Down-Regulation of Rice Glutelin by CRISPR-Cas9 Gene Editing Decreases Carbohydrate Content and Grain Weight and Modulates Synthesis of Seed Storage Proteins during Seed Maturation
Abstract
1. Introduction
2. Results
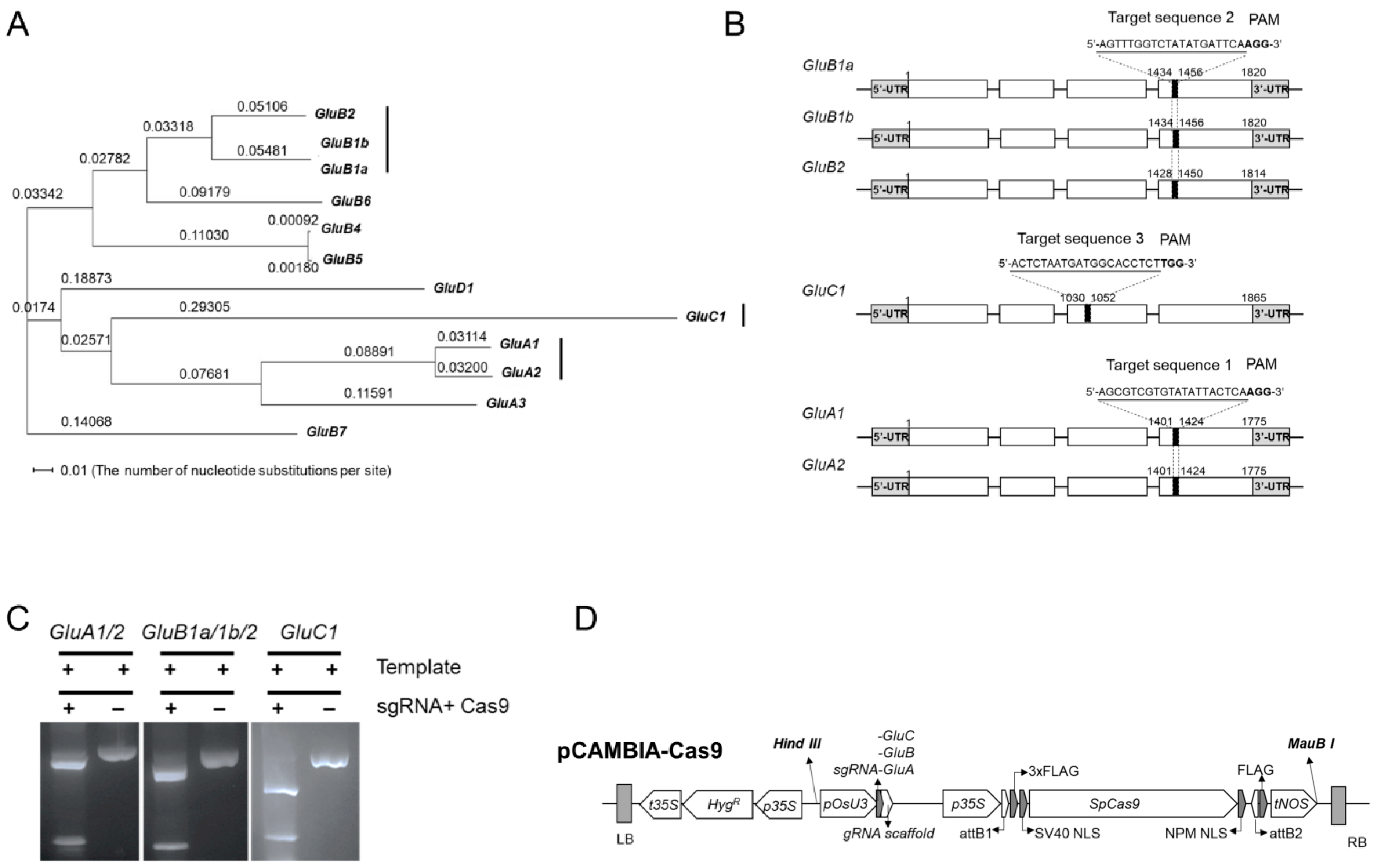
2.1. Design of sgRNA for CRISPR-Cas9-Mediated Inactivation of GluA, GluB or GluC Subsets of Rice Glutelin Genes
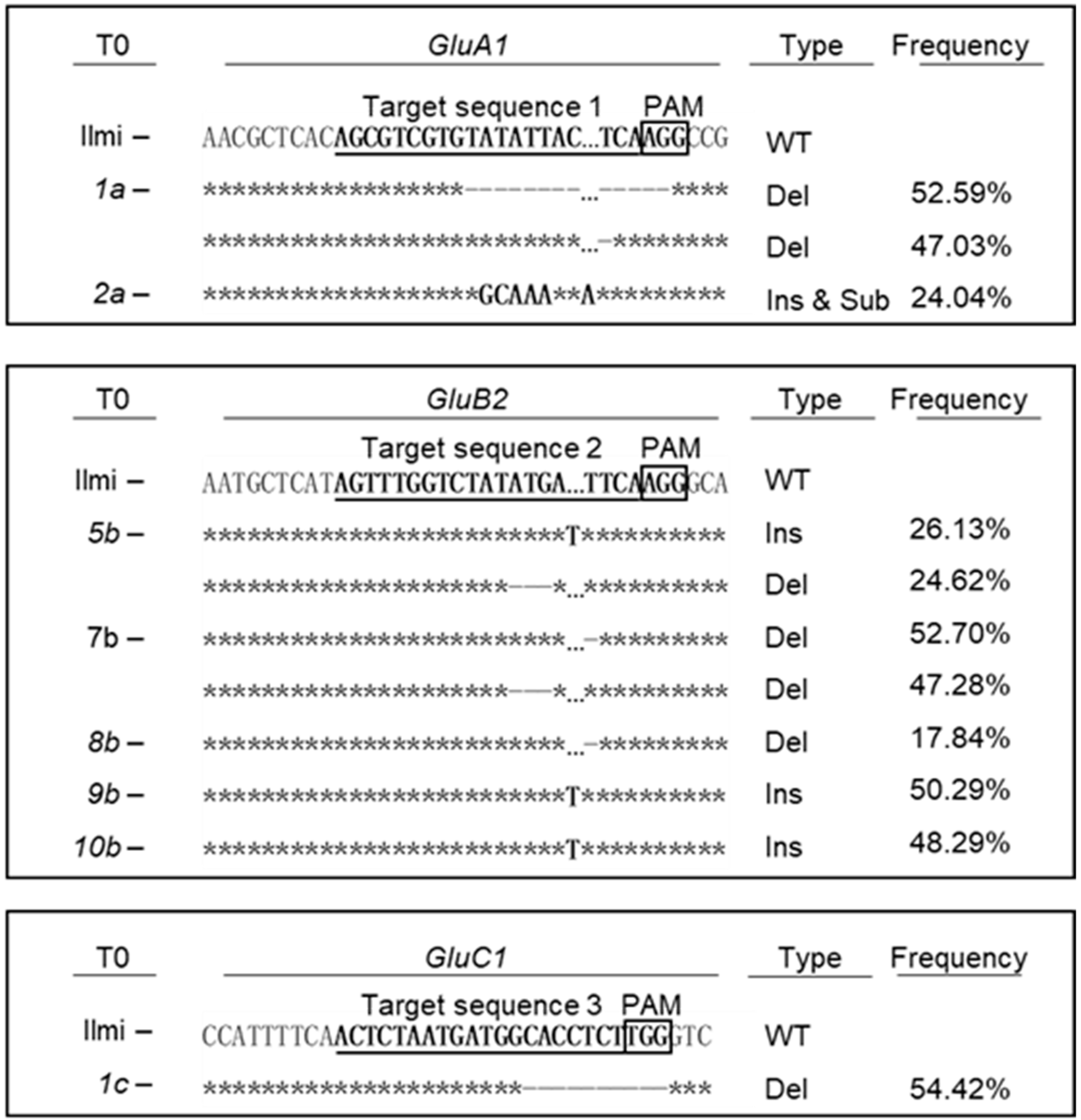
2.2. CRISPR/Cas9-Mediated Targeted Editing of All GluA, GluB or GluC Subgroup Rice Glutelin Genes
2.3. Transcriptome Analysis of T3 Transgenic Subgroup-Specific Glutelin-Gene Edited Rice Plants
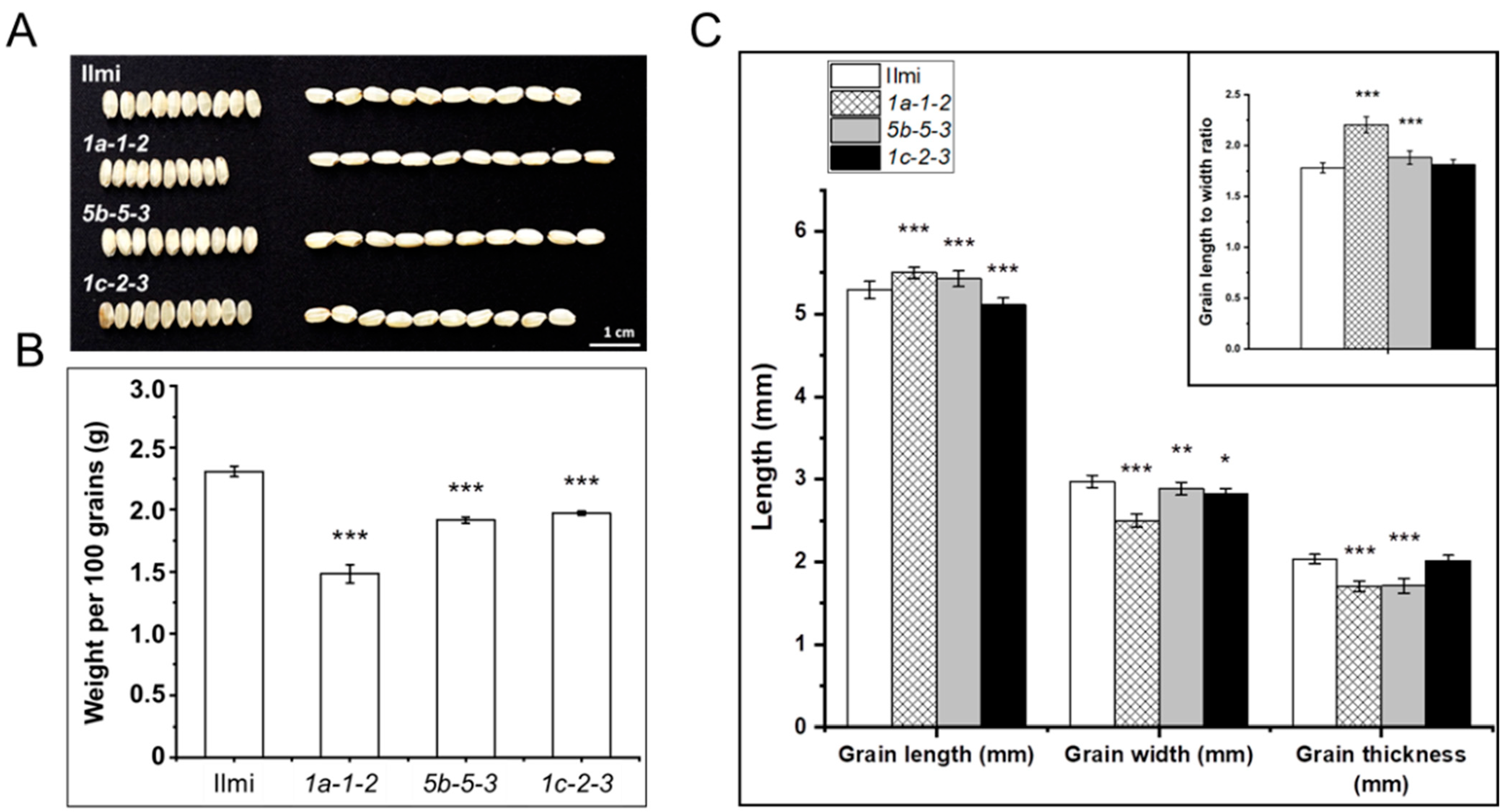
