OTX Genes in Adult Tissues
Abstract
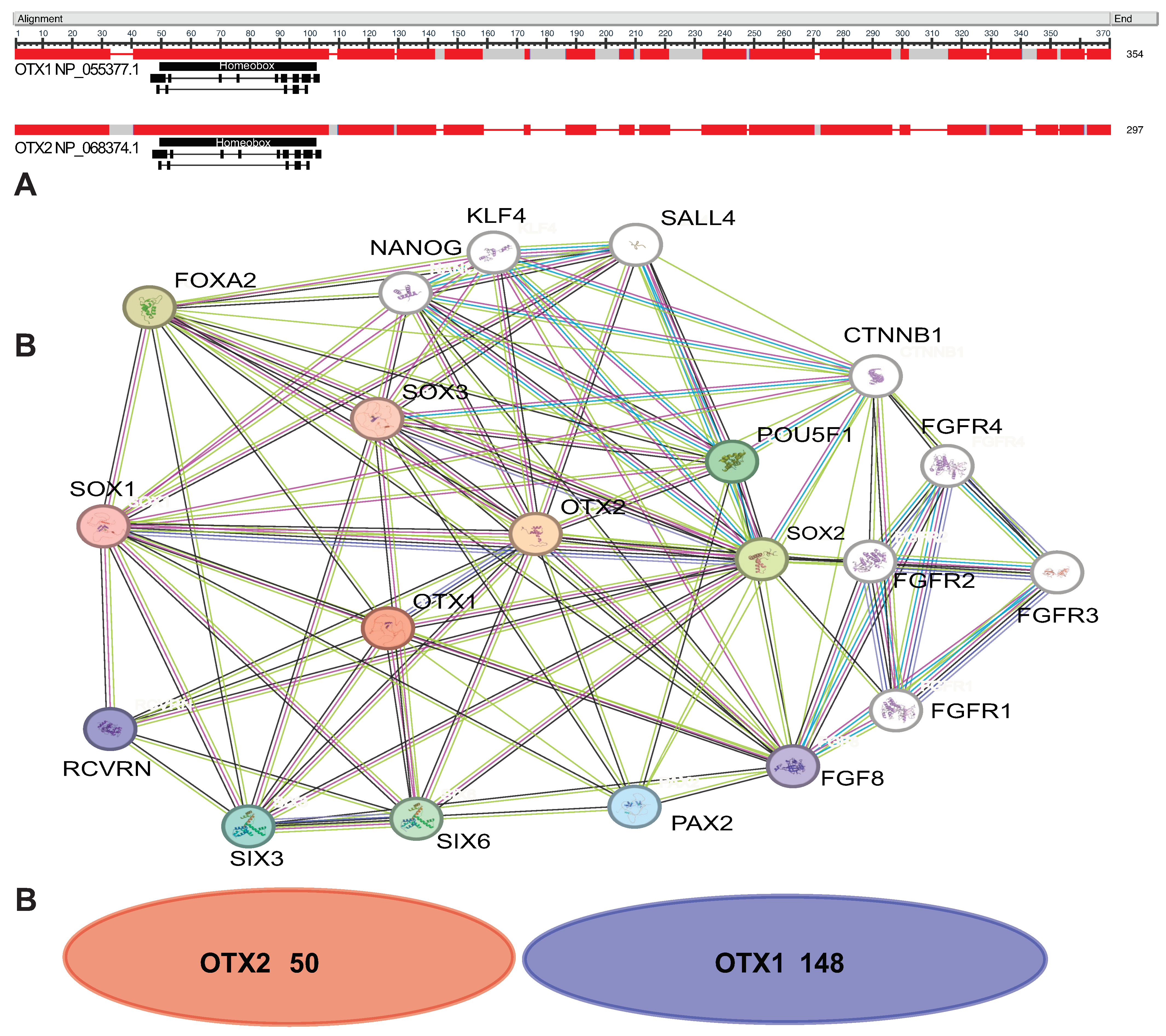
1. Introduction
2. Development
3. Embryonic and Adult Stem Cells
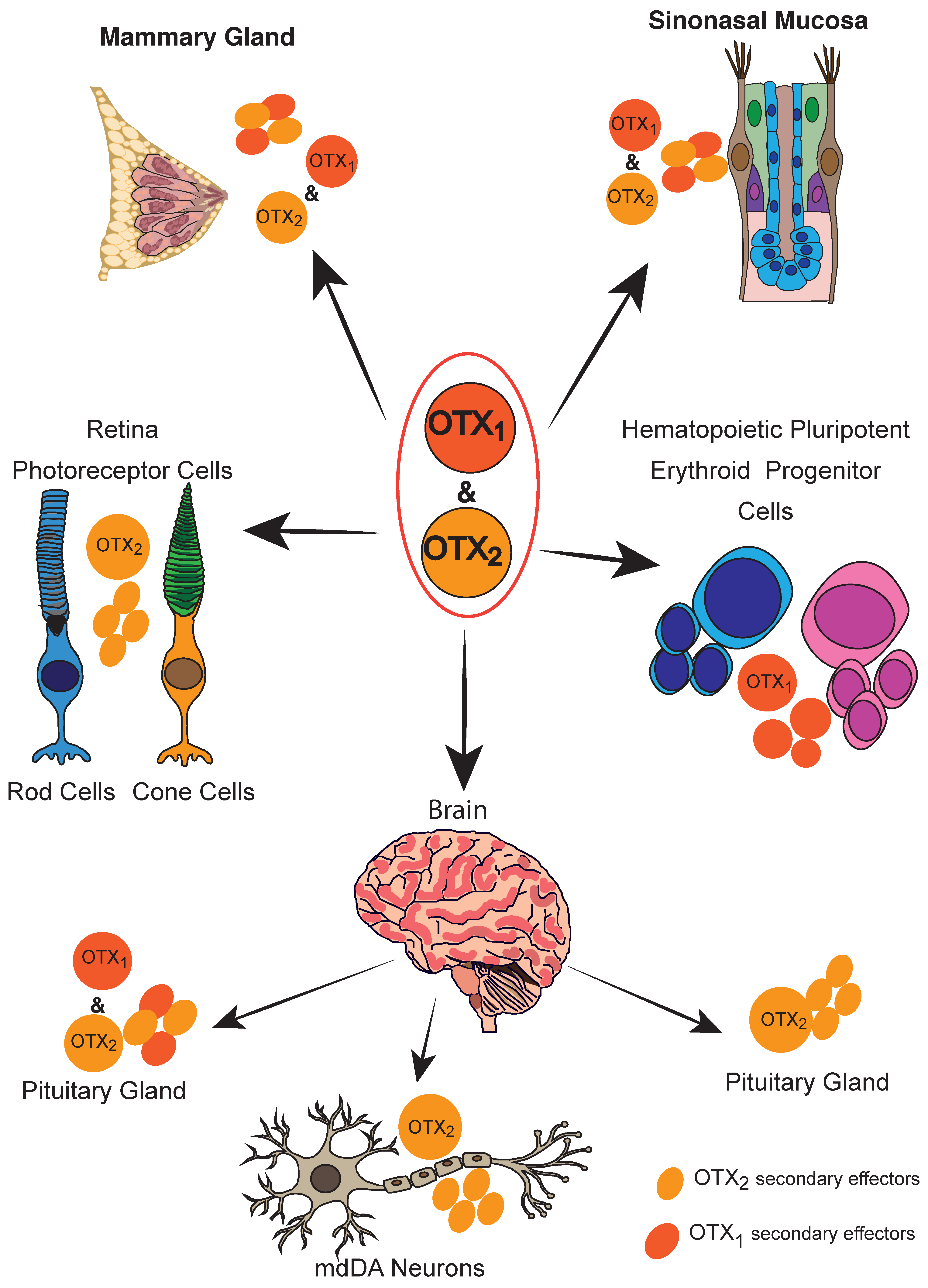
4. Adult Tissues
4.1. Neuronal Tissues
4.1.1. Brain
4.1.2. Dopaminergic Neurons
4.1.3. Retina and PVR
4.1.4. Pineal Gland
4.1.5. Pituitary Gland
4.1.6. Sinonasal Mucosae and Nasal Polyps
4.2. Breast
4.3. Hematopoiesis
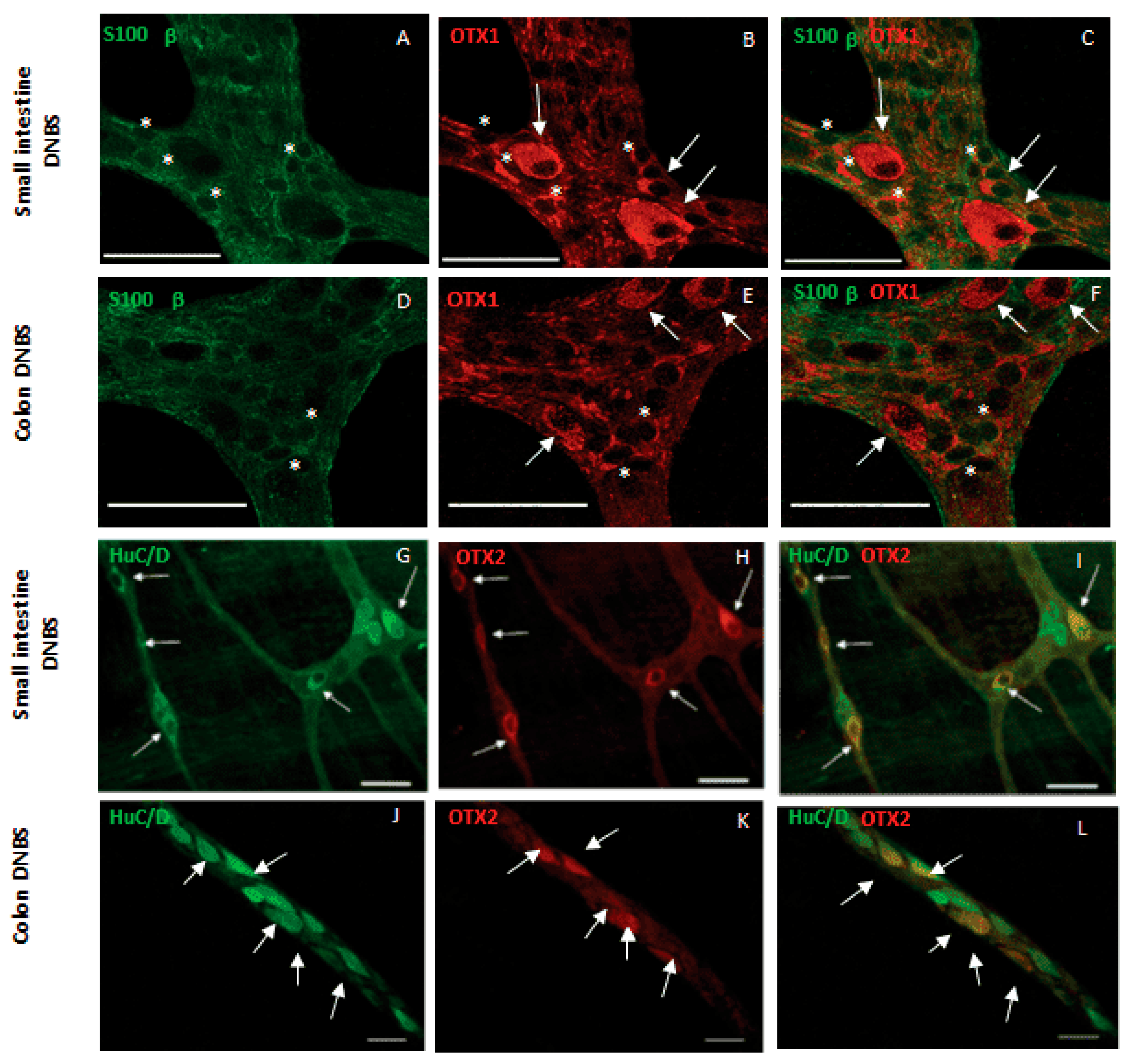
4.4. Myenteric Plexus/Intestine
4.4.1. Inflammation in Mice
4.4.2. Inflammation in Zebrafish
4.4.3. Ischemia/Reperfusion in Mice
4.5. Mastocytosis
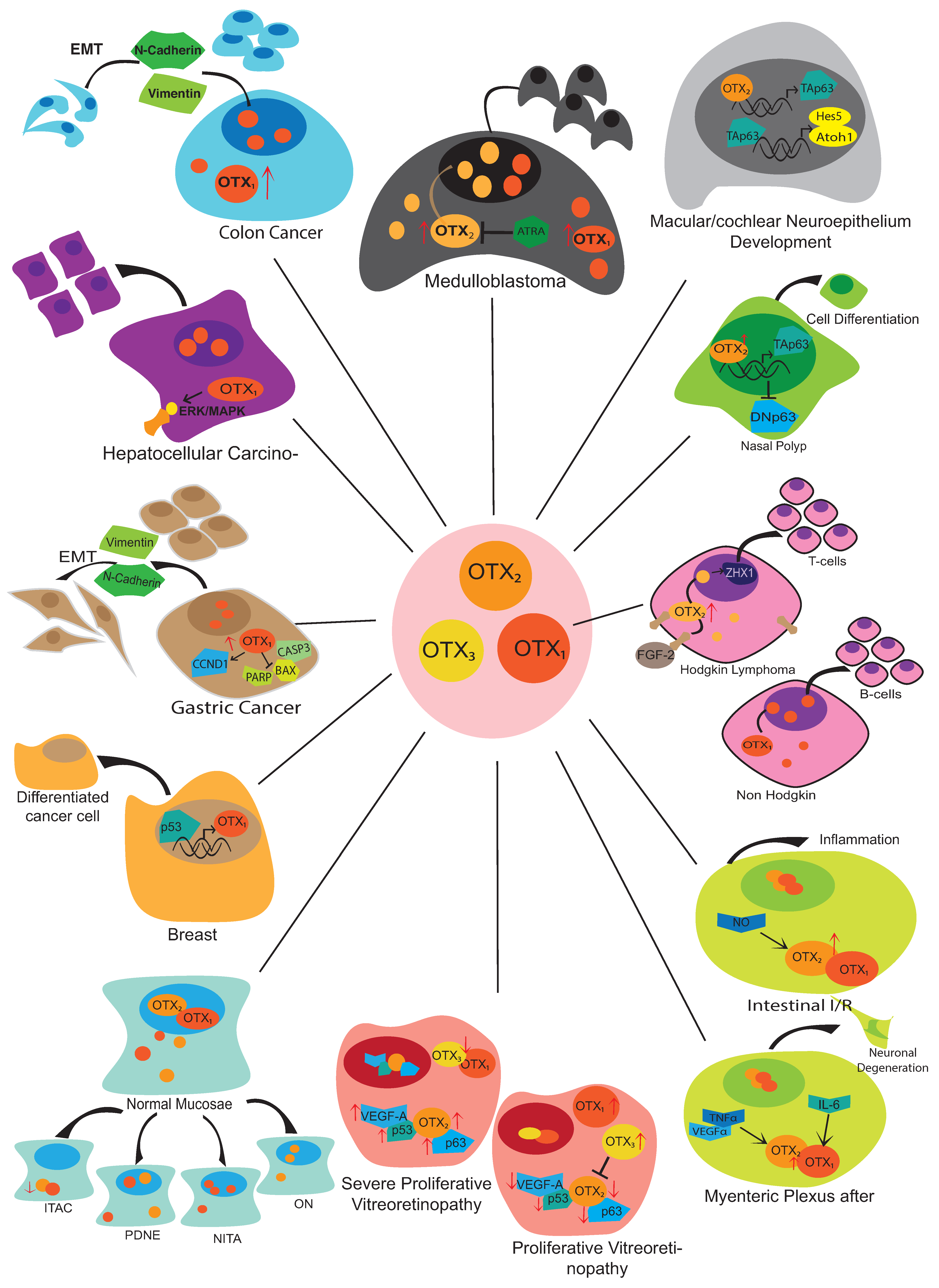
5. Tumor Progression
5.1. Medulloblastoma
5.2. Retinoblastoma
5.3. Sinonasal Neoplasms
5.4. Laryngeal Squamous Cell Carcinoma
5.5. Esophageal Squamous Cell Carcinoma
5.6. Gastric Cancer
5.7. Colon/Colorectal Cancer
5.8. Hepatocellular Carcinoma
5.9. Pancreatic Cancer
5.10. Breast Cancer
5.11. Lung
5.12. Bladder Cancer
5.13. Lymphoma
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kastury, K.; Druck, T.; Huebner, K.; Barletta, C.; Acampora, D.; Simeone, A.; Faiella, A.; Boncinelli, E. Chromosome Locations of Human EMX and OTX Genes. Genomics 1994, 22, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Simeone, A.; Puelles, E.; Acampora, D. The Otx Family. Curr. Opin. Genet. Dev. 2002, 12, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Boncinelli, E.; Simeone, A.; Acampora, D.; Gulisano, M. Homeobox Genes in the Developing Central Nervous System. Ann. Genet. 1993, 36, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Larsen, K.B.; Lutterodt, M.C.; Møllgård, K.; Møller, M. Expression of the Homeobox Genes OTX2 and OTX1 in the Early Developing Human Brain. J. Histochem. Cytochem. 2010, 58, 669–678. [Google Scholar] [CrossRef] [PubMed]
- Acampora, D.; Mazan, S.; Lallemand, Y.; Avantaggiato, V.; Maury, M.; Simeone, A.; Brulet, P. Forebrain and Midbrain Regions Are Deleted in Otx2−/− Mutants Due to a Defective Anterior Neuroectoderm Specification during Gastrulation. Development 1995, 121, 3279–3290. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, I.; Kuratani, S.; Kimura, C.; Takeda, N.; Aizawa, S. Mouse Otx2 Functions in the Formation and Patterning of Rostral Head. Genes Dev. 1995, 9, 2646–2658. [Google Scholar] [CrossRef] [PubMed]
- Ang, S.L.; Jin, O.; Rhinn, M.; Daigle, N.; Stevenson, L.; Rossant, J. A Targeted Mouse Otx2 Mutation Leads to Severe Defects in Gastrulation and Formation of Axial Mesoderm and to Deletion of Rostral Brain. Development 1996, 122, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Acampora, D.; Mazan, S.; Avantaggiato, V.; Barone, P.; Tuorto, F.; Lallemand, Y.; Brulet, P.; Simeone, A. Epilepsy and Brain Abnormalities in Mice Lacking the Otx1 Gene. Nat. Genet. 1996, 14, 218–222. [Google Scholar] [CrossRef]
- Bjornsson, C.S.; Apostolopoulou, M.; Tian, Y.; Temple, S. It Takes a Village: Constructing the Neurogenic Niche. Dev. Cell 2015, 32, 435–446. [Google Scholar] [CrossRef]
- Huang, B.; Li, X.; Tu, X.; Zhao, W.; Zhu, D.; Feng, Y.; Si, X.; Chen, J.G. OTX1 Regulates Cell Cycle Progression of Neural Progenitors in the Developing Cerebral Cortex. J. Biol. Chem. 2018, 293, 2137–2148. [Google Scholar] [CrossRef]
- Pantò, M.R.; Zappalà, A.; Tuorto, F.; Cicirata, F. Role of the Otx1 Gene in Cell Differentiation of Mammalian Cortex. Eur. J. Neurosci. 2004, 19, 2893–2902. [Google Scholar] [CrossRef] [PubMed]
- Hoch, R.V.; Lindtner, S.; Price, J.D.; Rubenstein, J.L.R. OTX2 Transcription Factor Controls Regional Patterning within the Medial Ganglionic Eminence and Regional Identity of the Septum. Cell Rep. 2015, 12, 482–494. [Google Scholar] [CrossRef] [PubMed]
- Silbereis, J.C.; Nobuta, H.; Tsai, H.H.; Heine, V.M.; McKinsey, G.L.; Meijer, D.H.; Howard, M.A.; Petryniak, M.A.; Potter, G.B.; Alberta, J.A.; et al. Olig1 Function Is Required to Repress Dlx1/2 and Interneuron Production in Mammalian Brain. Neuron 2014, 81, 574–587. [Google Scholar] [CrossRef] [PubMed]
- Puelles, E.; Acampora, D.; Gogoi, R.; Tuorto, F.; Papalia, A.; Guillemot, F.; Ang, S.L.; Simeone, A. Otx2 Controls Identity and Fate of Glutamatergic Progenitors of the Thalamus by Repressing GABAergic Differentiation. J. Neurosci. 2006, 26, 5955–5964. [Google Scholar] [CrossRef] [PubMed]
- Simeone, A.; Acampora, D.; Mallamaci, A.; Stornaiuolo, A.; D’Apice, M.R.; Nigro, V.; Boncinelli, E. A Vertebrate Gene Related to Orthodenticle Contains a Homeodomain of the Bicoid Class and Demarcates Anterior Neuroectoderm in the Gastrulating Mouse Embryo. EMBO J. 1993, 12, 2735–2747. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Morales, J.R.; Signore, M.; Acampora, D.; Simeone, A.; Bovolenta, P. Otx Genes Are Required for Tissue Specification in the Developing Eye. Development 2001, 128, 2019–2030. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Morales, J.R.; Dolez, V.; Rodrigo, I.; Zaccarini, R.; Leconte, L.; Bovolenta, P.; Saule, S. OTX2 Activates the Molecular Network Underlying Retina Pigment Epithelium Differentiation. J. Biol. Chem. 2003, 278, 21721–21731. [Google Scholar] [CrossRef] [PubMed]
- Nishida, A.; Furukawa, A.; Koike, C.; Tano, Y.; Aizawa, S.; Matsuo, I.; Furukawa, T. Otx2 Homeobox Gene Controls Retinal Photoreceptor Cell Fate and Pineal Gland Development. Nat. Neurosci. 2003, 6, 1255–1263. [Google Scholar] [CrossRef]
- Larsen, K.B.; Lutterodt, M.; Rath, M.F.; Møller, M. Expression of the Homeobox Genes PAX6, OTX2, and OTX1 in the Early Human Fetal Retina. Int. J. Dev. Neurosci. 2009, 27, 485–492. [Google Scholar] [CrossRef]
