed]
- Elias, M.; Tawfik, D.S. Divergence and convergence in enzyme evolution: Parallel evolution of paraoxonases from quorum-quenching lactonases. J. Biol. Chem. 2012, 287, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Teiber, J.F.; Horke, S.; Haines, D.C.; Chowdhary, P.K.; Xiao, J.; Kramer, G.L.; Haley, R.W.; Draganov, D.I. Dominant role of paraoxonases in inactivation of the Pseudomonas aeruginosa quorum-sensing signal N-(3-oxododecanoyl)-L-homoserine lactone. Infect. Immun. 2008, 76, 2512–2519. [Google Scholar] [CrossRef] [PubMed]
- Ozer, E.A.; Pezzulo, A.; Shih, D.M.; Chun, C.; Furlong, C.; Lusis, A.J.; Greenberg, E.P.; Zabner, J. Human and murine paraoxonase 1 are host modulators of Pseudomonas aeruginosa quorum-sensing. FEMS Microbiol. Lett. 2005, 253, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Stoltz, D.A.; Ozer, E.A.; Taft, P.J.; Barry, M.; Liu, L.; Kiss, P.J.; Moninger, T.O.; Parsek, M.R.; Zabner, J. Drosophila are protected from Pseudomonas aeruginosa lethality by transgenic expression of paraoxonase-1. J. Clin. Investig. 2008, 118, 3123–3131. [Google Scholar] [CrossRef]
- Porzio, E.; Di Gennaro, S.; Palma, A.; Manco, G. Mn2+ modulates the kinetic properties of an archaeal member of the PLL family. Chem. Biol. Interact. 2013, 203, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Lutter, E.I.; Faria, M.M.; Rabin, H.R.; Storey, D.G. Pseudomonas aeruginosa cystic fibrosis isolates from individual patients demonstrate a range of levels of lethality in two Drosophila melanogaster infection models. Infect. Immun. 2008, 76, 1877–1888. [Google Scholar] [CrossRef]
- Apidianakis, Y.; Rahme, L.G. Drosophila melanogaster as a model host for studying Pseudomonas aeruginosa infection. Nat. Protoc. 2009, 4, 1285–1294. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, B.; Hoffmann, J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007, 25, 697–743. [Google Scholar] [CrossRef] [PubMed]
- Merone, L.; Mandrich, L.; Rossi, M.; Manco, G. A thermostable phosphotriesterase from the archaeon Sulfolobus solfataricus: Cloning, overexpression and properties. Extremophiles 2005, 9, 297–305. [Google Scholar] [CrossRef]
- Ng, F.S.; Wright, D.M.; Seah, S.Y. Characterization of a phosphotriesterase-like lactonase from Sulfolobus solfataricus and its immobilization for disruption of quorum sensing. Appl. Environ. Microbiol. 2011, 77, 1181–1186. [Google Scholar] [CrossRef]
- Hiblot, J.; Gotthard, G.; Elias, M.; Chabriere, E. Differential active site loop conformations mediate promiscuous activities in the lactonase SsoPox. PLoS ONE 2013, 8, e75272. [Google Scholar] [CrossRef]
- Mandrich, L.; Cerreta, M.; Manco, G. An Engineered Version of Human PON2 Opens the Way to Understand the Role of Its Post-Translational Modifications in Modulating Catalytic Activity. PLoS ONE 2015, 10, e0144579. [Google Scholar] [CrossRef]
- Wang, Y.; Dai, Y.; Zhang, Y.; Hu, Y.; Yang, B.; Chen, S. Effects of quorum sensing autoinducer degradation gene on virulence and biofilm formation of Pseudomonas aeruginosa. Sci. China Ser. C 2007, 50, 385–391. [Google Scholar] [CrossRef]
- King, E.O.; Ward, M.K.; Raney, D.E. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 1954, 44, 301–307. [Google Scholar]
- Bhargava, H.N.; Leonard, P.A. Triclosan: Applications and safety. Am. J. Infect. Control 1996, 24, 209–218. [Google Scholar] [CrossRef]
- Alamu, J.; Kakithakara Vajravelu, L.; Venkatesan, B.; Thulukanam, J. Correlation of phenotypic and genotypic virulence markers, antimicrobial susceptibility pattern, and outcome of Pseudomonas aeruginosa sepsis infection. Microb. Pathog. 2022, 170, 105716. [Google Scholar] [CrossRef]
- Merz, A.J.; So, M.; Sheetz, M.P. Pilus retraction powers bacterial twitching motility. Nature 2000, 407, 98–102. [Google Scholar] [CrossRef]
- Chuchalin, A.; Csiszér, E.; Gyurkovics, K.; Bartnicka, M.T.; Sands, D.; Kapranov, N.; Varoli, G.; Monici Preti, P.A.; Mazurek, H. A formulation of aerosolized tobramycin (Bramitob) in the treatment of patients with cystic fibrosis and Pseudomonas aeruginosa infection: A double-blind, placebo-controlled, multicenter study. Pediatr. Drugs 2007, 9 (Suppl. S1), 21–31. [Google Scholar] [CrossRef]