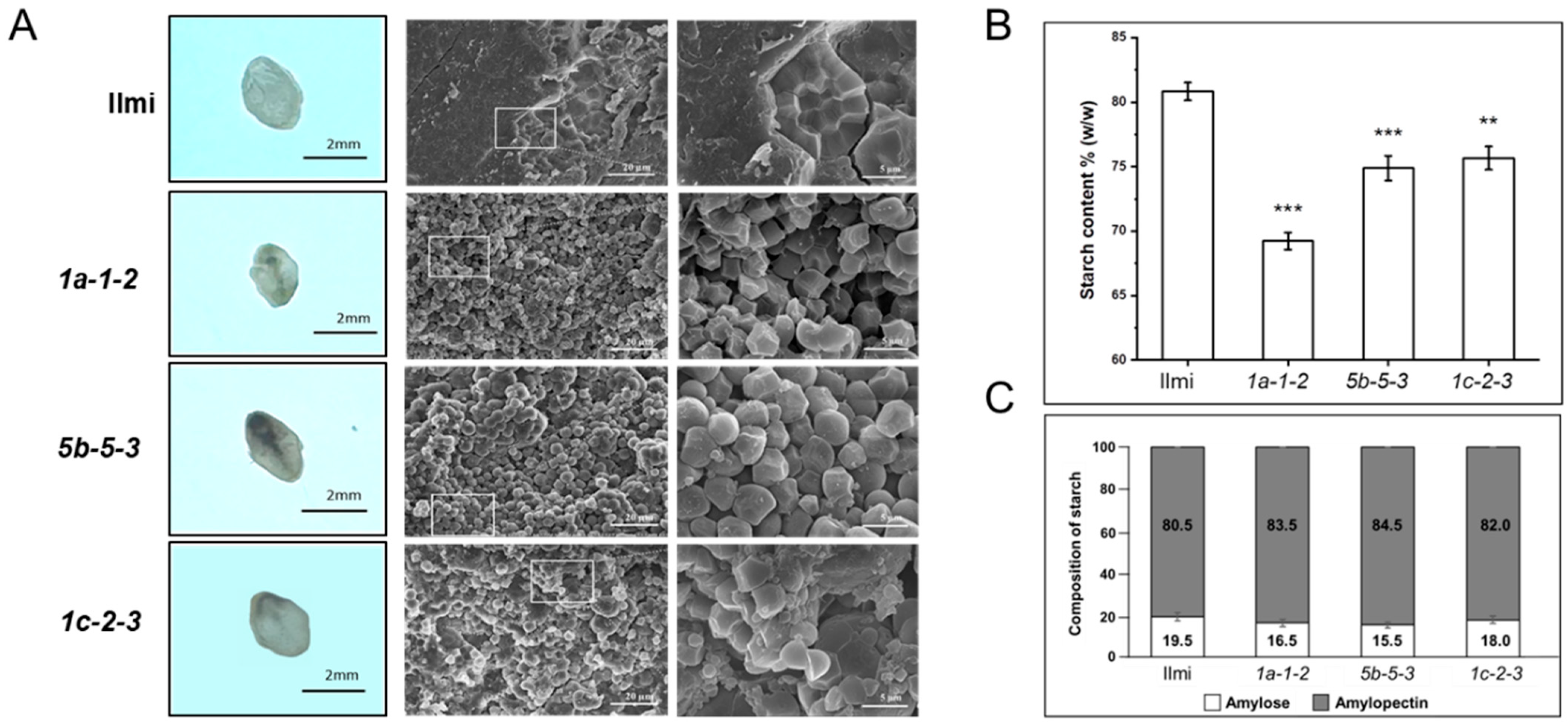
2.4. Morphology of Seeds from T2 Glutelin Gene-Edited Transgenic Rice Plants
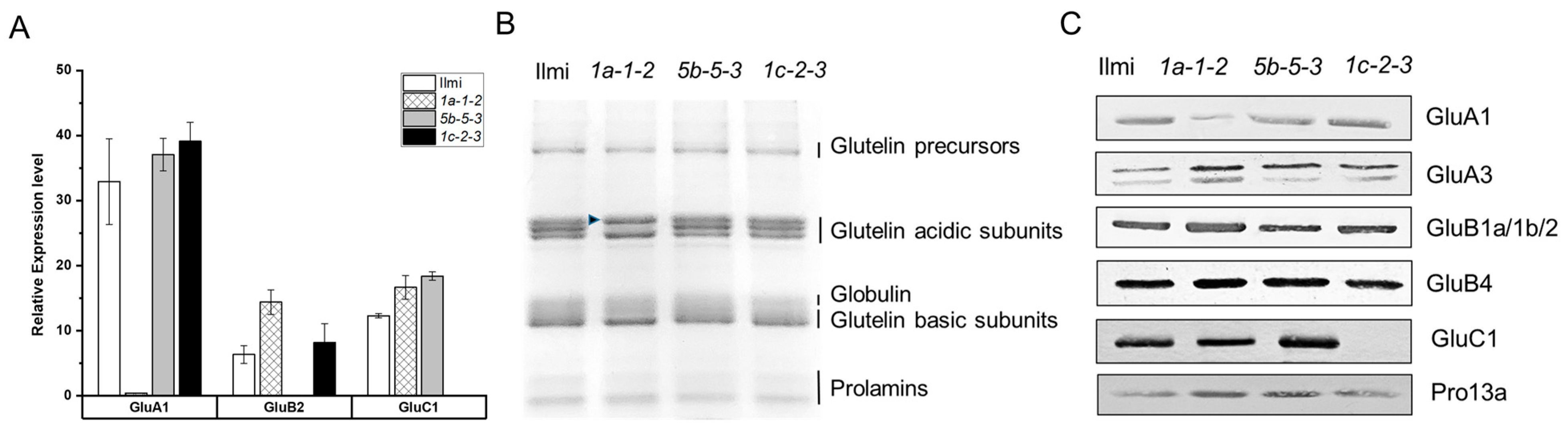
2.5. SSPs and Protein Bodies in T3 Glutelin Gene-Edited Transgenic Rice Seeds
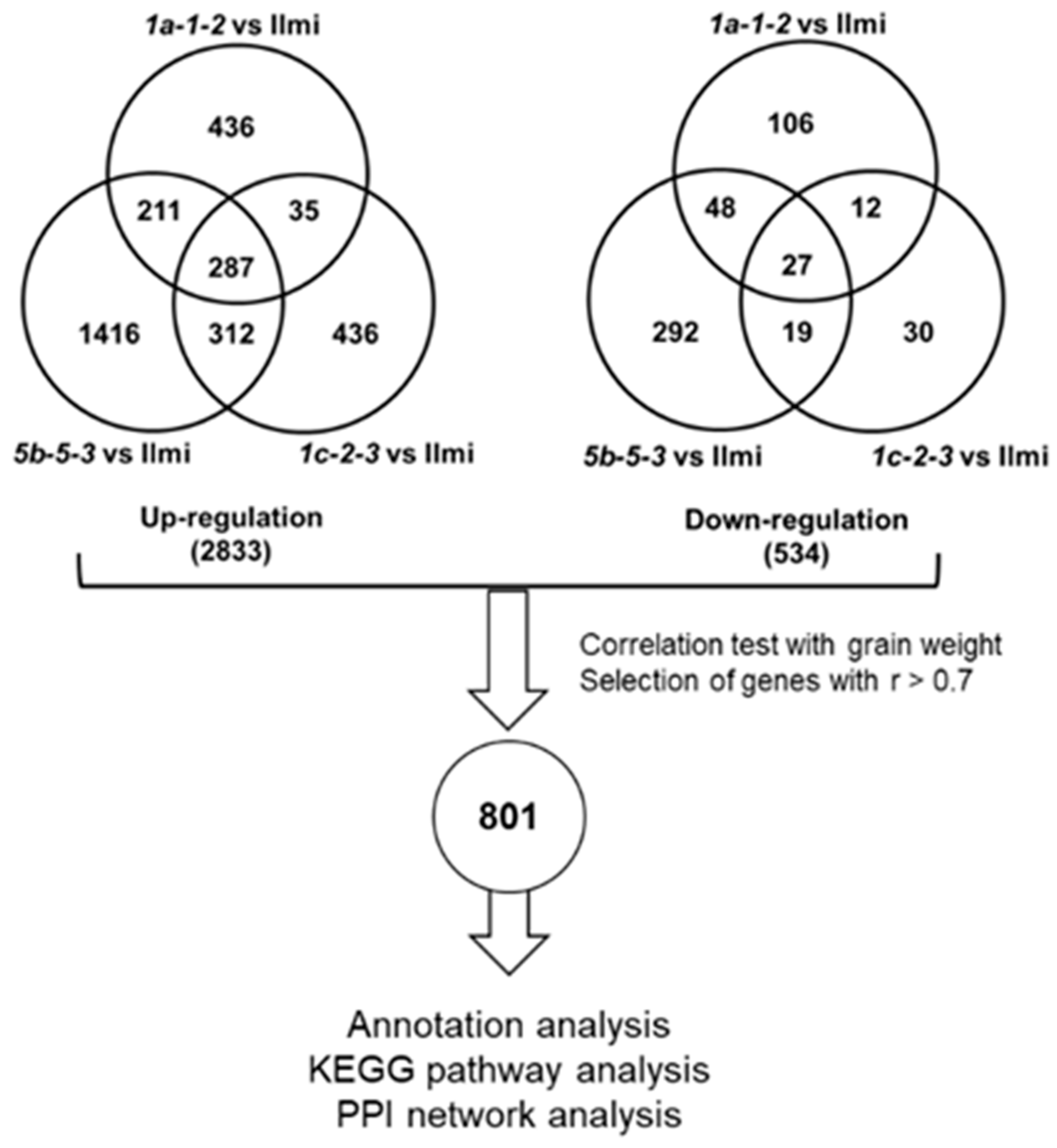
2.6. RNA-Seq/Global Transcriptome Analysis in Glutelin Gene-Edited Rice Seeds
2.7. Correlation between Differential Gene Expression and Reduced Rice Seed Weight
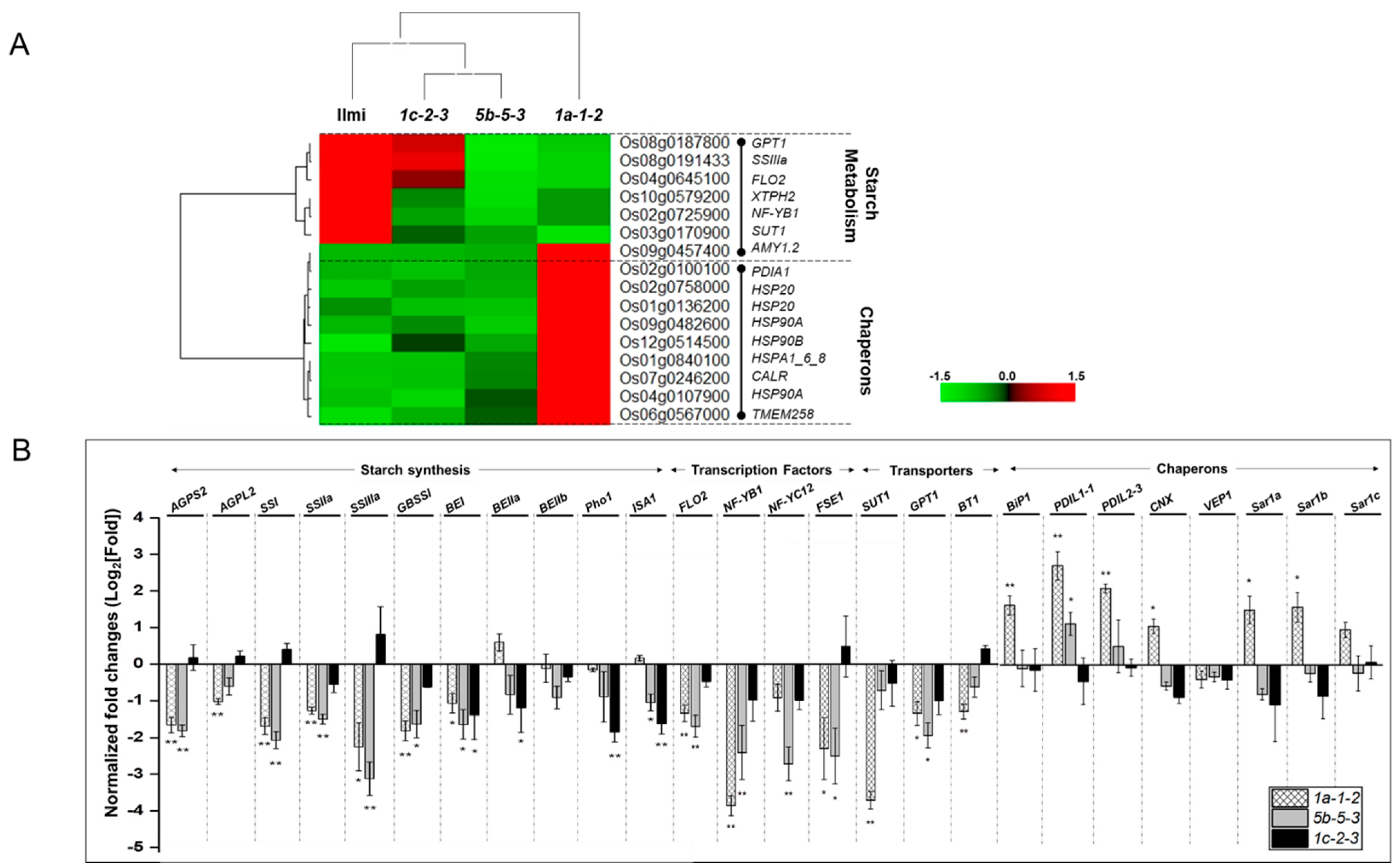
2.8. Starch Synthesis and ER Stress in Glutelin-Related Genes
3. Discussion
3.1. Atypical Morphology of Protein Bodies in the Endosperm of Seeds from Glutelin Gene-Edited Transgenic Rice