- Bovolenta, P.; Mallamaci, A.; Briata, P.; Corte, G.; Boncinelli, E. Implication of OTX2 in Pigment Epithelium Determination and Neural Retina Differentiation. J. Neurosci. 1997, 17, 4243–4252. [Google Scholar] [CrossRef]
- Ragge, N.K.; Brown, A.G.; Poloschek, C.M.; Lorenz, B.; Henderson, R.A.; Clarke, M.P.; Russell-Eggitt, I.; Fielder, A.; Gerrelli, D.; Martinez-Barbera, J.P.; et al. Heterozygous Mutations of OTX2 Cause Severe Ocular Malformations. Am. J. Hum. Genet. 2005, 76, 1008–1022. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, A.; Bakrania, P.; Bunyan, D.J.; Osborne, R.J.; Crolla, J.A.; Salt, A.; Ayuso, C.; Newbury-Ecob, R.; Abou-Rayyah, Y.; Collin, J.R.O.; et al. Novel Heterozygous OTX2 Mutations and Whole Gene Deletions in Anophthalmia, Microphthalmia and Coloboma. Hum. Mutat. 2008, 29, E278–E283. [Google Scholar] [CrossRef] [PubMed]
- Schilter, K.F.; Schneider, A.; Bardakjian, T.; Soucy, J.F.; Tyler, R.C.; Reis, L.M.; Semina, E.V. OTX2 Microphthalmia Syndrome: Four Novel Mutations and Delineation of a Phenotype. Clin. Genet. 2011, 79, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Rodriguez, J.; Pelcastre, E.L.; Tovilla-Canales, J.L.; Garcia-Ortiz, J.E.; Amato-Almanza, M.; Villanueva-Mendoza, C.; Espinosa-Mattar, Z.; Zenteno, J.C. Mutational Screening of CHX10, GDF6, OTX2, RAX and SOX2 Genes in 50 Unrelated Microphthalmia-Anophthalmia-Coloboma (MAC) Spectrum Cases. Br. J. Ophthalmol. 2010, 94, 1100–1104. [Google Scholar] [CrossRef] [PubMed]
- Morsli, H.; Tuorto, F.; Choo, D.; Postiglione, M.P.; Simeone, A.; Wu, D.K. Otx1 and Otx2 Activities Are Required for the Normal Development of the Mouse Inner Ear. Development 1999, 126, 2335–2343. [Google Scholar] [CrossRef] [PubMed]
- Palombo, R.; Porta, G.; Bruno, E.; Provero, P.; Serra, V.; Neduri, K.; Viziano, A.; Alessandrini, M.; Micarelli, A.; Ottaviani, F.; et al. OTX2 Regulates the Expression of TAp63 Leading to Macular and Cochlear Neuroepithelium Development. Aging 2015, 7, 928–936. [Google Scholar] [CrossRef][Green Version]
- Vendrell, V.; Lopez-Hernandez, I.; Alonso, M.B.D.; Feijoo-Redondo, A.; Abello, G.; Galvez, H.; Giraldez, F.; Lamonerie, T.; Schimmang, T. Otx2 Is a Target of N-Myc and Acts as a Suppressor of Sensory Development in the Mammalian Cochlea. Development 2015, 142, 2792–2800. [Google Scholar] [CrossRef]
- Acampora, D.; Di Giovannantonio, L.G.; Simeone, A. Otx2 Is an Intrinsic Determinant of the Embryonic Stem Cell State and Is Required for Transition to a Stable Epiblast Stem Cell Condition. Development 2013, 140, 43–55. [Google Scholar] [CrossRef]
- Kole, C.; Klipfel, L.; Yang, Y.; Ferracane, V.; Blond, F.; Reichman, S.; Millet-Puel, G.; Clérin, E.; Aït-Ali, N.; Pagan, D.; et al. Otx2-Genetically Modified Retinal Pigment Epithelial Cells Rescue Photoreceptors after Transplantation. Mol. Ther. 2018, 26, 219–237. [Google Scholar] [CrossRef]
- Prochiantz, A.; Di Nardo, A.A. Homeoprotein Signaling in the Developing and Adult Nervous System. Neuron 2015, 85, 911–925. [Google Scholar] [CrossRef]
- Johansson, P.A.; Irmler, M.; Acampora, D.; Beckers, J.; Simeone, A.; Götz, M. The Transcription Factor Otx2 Regulates Choroid Plexus Development and Function. Development 2013, 140, 1055–1066. [Google Scholar] [CrossRef]
- Spatazza, J.; Lee, H.H.C.; DiNardo, A.A.; Tibaldi, L.; Joliot, A.; Hensch, T.K.; Prochiantz, A. Choroid-Plexus-Derived Otx2 Homeoprotein Constrains Adult Cortical Plasticity. Cell Rep. 2013, 3, 1815–1823. [Google Scholar] [CrossRef] [PubMed]
- Planques, A.; Moreira, V.O.; Benacom, D.; Bernard, C.; Jourdren, L.; Blugeon, C.; Dingli, F.; Masson, V.; Loew, D.; Prochiantz, A.; et al. Otx2 Homeoprotein Functions in Adult Choroid Plexus. Int. J. Mol. Sci. 2021, 22, 8951. [Google Scholar] [CrossRef]
- Sugiyama, S.; Di Nardo, A.A.; Aizawa, S.; Matsuo, I.; Volovitch, M.; Prochiantz, A.; Hensch, T.K. Experience-Dependent Transfer of Otx2 Homeoprotein into the Visual Cortex Activates Postnatal Plasticity. Cell 2008, 134, 508–520. [Google Scholar] [CrossRef]
- Kim, N.; Acampora, D.; Dingli, F.; Loew, D.; Simeone, A.; Prochiantz, A.; Nardo, A. Di Immunoprecipitation and Mass Spectrometry Identify Non-Cell Autonomous Otx2 Homeoprotein in the Granular and Supragranular Layers of Mouse Visual Cortex. F1000Research 2014, 3, 178. [Google Scholar] [CrossRef]
- Vincent, C.; Gilabert-Juan, J.; Gibel-Russo, R.; Alvarez-Fischer, D.; Krebs, M.O.; Le Pen, G.; Prochiantz, A.; Di Nardo, A.A. Correction: Non-Cell-Autonomous OTX2 Transcription Factor Regulates Anxiety-Related Behavior in the Mouse. Mol. Psychiatry 2021, 26, 6481. [Google Scholar] [CrossRef]
- Beurdeley, M.; Spatazza, J.; Lee, H.H.C.; Sugiyama, S.; Bernard, C.; Di Nardo, A.A.; Hensch, T.K.; Prochiantz, A. Otx2 Binding to Perineuronal Nets Persistently Regulates Plasticity in the Mature Visual Cortex. J. Neurosci. 2012, 32, 9429–9437. [Google Scholar] [CrossRef]
- Apulei, J.; Kim, N.; Testa, D.; Ribot, J.; Morizet, D.; Bernard, C.; Jourdren, L.; Blugeon, C.; Di Nardo, A.A.; Prochiantz, A. Non-Cell Autonomous OTX2 Homeoprotein Regulates Visual Cortex Plasticity Through Gadd45b/g. Cereb. Cortex 2019, 29, 2384–2395. [Google Scholar] [CrossRef] [PubMed]
- Bernard, C.; Vincent, C.; Testa, D.; Bertini, E.; Ribot, J.; Di Nardo, A.A.; Volovitch, M.; Prochiantz, A. A Mouse Model for Conditional Secretion of Specific Single-Chain Antibodies Provides Genetic Evidence for Regulation of Cortical Plasticity by a Non-Cell Autonomous Homeoprotein Transcription Factor. PLoS Genet. 2016, 12, e1006035. [Google Scholar] [CrossRef] [PubMed]