- Horstmann, J.C.; Laric, A.; Boese, A.; Yildiz, D.; Röhrig, T.; Empting, M.; Frank, N.; Krug, D.; Müller, R.; Schneider-Daum, N.; et al. Transferring Microclusters of P. aeruginosa Biofilms to the Air-Liquid Interface of Bronchial Epithelial Cells for Repeated Deposition of Aerosolized Tobramycin. ACS Infect. Dis. 2022, 8, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Thorn, C.R.; Wignall, A.; Kopecki, Z.; Kral, A.; Prestidge, C.A.; Thomas, N. Liquid Crystal Nanoparticles Enhance Tobramycin Efficacy in a Murine Model of Pseudomonas aeruginosa Biofilm Wound Infection. ACS Infect. Dis. 2022, 8, 841–854. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Cheng, K.J.; Geesey, G.G.; Ladd, T.I.; Nickel, J.C.; Dasgupta, M.; Marrie, T.J. Bacterial Biofilms in Nature and Disease. Annu. Rev. Microbiol. 1987, 41, 435–464. [Google Scholar] [CrossRef]
- Lewis, K. Riddle of Biofilm Resistance. Antimicrob. Agents Chemother. 2001, 45, 999–1007. [Google Scholar] [CrossRef]
- Lebeaux, D.; Ghigo, J.M.; Beloin, C. Biofilm-Related Infections: Bridging the Gap Between Clinical Management and Fundamental Aspects of Recalcitrance Toward Antibiotics. Microbiol. Mol. Biol. Rev. 2014, 78, 510–543. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.W.; Mah, T.F. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017, 41, 276–301. [Google Scholar] [CrossRef] [PubMed]
- Ciofu, O.; Tolker-Nielsen, T. Tolerance and Resistance of Pseudomonas Aeruginosa Biofilms to Antimicrobial Agents—How, P. Aeruginosa Can Escape Antibiotics. Front. Microbiol. 2019, 10, 913. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, L.R.; D’Argenio, D.A.; MacCoss, M.J.; Zhang, Z.; Jones, R.A.; Miller, S.I. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 2005, 436, 1171–1175. [Google Scholar] [CrossRef] [PubMed]
- Harms, A.; Maisonneuve, E.; Gerdes, K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 2016, 354, aaf4268. [Google Scholar] [CrossRef]
- Liu, Q.; Yin, L.; Zhang, X.; Zhu, G.; Liu, H.; Bai, F.; Cheng, Z.; Wu, W.; Jin, Y. Reversion of Ceftazidime Resistance in Pseudomonas aeruginosa under Clinical Setting. Microorganisms 2022, 10, 2395. [Google Scholar] [CrossRef]
- Laemmli, U. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Nagant, C.; Feng, Y.; Lucas, B.; Braeckmans, K.; Savage, P.; Dehaye, J.P. Effect of a low concentration of a cationic steroid antibiotic (CSA-13) on the formation of a biofilm by Pseudomonas aeruginosa. J. Appl. Microbiol. 2011, 111, 763–772. [Google Scholar] [CrossRef]

| Substrate | Enzyme | Specific Activity (U/mg) [S] = (300 μM) | kcat (s−1) | KM (mM) | kcat/KM (M−1 s−1) |
|---|---|---|---|---|---|
TBBL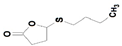 | SacPox | 58.3 ± 0.8 | 38.5 ± 0.74 | 0.024 ± 0.0005 | 1.86 ± 0.07 × 106 |
| SsoPox | 49.0 ± 11.2 | 29.0 ± 6.6 | 0.08 ± 0.003 | 3.6 ± 0.1 × 105 | |
| recPON2 b | 1.40 ± 0.07 | 1.10 ± 0.10 | 0.50 ± 0.15 | 2.20 ± 0.50 × 103 | |
4-undecanolide *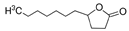 | SacPox | 1016 ± 33.3 | 912.8 ± 48.9 | 0.120 ± 0.010 | 7.6 ± 0.9 × 106 |
3-Oxo-C12-HSL *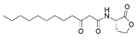 | SacPox | 2337 ± 9.5 | 2027.9 ± 5.5 | 0.120 ± 0.012 | 16.2 ± 2.2 × 106 |
| SsoPox a | 1.01 ± 0.13 | 0.456 ± 0.128 | 2.2 ± 0.68 × 103 | ||
| recPON2 b | 4.1 ± 0.4 | 2.7 | 0.50 ± 0.1 | 5.4 × 103 | |
C4-HSL *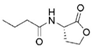 | SacPox | n.d. | n.d. | n.d. | n.d. |
| SsoPox a | 2.3 ± 0.2 | n.d. | n.d. | 11.62 ± 0.72 | |
| recPON2 b | 1.53 | 0.6 ± 0.1 | 2.55 × 103 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Porzio, E.; Andrenacci, D.; Manco, G. Thermostable Lactonases Inhibit Pseudomonas aeruginosa Biofilm: Effect In Vitro and in Drosophila melanogaster Model of Chronic Infection. Int. J. Mol. Sci. 2023, 24, 17028. https://doi.org/10.3390/ijms242317028
Porzio E, Andrenacci D, Manco G. Thermostable Lactonases Inhibit Pseudomonas aeruginosa Biofilm: Effect In Vitro and in Drosophila melanogaster Model of Chronic Infection. International Journal of Molecular Sciences. 2023; 24(23):17028. https://doi.org/10.3390/ijms242317028
Chicago/Turabian StylePorzio, Elena, Davide Andrenacci, and Giuseppe Manco. 2023. "Thermostable Lactonases Inhibit Pseudomonas aeruginosa Biofilm: Effect In Vitro and in Drosophila melanogaster Model of Chronic Infection" International Journal of Molecular Sciences 24, no. 23: 17028. https://doi.org/10.3390/ijms242317028
APA StylePorzio, E., Andrenacci, D., & Manco, G. (2023). Thermostable Lactonases Inhibit Pseudomonas aeruginosa Biofilm: Effect In Vitro and in Drosophila melanogaster Model of Chronic Infection. International Journal of Molecular Sciences, 24(23), 17028. https://doi.org/10.3390/ijms242317028