3.2. Differential Gene Expression in Transgenic Rice Correlated with Low Seed Weight and Reduced Seed Starch Content
4. Materials and Methods
4.1. Designing gRNA for Different Glutelin Genes
4.2. Testing the Efficacy of sgRNAs In Vitro Using Takara Guide-it™ sgRNA In Vitro Transcription and Screening System
4.3. Cloning of Two Different Cas9-sgRNA Vectors
4.3.1. Using NEB DNA Assembly System sgRNAs Are Introduced into pRGE31-Cas9
4.3.2. Cas9 Binary Vector: Cloning Cas9 Gene and gRNA Scaffold from pRGE31 to pCAMBIA
4.4. Agrobacterium-Mediated Transformation of Rice Callus
4.5. Sequencing of Glutelin Genes of Potential Transgenic Plants
4.6. Phenotypic and Physiological Characterization of Transgenic Seeds
4.7. SDS-PAGE and Western Blot Analysis
4.8. RNA Extraction and Real Time qRT-PCR
4.9. Fractionation of Seed Storage Proteins (SSP)
4.10. Starch and Amylose Content
4.11. Transcriptome Data Analysis
4.12. Selection of Transcripts Correlated with Grain Quality and Establishment of PPI Networks
4.13. Scanning Electron Microscope (SEM)
4.14. Transmission Electron Microscope (TEM)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sasaki, T.; Burr, B. International Rice Genome Sequencing Project: The effort