- Bernard, C.; Prochiantz, A. Otx2-PNN Interaction to Regulate Cortical Plasticity. Neural Plast. 2016, 2016, 7931693. [Google Scholar] [CrossRef] [PubMed]
- Planques, A.; Moreira, V.O.; Dubreuil, C.; Prochiantz, A.; Di Nardo, A.A. OTX2 Signals from the Choroid Plexus to Regulate Adult Neurogenesis. eNeuro 2019, 6, ENEURO.0262-18.2019. [Google Scholar] [CrossRef] [PubMed]
- Sen, S.; Reichert, H.; VijayRaghavan, K. Conserved Roles of Ems/Emx and Otd/Otx Genes in Olfactory and Visual System Development in Drosophila and Mouse. Open Biol. 2013, 3, 120177. [Google Scholar] [CrossRef][Green Version]
- Lee, H.H.C.; Bernard, C.; Ye, Z.; Acampora, D.; Simeone, A.; Prochiantz, A.; Di Nardo, A.A.; Hensch, T.K. Genetic Otx2 Mis-Localization Delays Critical Period Plasticity across Brain Regions. Mol. Psychiatry 2017, 22, 680–688. [Google Scholar] [CrossRef] [PubMed]
- Puelles, E.; Annino, A.; Tuorto, F.; Usiello, A.; Acampora, D.; Czerny, T.; Brodski, C.; Ang, S.L.; Wurst, W.; Simeone, A. Otx2 Regulates the Extent, Identity and Fate of Neuronal Progenitor Domains in the Ventral Midbrain. Development 2004, 131, 2037–2048. [Google Scholar] [CrossRef] [PubMed]
- Prakash, N.; Wurst, W. Development of Dopaminergic Neurons in the Mammalian Brain. Cell Mol. Life Sci. 2006, 63, 187–206. [Google Scholar] [CrossRef]
- Ang, S.L. Transcriptional Control of Midbrain Dopaminergic Neuron Development. Development 2006, 133, 3499–3506. [Google Scholar] [CrossRef]
- Borgkvist, A.; Puelles, E.; Carta, M.; Acampora, D.; Ang, S.L.; Wurst, W.; Goiny, M.; Fisone, G.; Simeone, A.; Usiello, A. Altered Dopaminergic Innervation and Amphetamine Response in Adult Otx2 Conditional Mutant Mice. Mol. Cell Neurosci. 2006, 31, 293–302. [Google Scholar] [CrossRef]
- Chung, C.Y.; Licznerski, P.; Alavian, K.N.; Simeone, A.; Lin, Z.; Martin, E.; Vance, J.; Isacson, O. The Transcription Factor Orthodenticle Homeobox 2 Influences Axonal Projections and Vulnerability of Midbrain Dopaminergic Neurons. Brain 2010, 133, 2022–2031. [Google Scholar] [CrossRef]
- Tripathi, P.P.; Di Giovannantonio, L.G.; Sanguinetti, E.; Acampora, D.; Allegra, M.; Caleo, M.; Wurst, W.; Simeone, A.; Bozzi, Y. Increased Dopaminergic Innervation in the Brain of Conditional Mutant Mice Overexpressing Otx2: Effects on Locomotor Behavior and Seizure Susceptibility. Neuroscience 2014, 261, 173–183. [Google Scholar] [CrossRef]
- Simeone, A.; Di Salvio, M.; Di Giovannantonio, L.G.; Acampora, D.; Omodei, D.; Tomasetti, C. The Role of Otx2 in Adult Mesencephalic-Diencephalic Dopaminergic Neurons. Mol. Neurobiol. 2011, 43, 107–113. [Google Scholar] [CrossRef]
- Azzolini, C.; Pagani, I.S.; Pirrone, C.; Borroni, D.; Donati, S.; Al Oum, M.; Pigni, D.; Chiaravalli, A.M.; Vinciguerra, R.; Simonelli, F.; et al. Expression of VEGF-A, Otx Homeobox and P53 Family Genes in Proliferative Vitreoretinopathy. Mediat. Inflamm. 2013, 2013, 857380. [Google Scholar] [CrossRef]
- Glubrecht, D.D.; Kim, J.H.; Russell, L.; Bamforth, J.S.; Godbout, R. Differential CRX and OTX2 Expression in Human Retina and Retinoblastoma. J. Neurochem. 2009, 111, 250–263. [Google Scholar] [CrossRef]
- Kazlauskas, A. Advances in a Gene Therapy-Based Approach to Treat Proliferative Vitreoretinopathy. Arch. Soc. Esp. Oftalmol. 2003, 78, 3–5. [Google Scholar]
- Azzolini, C.; Donati, S.; Micheloni, G.; Moretti, V.; Valli, R.; Acquati, F.; Costantino, L.; Ferrara, F.; Borroni, D.; Premi, E.; et al. Expression of Otx Genes in Müller Cells Using an In Vitro Experimental Model of Retinal Hypoxia. J. Ophthalmol. 2021, 2021, 6265553. [Google Scholar] [CrossRef]
- Staudt, N.; Giger, F.A.; Fielding, T.; Hutt, J.A.; Foucher, I.; Snowden, V.; Hellich, A.; Kiecker, C.; Houart, C. Pineal Progenitors Originate from a Non-Neural Territory Limited by FGF Signalling. Development 2019, 146, dev171405. [Google Scholar] [CrossRef]
- Rohde, K.; Hertz, H.; Rath, M.F. Homeobox Genes in Melatonin-Producing Pinealocytes: Otx2 and Crx Act to Promote Hormone Synthesis in the Mature Rat Pineal Gland. J. Pineal Res. 2019, 66, e12567. [Google Scholar] [CrossRef]
- Kelberman, D.; Rizzoti, K.; Lovell-Badge, R.; Robinson, I.C.A.F.; Dattani, M.T. Genetic Regulation of Pituitary Gland Development in Human and Mouse. Endocr. Rev. 2009, 30, 790–829. [Google Scholar] [CrossRef]
- Dateki, S.; Fukami, M.; Sato, N.; Muroya, K.; Adachi, M.; Ogata, T. OTX2 Mutation in a Patient with Anophthalmia, Short Stature, and Partial Growth Hormone Deficiency: Functional Studies Using the IRBP, HESX1, and POU1F1 Promoters. J. Clin. Endocrinol. Metab. 2008, 93, 3697–3702. [Google Scholar] [CrossRef]
- Nolen, L.D.; Amor, D.; Haywood, A.; St. Heaps, L.; Willcock, C.; Mihelec, M.; Tam, P.; Billson, F.; Grigg, J.; Peters, G.; et al. Deletion at 14q22-23 Indicates a Contiguous Gene Syndrome Comprising Anophthalmia, Pituitary Hypoplasia, and Ear Anomalies. Am. J. Med. Genet. A 2006, 140, 1711–1718. [Google Scholar] [CrossRef]
- Tajima, T.; Ohtake, A.; Hoshino, M.; Amemiya, S.; Sasaki, N.; Ishizu, K.; Fujieda, K. OTX2 Loss of Function Mutation Causes Anophthalmia and Combined Pituitary Hormone Deficiency with a Small Anterior and Ectopic Posterior Pituitary. J. Clin. Endocrinol. Metab. 2009, 94, 314–319. [Google Scholar] [CrossRef]
- Diaczok, D.; Romero, C.; Zunich, J.; Marshall, I.; Radovick, S. A Novel Dominant Negative Mutation of OTX2 Associated with Combined Pituitary Hormone Deficiency. J. Clin. Endocrinol. Metab. 2008, 93, 4351–4359. [Google Scholar] [CrossRef]
- Dateki, S.; Kosaka, K.; Hasegawa, K.; Tanaka, H.; Azuma, N.; Yokoya, S.; Muroya, K.; Adachi, M.; Tajima, T.; Motomura, K.; et al. Heterozygous Orthodenticle Homeobox 2 Mutations Are Associated with Variable Pituitary Phenotype. J. Clin. Endocrinol. Metab. 2010, 95, 756–764. [Google Scholar] [CrossRef]
- Gorbenko Del Blanco, D.; Romero, C.J.; Diaczok, D.; De Graaff, L.C.G.; Radovick, S.; Hokken-Koelega, A.C.S. A Novel OTX2 Mutation in a Patient with Combined Pituitary Hormone Deficiency, Pituitary Malformation, and an Underdeveloped Left Optic Nerve. Eur. J. Endocrinol. 2012, 167, 441–452. [Google Scholar] [CrossRef]
- Kelley, C.G.; Lavorgna, G.; Clark, M.E.; Boncinelli, E.; Mellon, P.L. The Otx2 Homeoprotein Regulates Expression from the Gonadotropin-Releasing Hormone Proximal Promoter. Mol. Endocrinol. 2000, 14, 1246–1256. [Google Scholar] [CrossRef]
- Larder, R.; Mellon, P.L. Otx2 Induction of the Gonadotropin-Releasing Hormone Promoter Is Modulated by Direct Interactions with Grg Co-Repressors. J. Biol. Chem. 2009, 284, 16966–16978. [Google Scholar] [CrossRef]
- Terrinoni, A.; Palombo, R.; Pitolli, C.; Caporali, S.; De Berardinis, R.; Ciccarone, S.; Lanzillotta, A.; Mauramati, S.; Porta, G.; Minieri, M.; et al. Role of the TAp63 Isoform in Recurrent Nasal Polyps. Folia Biol. 2019, 65, 170–180. [Google Scholar] [CrossRef]
- Stevens, W.W.; Peters, A.T.; Suh, L.; Norton, J.E.; Kern, R.C.; Conley, D.B.; Chandra, R.K.; Tan, B.K.; Grammer, L.C.; Harris, K.E.; et al. A Retrospective, Cross-Sectional Study Reveals That Women with Crswnp Have More Severe Disease than Men. Immun. Inflamm. Dis. 2015, 3, 14–22. [Google Scholar] [CrossRef]
- Hackett, T.L.; Warner, S.M.; Stefanowicz, D.; Shaheen, F.; Pechkovsky, D.V.; Murray, L.A.; Argentieri, R.; Kicic, A.; Stick, S.M.; Bai, T.R.; et al. Induction of Epithelial-Mesenchymal Transition in Primary Airway Epithelial Cells from Patients with Asthma by Transforming Growth Factor-Β1. Am. J. Respir. Crit. Care Med. 2009, 180, 122–133. [Google Scholar] [CrossRef]
- Warner, S.M.B.; Hackett, T.L.; Shaheen, F.; Hallstrand, T.S.; Kicic, A.; Stick, S.M.; Knight, D.A. Transcription Factor P63 Regulates Key Genes and Wound Repair in Human Airway Epithelial Basal Cells. Am. J. Respir. Cell Mol. Biol. 2013, 49, 978–988. [Google Scholar] [CrossRef]
- Mills, A.A. P63: Oncogene or Tumor Suppressor? Curr. Opin. Genet. Dev. 2006, 16, 38–44. [Google Scholar] [CrossRef]
- Candi, E.; Dinsdale, D.; Rufini, A.; Salomoni, P.; Knight, R.A.; Mueller, M.; Krammer, P.H.; Melino, G. TAp63 and ΔNp63 in Cancer and Epidermal Development. Cell Cycle 2007, 6, 274–284. [Google Scholar] [CrossRef]
- Lewis, M.T. Homeobox Genes in Mammary Gland Development and Neoplasia. Breast Cancer Res. 2000, 2, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Sukumar, S. Role of Homeobox Genes in Normal Mammary Gland Development and Breast Tumorigenesis. J. Mammary Gland. Biol. Neoplasia 2003, 8, 159–175. [Google Scholar] [CrossRef] [PubMed]
- Terrinoni, A.; Pagani, I.S.; Zucchi, I.; Chiaravalli, A.M.; Serra, V.; Rovera, F.; Sirchia, S.; Dionigi, G.; Miozzo, M.; Frattini, A.; et al. OTX1 Expression in Breast Cancer Is Regulated by P53. Oncogene 2011, 30, 3096–3103. [Google Scholar] [CrossRef]
- Pagani, I.S.; Terrinoni, A.; Marenghi, L.; Zucchi, I.; Chiaravalli, A.M.; Serra, V.; Rovera, F.; Sirchia, S.; Dionigi, G.; Mozzo, M.; et al. The Mammary Gland and the Homeobox Gene Otx1. Breast J. 2010, 16, S53–S56. [Google Scholar] [CrossRef]
- Levantini, E.; Giorgetti, A.; Cerisoli, F.; Traggiai, E.; Guidi, A.; Martin, R.; Acampora, D.; Aplan, P.D.; Keller, G.; Simeone, A.; et al. Unsuspected Role of the Brain Morphogenetic Gene Otx1 in Hematopoiesis. Proc. Natl. Acad. Sci. USA 2003, 100, 10299–10303. [Google Scholar] [CrossRef]
- Morini, J.; Nacci, L.; Babini, G.; Cesaro, S.; Valli, R.; Ottolenghi, A.; Nicolis, E.; Pintani, E.; Maserati, E.; Cipolli, M.; et al. Whole Exome Sequencing Discloses Heterozygous Variants in the DNAJC21 and EFL1 Genes but Not in SRP54 in 6 out of 16 Patients with Shwachman-Diamond Syndrome Carrying Biallelic SBDS Mutations. Br. J. Haematol. 2019, 185, 627–630. [Google Scholar] [CrossRef]
- Hesling, C.; Oliveira, C.C.; Castilho, B.A.; Zanchin, N.I.T. The Shwachman-Bodian-Diamond Syndrome Associated Protein Interacts with HsNip7 and Its down-Regulation Affects Gene Expression at the Transcriptional and Translational Levels. Exp. Cell Res. 2007, 313, 4180–4195. [Google Scholar] [CrossRef]
- Bistoletti, M.; Micheloni, G.; Baranzini, N.; Bosi, A.; Conti, A.; Filpa, V.; Pirrone, C.; Millefanti, G.; Moro, E.; Grimaldi, A.; et al. Homeoprotein OTX1 and OTX2 Involvement in Rat Myenteric Neuron Adaptation after DNBS-Induced Colitis. PeerJ 2020, 8, e8442. [Google Scholar] [CrossRef]
- Mathis, T.; Housset, M.; Eandi, C.; Beguier, F.; Touhami, S.; Reichman, S.; Augustin, S.; Gondouin, P.; Sahel, J.A.; Kodjikian, L.; et al. Activated Monocytes Resist Elimination by Retinal Pigment Epithelium and Downregulate Their OTX2 Expression via TNF-α. Aging Cell 2017, 16, 173–182. [Google Scholar] [CrossRef]
- Zhang, T.; Chen, H.; Qi, L.; Zhang, J.; Wu, R.; Zhang, Y.; Sun, Y. Transcript Profiling Identifies Early Response Genes against FMDV Infection in PK-15 Cells. Viruses 2018, 10, 0364. [Google Scholar] [CrossRef]
- Neurath, M.F. Current and Emerging Therapeutic Targets for IBD. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 269–278. [Google Scholar] [CrossRef]
- Micheloni, G.; Carnovali, M.; Millefanti, G.; Rizzetto, M.; Moretti, V.; Montalbano, G.; Acquati, F.; Giaroni, C.; Valli, R.; Costantino, L.; et al. Soy Diet Induces Intestinal Inflammation in Adult Zebrafish: Role of OTX and P53 Family. Int. J. Exp. Pathol. 2022, 103, 13–22. [Google Scholar] [CrossRef]