to completely sequence the rice genome. Curr. Opin. Plant Biol. 2000, 2, 138–141. [Google Scholar] [CrossRef]
- Kawakatsu, T.; Yamamoto, M.P.; Hirose, S.; Yano, M.; Takaiwa, F. Characterization of a new rice glutelin gene GluD-1 expressed in the starchy endosperm. J. Exp. Bot. 2008, 59, 4233–4245. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Shigemitsu, T.; Yamasaki, R.; Sasou, A.; Goto, F.; Kishida, K.; Kuroda, M.; Tanaka, K.; Morita, S.; Satoh, S.; et al. Formation mechanism of the internal structure of type I protein bodies in rice endosperm: Relationship between the localization of prolamin species and the expression of individual genes. Plant J. 2012, 70, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Kumamaru, T.; Satoh, H.; Iwata, N.; Omura, T.; Kasai, Z.; Tanaka, K. Purification of Protein Body-I of Rice Seed and its Polypeptide Composition. Plant Cell Physiol. 1987, 28, 1517–1527. [Google Scholar] [CrossRef]
- Xu, J.H.; Messing, J. Amplification of prolamin storage protein genes in different subfamilies of the Poaceae. Theor. Appl. Genet. 2009, 119, 1397–1412. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Sugimoto, T.; Ogawa, M.; Kasai, Z. Isolation and Characterization of Two Types of Protein Bodies in the Rice Endosperm. Agric. Biol. Chem. 1980, 44, 1633–1639. [Google Scholar] [CrossRef]