- Filpa, V.; Carpanese, E.; Marchet, S.; Pirrone, C.; Conti, A.; Rainero, A.; Moro, E.; Chiaravalli, A.M.; Zucchi, I.; Moriondo, A.; et al. Nitric Oxide Regulates Homeoprotein OTX1 and OTX2 Expression in the Rat Myenteric Plexus after Intestinal Ischemia-Reperfusion Injury. Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 312, 374–389. [Google Scholar] [CrossRef]
- Rivera, L.R.; Pontell, L.; Cho, H.J.; Castelucci, P.; Thacker, M.; Poole, D.P.; Frugier, T.; Furness, J.B. Knock out of Neuronal Nitric Oxide Synthase Exacerbates Intestinal Ischemia/Reperfusion Injury in Mice. Cell Tissue Res. 2012, 349, 565–576. [Google Scholar] [CrossRef]
- Gonzalez, L.M.; Moeser, A.J.; Blikslager, A.T. Animal Models of Ischemia-Reperfusion-Induced Intestinal Injury: Progress and Promise for Translational Research. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 308, 63–75. [Google Scholar] [CrossRef]
- Nedoszytko, B.; Sobalska-Kwapis, M.; Strapagiel, D.; Lange, M.; Górska, A.; Oude Elberink, J.N.G.; van Doormaal, J.; Słomka, M.; Kalinowski, L.; Gruchała-Niedoszytko, M.; et al. Results from a Genome-Wide Association Study (GWAS) in Mastocytosis Reveal New Gene Polymorphisms Associated with WHO Subgroups. Int. J. Mol. Sci. 2020, 21, 5506. [Google Scholar] [CrossRef]
- Valent, P.; Horny, H.P.; Escribano, L.; Longley, B.J.; Li, C.Y.; Schwartz, L.B.; Marone, G.; Nuñez, R.; Akin, C.; Sotlar, K.; et al. Diagnostic Criteria and Classification of Mastocytosis: A Consensus Proposal. Leuk. Res. 2001, 25, 603–625. [Google Scholar] [CrossRef]
- Krappinger, J.C.; Bonstingl, L.; Pansy, K.; Sallinger, K.; Wreglesworth, N.I.; Grinninger, L.; Deutsch, A.; El-Heliebi, A.; Kroneis, T.; Mcfarlane, R.J.; et al. Non-Coding Natural Antisense Transcripts: Analysis and Application. J. Biotechnol. 2021, 340, 75–101. [Google Scholar] [CrossRef] [PubMed]
- Alfano, G.; Vitiello, C.; Caccioppoli, C.; Caramico, T.; Carola, A.; Szego, M.J.; McInnes, R.R.; Auricchio, A.; Banfi, S. Natural Antisense Transcripts Associated with Genes Involved in Eye Development. Hum. Mol. Genet. 2005, 14, 913–923. [Google Scholar] [CrossRef]
- De Haas, T.; Oussoren, E.; Grajkowska, W.; Perek-Polnik, M.; Popovic, M.; Zadravec-Zaletel, L.; Perera, M.; Corte, G.; Wirths, O.; Van Sluis, P.; et al. OTX1 and OTX2 Expression Correlates with the Clinicopathologic Classification of Medulloblastomas. J. Neuropathol. Exp. Neurol. 2006, 65, 176–186. [Google Scholar] [CrossRef] [PubMed]
- Schwalbe, E.C.; Lindsey, J.C.; Nakjang, S.; Crosier, S.; Smith, A.J.; Hicks, D.; Rafiee, G.; Hill, R.M.; Iliasova, A.; Stone, T.; et al. Novel Molecular Subgroups for Clinical Classification and Outcome Prediction in Childhood Medulloblastoma: A Cohort Study. Lancet Oncol. 2017, 18, 958–971. [Google Scholar] [CrossRef] [PubMed]
- Fossat, N.; Chatelain, G.; Brun, G.; Lamonerie, T. Temporal and Spatial Delineation of Mouse Otx2 Functions by Conditional Self-Knockout. EMBO Rep. 2006, 7, 824–830. [Google Scholar] [CrossRef]
- Cavalli, F.M.G.; Remke, M.; Rampasek, L.; Peacock, J.; Shih, D.J.H.; Luu, B.; Garzia, L.; Torchia, J.; Nor, C.; Morrissy, A.S.; et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell 2017, 31, 737–754.e6. [Google Scholar] [CrossRef] [PubMed]
- Stromecki, M.; Tatari, N.; Morrison, L.C.; Kaur, R.; Zagozewski, J.; Palidwor, G.; Ramaswamy, V.; Skowron, P.; Wölfl, M.; Milde, T.; et al. Characterization of a Novel OTX2-Driven Stem Cell Program in Group 3 and Group 4 Medulloblastoma. Mol. Oncol. 2018, 12, 495–513. [Google Scholar] [CrossRef] [PubMed]
- Bunt, J.; De Haas, T.G.; Hasselt, N.E.; Zwijnenburg, D.A.; Koster, J.; Versteeg, R.; Kool, M. Regulation of Cell Cycle Genes and Induction of Senescence by Overexpression of OTX2 in Medulloblastoma Cell Lines. Mol. Cancer Res. 2010, 8, 1344–1357. [Google Scholar] [CrossRef] [PubMed]
- Figueira Muoio, V.M.; Uno, M.; Oba-Shinjo, S.; da Silva, R.; Araújo Pereira, B.J.; Clara, C.; Matushita, H.; Marie, S.N.K. OTX1 and OTX2 Genes in Medulloblastoma. World Neurosurg. 2019, 127, e58–e64. [Google Scholar] [CrossRef]
- Yokota, N.; Mainprize, T.G.; Taylor, M.D.; Kohata, T.; Loreto, M.; Ueda, S.; Dura, W.; Grajkowska, W.; Kuo, J.S.; Rutka, J.T. Identification of Differentially Expressed and Developmentally Regulated Genes in Medulloblastoma Using Suppression Subtraction Hybridization. Oncogene 2004, 23, 3444–3453. [Google Scholar] [CrossRef]
- Di, C.; Liao, S.; Adamson, D.C.; Parrett, T.J.; Broderick, D.K.; Shi, Q.; Lengauer, C.; Cummins, J.M.; Velculescu, V.E.; Fults, D.W.; et al. Identification of OTX2 as a Medulloblastoma Oncogene Whose Product Can Be Targeted by All-Trans Retinoic Acid. Cancer Res. 2005, 65, 919–924. [Google Scholar] [CrossRef]
- Gumireddy, K.; Sutton, L.N.; Phillips, P.C.; Reddy, C.D. All-Trans-Retinoic Acid-Induced Apoptosis in Human Medulloblastoma: Activation of Caspase-3/Poly(ADP-Ribose) Polymerase 1 Pathway 1. Clin. Cancer Res. 2003, 9, 4052–4059. [Google Scholar]
- Bai, R.; Siu, I.M.; Tyler, B.M.; Staedtke, V.; Gallia, G.L.; Riggins, G.J. Evaluation of Retinoic Acid Therapy for OTX2-Positive Medulloblastomas. Neuro-Oncology 2010, 12, 655–663. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Northcott, P.A.; Nakahara, Y.; Wu, X.; Feuk, L.; Ellison, D.W.; Croul, S.; Mack, S.; Kongkham, P.N.; Peacock, J.; Dubuc, A.; et al. Multiple Recurrent Genetic Events Converge on Control of Histone Lysine Methylation in Medulloblastoma. Nat. Genet. 2009, 41, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Bunt, J.; Hasselt, N.E.; Zwijnenburg, D.A.; Hamdi, M.; Koster, J.; Versteeg, R.; Kool, M. OTX2 Directly Activates Cell Cycle Genes and Inhibits Differentiation in Medulloblastoma Cells. Int. J. Cancer 2012, 131, E21–E32. [Google Scholar] [CrossRef] [PubMed]
- Boon, K.; Edwards, J.B.; Siu, I.M.; Olschner, D.; Eberhart, C.G.; Marra, M.A.; Strausberg, R.L.; Riggins, G.J. Comparison of Medulloblastoma and Normal Neural Transcriptomes Identifies a Restricted Set of Activated Genes. Oncogene 2003, 22, 7687–7694. [Google Scholar] [CrossRef]
- Bunt, J.; Hasselt, N.E.; Zwijnenburg, D.A.; Koster, J.; Versteeg, R.; Kool, M. Joint Binding of OTX2 and MYC in Promotor Regions Is Associated with High Gene Expression in Medulloblastoma. PLoS ONE 2011, 6, e26058. [Google Scholar] [CrossRef]
- Ramalho-Santos, M.; Yoon, S.; Matsuzaki, Y.; Mulligan, R.C.; Melton, D.A. “Stemness”: Transcriptional Profiling of Embryonic and Adult Stem Cells. Science 2002, 298, 597–600. [Google Scholar] [CrossRef] [PubMed]
- Peng, G.H.; Chen, S. Crx Activates Opsin Transcription by Recruiting HAT-Containing Co-Activators and Promoting Histone Acetylation. Hum. Mol. Genet. 2007, 16, 2433–2452. [Google Scholar] [CrossRef] [PubMed]
- Robinson, G.; Parker, M.; Kranenburg, T.A.; Lu, C.; Chen, X.; Ding, L.; Phoenix, T.N.; Hedlund, E.; Wei, L.; Zhu, X.; et al. Novel Mutations Target Distinct Subgroups of Medulloblastoma. Nature 2012, 488, 43–48. [Google Scholar] [CrossRef]
- Bunt, J.; Hasselt, N.A.; Zwijnenburg, D.A.; Koster, J.; Versteeg, R.; Kool, M. OTX2 Sustains a Bivalent-like State of OTX2-Bound Promoters in Medulloblastoma by Maintaining Their H3K27me3 Levels. Acta Neuropathol. 2013, 125, 385–394. [Google Scholar] [CrossRef]
- Zagozewski, J.; Shahriary, G.M.; Morrison, L.C.; Saulnier, O.; Stromecki, M.; Fresnoza, A.; Palidwor, G.; Porter, C.J.; Forget, A.; Ayrault, O.; et al. An OTX2-PAX3 Signaling Axis Regulates Group 3 Medulloblastoma Cell Fate. Nat. Commun. 2020, 11, 3627. [Google Scholar] [CrossRef]
- Gallie, B.L.; Campbell, C.; Devlin, H.; Duckett, A.; Squire, J.A. Developmental Basis of Retinal-Specific Induction of Cancer by RB Mutation I. Cancer Res. 1999, 59, 1731s–1735s. [Google Scholar]
- Indovina, P.; Acquaviva, A.; De Falco, G.; Rizzo, V.; Onnis, A.; Luzzi, A.; Giorgi, F.; Hadjistilianou, T.; Toti, P.; Tomei, V.; et al. Downregulation and Aberrant Promoter Methylation of P16INK4A: A Possible Novel Heritable Susceptibility Marker to Retinoblastoma. J. Cell Physiol. 2010, 223, 143–150. [Google Scholar] [CrossRef]
- Vitale, I.; Pietrocola, F.; Guilbaud, E.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostini, M.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; et al. Apoptotic Cell Death in Disease-Current Understanding of the NCCD 2023. Cell Death Differ. 2023, 30, 1097–1154. [Google Scholar] [CrossRef]
- Dalgard, C.L.; Van Quill, K.R.; O’Brien, J.M. Evaluation of the in Vitro and in Vivo Antitumor Activity of Histone Deacetylase Inhibitors for the Therapy of Retinoblastoma. Clin. Cancer Res. 2008, 14, 3113–3123. [Google Scholar] [CrossRef]
- Chintagumpala, M.; Chevez-Barrios, P.; Paysse, E.A.; Plon, S.E.; Hurwitz, R. Retinoblastoma: Review of Current Management. Oncologist 2007, 12, 1237–1246. [Google Scholar] [CrossRef]
- Bao, B.; Ahmad, A.; Azmi, A.S.; Ali, S.; Sarkar, F.H. Overview of Cancer Stem Cells (CSCs) and Mechanisms of Their Regulation: Implications for Cancer Therapy. Curr. Protoc. Pharmacol. 2013, 61, 14.25.1–14.25.14. [Google Scholar] [CrossRef]
- Juliano, C.; Wang, J.; Lin, H. Uniting Germline and Stem Cells: The Function of Piwi Proteins and the PiRNA Pathway in Diverse Organisms. Annu. Rev. Genet. 2011, 45, 447–469. [Google Scholar] [CrossRef]
- Mol, B.M.; Massink, M.P.G.; van der Hout, A.H.; Dommering, C.J.; Zaman, J.M.A.; Bosscha, M.I.; Kors, W.A.; Meijers-Heijboer, H.E.; Kaspers, G.J.L.; Riele, H.t.; et al. High Resolution SNP Array Profiling Identifies Variability in Retinoblastoma Genome Stability. Genes Chromosomes Cancer 2014, 53, 1–14. [Google Scholar] [CrossRef]
- Sivagurunathan, S.; Arunachalam, J.P.; Chidambaram, S. PIWI-like Protein, HIWI2 Is Aberrantly Expressed in Retinoblastoma Cells and Affects Cell-Cycle Potentially through OTX2. Cell Mol. Biol. Lett. 2017, 22, 17. [Google Scholar] [CrossRef]
- Peng, Y.; Jiang, B.H.; Yang, P.H.; Cao, Z.; Shi, X.; Lin, M.C.M.; He, M.L.; Kung, H.F. Phosphatidylinositol 3-Kinase Signaling Is Involved in Neurogenesis during Xenopus Embryonic Development. J. Biol. Chem. 2004, 279, 28509–28514. [Google Scholar] [CrossRef]
- Koike, C.; Nishida, A.; Ueno, S.; Saito, H.; Sanuki, R.; Sato, S.; Furukawa, A.; Aizawa, S.; Matsuo, I.; Suzuki, N.; et al. Functional Roles of Otx2 Transcription Factor in Postnatal Mouse Retinal Development. Mol. Cell Biol. 2007, 27, 8318–8329. [Google Scholar] [CrossRef]
- Béby, F.; Housset, M.; Fossat, N.; Le Greneur, C.; Flamant, F.; Godement, P.; Lamonerie, T. Otx2 Gene Deletion in Adult Mouse Retina Induces Rapid RPE Dystrophy and Slow Photoreceptor Degeneration. PLoS ONE 2010, 5, e11673. [Google Scholar] [CrossRef]
- Li, J.; Di, C.; Jing, J.; Di, Q.; Nakhla, J.; Adamson, D.C. OTX2 Is a Therapeutic Target for Retinoblastoma and May Function as a Common Factor between C-MYC, CRX, and Phosphorylated RB Pathways. Int. J. Oncol. 2015, 47, 1703–1710. [Google Scholar] [CrossRef]
- Pirrone, C.; Chiaravalli, A.M.; Marando, A.; Conti, A.; Rainero, A.; Pistochini, A.; Lo Curto, F.; Pasquali, F.; Castelnuovo, P.; Capella, C.; et al. OTX1 and OTX2 as Possible Molecular Markers of Sinonasal Carcinomas and Olfactory Neuroblastomas. Eur. J. Histochem. 2017, 61, 2730. [Google Scholar] [CrossRef][Green Version]
- Micheloni, G.; Millefanti, G.; Conti, A.; Pirrone, C.; Marando, A.; Rainero, A.; Tararà, L.; Pistochini, A.; Curto, F.L.; Pasquali, F.; et al. Identification of OTX1 and OTX2 as Two Possible Molecular Markers for Sinonasal Carcinomas and Olfactory Neuroblastomas. J. Vis. Exp. 2019, 2019, e56880. [Google Scholar] [CrossRef]
- Valli, R.; De Bernardi, F.; Frattini, A.; Volpi, L.; Bignami, M.; Facchetti, F.; Pasquali, F.; Castelnuovo, P.; Maserati, E. Comparative Genomic Hybridization on Microarray (a-CGH) in Olfactory Neuroblastoma: Analysis of Ten Cases and Review of the Literature. Genes Chromosomes Cancer 2015, 54, 771–775. [Google Scholar] [CrossRef]
- Chen, W.; Zheng, R.; Baade, P.D.; Zhang, S.; Zeng, H.; Bray, F.; Jemal, A.; Yu, X.Q.; He, J. Cancer Statistics in China, 2015. CA Cancer J. Clin. 2016, 66, 115–132. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef]
- Tu, X.P.; Li, H.; Chen, L.S.; Luo, X.N.; Lu, Z.M.; Zhang, S.Y.; Chen, S.H. OTX1 Exerts an Oncogenic Role and Is Negatively Regulated by MiR129-5p in Laryngeal Squamous Cell Carcinoma. BMC Cancer 2020, 20, 794. [Google Scholar] [CrossRef]
- Meng, R.; Fang, J.; Yu, Y.; Hou, L.K.; Chi, J.R.; Chen, A.X.; Zhao, Y.; Cao, X.C. MiR-129-5p Suppresses Breast Cancer Proliferation by Targeting CBX4. Neoplasma 2018, 65, 572–578. [Google Scholar] [CrossRef]
- Døssing, K.B.V.; Binderup, T.; Kaczkowski, B.; Jacobsen, A.; Rossing, M.; Winther, O.; Federspiel, B.; Knigge, U.; Kjær, A.; Friis-Hansen, L. Down-Regulation of MiR-129-5p and the Let-7 Family in Neuroendocrine Tumors and Metastases Leads to Up-Regulation of Their Targets Egr1, G3bp1, Hmga2 and Bach1. Genes 2014, 6, 1–21. [Google Scholar] [CrossRef]
- Liu, Q.; Jiang, J.; Fu, Y.; Liu, T.; Yu, Y.; Zhang, X. MiR-129-5p Functions as a Tumor Suppressor in Gastric Cancer Progression through Targeting ADAM9. Biomed. Pharmacother. 2018, 105, 420–427. [Google Scholar] [CrossRef]
- Kawasaki, H.; Takeuchi, T.; Ricciardiello, F.; Lombardi, A.; Biganzoli, E.; Fornili, M.; De Bortoli, D.; Mesolella, M.; Cossu, A.M.; Scrima, M.; et al. Definition of MiRNA Signatures of Nodal Metastasis in LCa: MiR-449a Targets Notch Genes and Suppresses Cell Migration and Invasion. Mol. Ther. Nucleic Acids 2020, 20, 711–724. [Google Scholar] [CrossRef]
- Takeuchi, T.; Kawasaki, H.; Luce, A.; Cossu, A.M.; Misso, G.; Scrima, M.; Bocchetti, M.; Ricciardiello, F.; Caraglia, M.; Zappavigna, S. Insight toward the MicroRNA Profiling of Laryngeal Cancers: Biological Role and Clinical Impact. Int. J. Mol. Sci. 2020, 21, 3693. [Google Scholar] [CrossRef]
- Miyata, H.; Yamasaki, M.; Kurokawa, Y.; Takiguchi, S.; Nakajima, K.; Fujiwara, Y.; Mori, M.; Doki, Y. Multimodal Treatment for Resectable Esophageal Cancer. Gen. Thorac. Cardiovasc. Surg. 2011, 59, 461–466. [Google Scholar] [CrossRef]
- Chai, J.; Xu, T.; Yang, Y.; Yuan, Y.; Xu, J.; Liu, J.; Wang, K.; Lv, Y.; Chai, J.; Kang, Y.; et al. Overexpression of OTX1 Promotes Tumorigenesis in Patients with Esophageal Squamous Cell Carcinoma. Pathol. Res. Pract. 2022, 232, 153841. [Google Scholar] [CrossRef]
- Mao, Y.; Zhao, Q.; Yin, S.; Ding, X.; Wang, H. Genome-Wide Expression Profiling and Bioinformatics Analysis of Deregulated Genes in Human Gastric Cancer Tissue after Gastroscopy. Asia Pac. J. Clin. Oncol. 2018, 14, e29–e36. [Google Scholar] [CrossRef]
- Qin, S.C.; Zhao, Z.; Sheng, J.X.; Xu, X.H.; Yao, J.; Lu, J.J.; Chen, B.; Zhao, G.D.; Wang, X.Y.; Yang, Y.D. Dowregulation of OTX1 Attenuates Gastric Cancer Cell Proliferation, Migration and Invasion. Oncol. Rep. 2018, 40, 1907–1916. [Google Scholar] [CrossRef]
- Nieto, M.A. Epithelial-Mesenchymal Transitions in Development and Disease: Old Views and New Perspectives. Int. J. Dev. Biol. 2009, 53, 1541–1547. [Google Scholar] [CrossRef]
- Chen, G.; Wan, J.; Wang, Z.; Li, L.; Jia, H.; Xing, S.; Chen, S.; Fan, X.; Li, R. MiR-3196 Acts as a Tumor Suppressor and Predicts Survival Outcomes in Patients With Gastric Cancer. Technol. Cancer Res. Treat. 2020, 19, 1533033820923427. [Google Scholar] [CrossRef]
- Yu, K.; Cai, X.Y.; Li, Q.; Yang, Z.B.; Xiong, W.; Shen, T.; Wang, W.Y.; Li, Y.F. OTX1 Promotes Colorectal Cancer Progression through Epithelial-Mesenchymal Transition. Biochem. Biophys. Res. Commun. 2014, 444, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhao, L.M.; Zhang, C.; Li, M.; Gao, B.; Hu, X.H.; Cao, J.; Wang, G.Y. The LncRNA FEZF1-AS1 Promotes the Progression of Colorectal Cancer Through Regulating OTX1 and Targeting MiR-30a-5p. Oncol. Res. 2020, 28, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Meng, X.; Jia, Y.; Chai, J.; Wang, J.; Xue, X.; Dang, T. Long Non-Coding RNA HNF1A-AS1 Upregulates OTX1 to Enhance Angiogenesis in Colon Cancer via the Binding of Transcription Factor PBX3. Exp. Cell Res. 2020, 393, 112025. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Miao, Q.; Xu, C.W.; Huang, J.H.; Zhou, Y.F.; Wu, M.J. OTX1 Contributes to Hepatocellular Carcinoma Progression by Regulation of ERK/MAPK Pathway. J. Korean Med. Sci. 2016, 31, 1215–1223. [Google Scholar] [CrossRef] [PubMed]
- Burotto, M.; Chiou, V.L.; Lee, J.M.; Kohn, E.C. The MAPK Pathway across Different Malignancies: A New Perspective. Cancer 2014, 120, 3446–3456. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.Q.; Li, H.C.; Teng, F.; Chang, Q.M.; Wu, X.B.; Feng, J.F.; Zhang, Z.P. Long Noncoding RNA MAFG-AS1 Facilitates the Progression of Hepatocellular Carcinoma via Targeting MiR-3196/OTX1 Axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 12131–12143. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Chen, S.; Xu, M.; Zhao, J.; Shen, J.; Li, J.; Liu, Y.; Cao, G.; Ma, J.; He, W.; Chen, X.; et al. MicroRNA-4516 Suppresses Pancreatic Cancer Development via Negatively Regulating Orthodenticle Homeobox 1. Int. J. Biol. Sci. 2020, 16, 2159–2169. [Google Scholar] [CrossRef]
- Sui, X.; Sui, Z. MiR-4269 Suppresses the Tumorigenesis and Development of Pancreatic Cancer by Targeting ZEB1/OTX1 Pathway. Biosci. Rep. 2020, 40, BSR20200010. [Google Scholar] [CrossRef]
- Funahashi, J.I.; Sekido, R.; Murai, K.; Kamachi, Y.; Kondoh, H. Delta-Crystallin Enhancer Binding Protein Delta EF1 Is a Zinc Finger-Homeodomain Protein Implicated in Postgastrulation Embryogenesis. Development 1993, 119, 433–446. [Google Scholar] [CrossRef]
- Ponticos, M.; Partridge, T.; Black, C.M.; Abraham, D.J.; Bou-Gharios, G. Regulation of Collagen Type I in Vascular Smooth Muscle Cells by Competition between Nkx2.5 and DeltaEF1/ZEB1. Mol. Cell Biol. 2004, 24, 6151–6161. [Google Scholar] [CrossRef] [PubMed]
- Cicalese, A.; Bonizzi, G.; Pasi, C.E.; Faretta, M.; Ronzoni, S.; Giulini, B.; Brisken, C.; Minucci, S.; Di Fiore, P.P.; Pelicci, P.G. The Tumor Suppressor P53 Regulates Polarity of Self-Renewing Divisions in Mammary Stem Cells. Cell 2009, 138, 1083–1095. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wu, W.; Wu, M.; Ding, J. Long Noncoding RNA ADPGK-AS1 Promotes Cell Proliferation, Migration, and EMT Process through Regulating MiR-3196/OTX1 Axis in Breast Cancer. In Vitro Cell Dev. Biol. Anim. 2019, 55, 522–532. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Liu, K.; Yao, Y.; Sun, Q.; Zheng, X.; Zhu, B.; Zhang, Q.; Xu, L.; Shen, Y.; Ren, B. DMBX1 Promotes Tumor Proliferation and Regulates Cell Cycle Progression via Repressing OTX2-Mediated Transcription of P21 in Lung Adenocarcinoma Cell. Cancer Lett. 2019, 453, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.Y.; Wang, L.; Mu, D.C.; Li, F.F.; Ran, P.Z.; Shen, H.; Li, W.Y.; Ma, J.; Wu, J.H.; Yang, X.R.; et al. OTX1 Is a Novel Regulator of Proliferation, Migration, Invasion and Apoptosis in Lung Adenocarcinoma. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 9497–9510. [Google Scholar] [CrossRef]
- Lenis, A.T.; Lec, P.M.; Chamie, K. Bladder Cancer: A Review. JAMA 2020, 324, 1980–1991. [Google Scholar] [CrossRef] [PubMed]
- Kandimalla, R.; Van Tilborg, A.A.G.; Kompier, L.C.; Stumpel, D.J.P.M.; Stam, R.W.; Bangma, C.H.; Zwarthoff, E.C. Genome-Wide Analysis of CpG Island Methylation in Bladder Cancer Identified TBX2, TBX3, GATA2, and ZIC4 as PTa-Specific Prognostic Markers. Eur. Urol. 2012, 61, 1245–1256. [Google Scholar] [CrossRef] [PubMed]
- Beukers, W.; van der Keur, K.A.; Kandimalla, R.; Vergouwe, Y.; Steyerberg, E.W.; Boormans, J.L.; Jensen, J.B.; Lorente, J.A.; Real, F.X.; Segersten, U.; et al. FGFR3, TERT and OTX1 as a Urinary Biomarker Combination for Surveillance of Patients with Bladder Cancer in a Large Prospective Multicenter Study. J. Urol. 2017, 197, 1410–1418. [Google Scholar] [CrossRef]
- Jiang, L.; Zuo, Z.; Lin, J.; Yang, C. Orthodenticle Homeobox OTX1 Is a Potential Prognostic Biomarker for Bladder Cancer. Bioengineered 2021, 12, 6559–6571. [Google Scholar] [CrossRef]
- Berrondo, C.; Flax, J.; Kucherov, V.; Siebert, A.; Osinski, T.; Rosenberg, A.; Fucile, C.; Richheimer, S.; Beckham, C.J. Expression of the Long Non-Coding RNA HOTAIR Correlates with Disease Progression in Bladder Cancer and Is Contained in Bladder Cancer Patient Urinary Exosomes. PLoS ONE 2016, 11, e0147236. [Google Scholar] [CrossRef]
- Nagel, S.; Ehrentraut, S.; Meyer, C.; Kaufmann, M.; Drexler, H.G.; MacLeod, R.A.F. Aberrantly Expressed OTX Homeobox Genes Deregulate B-Cell Differentiation in Hodgkin Lymphoma. PLoS ONE 2015, 10, e0138416. [Google Scholar] [CrossRef] [PubMed]
- Omodei, D.; Acampora, D.; Russo, F.; De Filippi, R.; Severino, V.; Di Francia, R.; Frigeri, F.; Mancuso, P.; De Chiara, A.; Pinto, A.; et al. Expression of the Brain Transcription Factor OTX1 Occurs in a Subset of Normal Germinal-Center B Cells and in Aggressive Non-Hodgkin Lymphoma. Am. J. Pathol. 2009, 175, 2609–2617. [Google Scholar] [CrossRef] [PubMed]
- Duclos, K.K.; Hendrikse, J.L.; Jamniczky, H.A. Investigating the Evolution and Development of Biological Complexity under the Framework of Epigenetics. Evol. Dev. 2019, 21, 247–264. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Terrinoni, A.; Micheloni, G.; Moretti, V.; Caporali, S.; Bernardini, S.; Minieri, M.; Pieri, M.; Giaroni, C.; Acquati, F.; Costantino, L.; et al. OTX Genes in Adult Tissues. Int. J. Mol. Sci. 2023, 24, 16962. https://doi.org/10.3390/ijms242316962
Terrinoni A, Micheloni G, Moretti V, Caporali S, Bernardini S, Minieri M, Pieri M, Giaroni C, Acquati F, Costantino L, et al. OTX Genes in Adult Tissues. International Journal of Molecular Sciences. 2023; 24(23):16962. https://doi.org/10.3390/ijms242316962
Chicago/Turabian StyleTerrinoni, Alessandro, Giovanni Micheloni, Vittoria Moretti, Sabrina Caporali, Sergio Bernardini, Marilena Minieri, Massimo Pieri, Cristina Giaroni, Francesco Acquati, Lucy Costantino, and et al. 2023. "OTX Genes in Adult Tissues" International Journal of Molecular Sciences 24, no. 23: 16962. https://doi.org/10.3390/ijms242316962
APA StyleTerrinoni, A., Micheloni, G., Moretti, V., Caporali, S., Bernardini, S., Minieri, M., Pieri, M., Giaroni, C., Acquati, F., Costantino, L., Ferrara, F., Valli, R., & Porta, G. (2023). OTX Genes in Adult Tissues. International Journal of Molecular Sciences, 24(23), 16962. https://doi.org/10.3390/ijms242316962