- Yamagata, H.; Tanaka, K. The Site of Synthesis and Accumulation of Rice Storage Proteins. Plant Cell Physiol. 1986, 27, 135–145. [Google Scholar] [CrossRef]
- Bechtel, D.B.; Juliano, B.O. Formation of Protein Bodies in the Starchy Endosperm of Rice (Oryza sativa L.): A Re-investigation. Ann. Bot. 1980, 45, 503–509. [Google Scholar] [CrossRef]
- Krishnan, H.B.; Franceschi, V.R.; Okita, T.W. Immunochemical studies on the role of the Golgi complex in protein-body formation in rice seeds. Planta 1986, 169, 71–80. [Google Scholar] [CrossRef]
- Krishnan, H.B.; White, J.A.; Pueppke, S.G. Characterization and localization of rice (Oryza sativa L.) seed globulins. Plant Sci. 1992, 81, 1–11. [Google Scholar] [CrossRef]
- Giroux, M.J.; Boyer, C.; Feix, G.; Hannah, L.C. Coordinated Transcriptional Regulation of Storage Product Genes in the Maize Endosperm. Plant Physiol. 1994, 106, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.; Smith, A.M. Starch biosynthesis. Plant Cell. 1995, 7, 971–985. [Google Scholar] [CrossRef]
- López-González, C.; Juárez-Colunga, S.; Morales-Elías, N.C.; Tiessen, A. Exploring regulatory networks in plants: Transcription factors of starch metabolism. PeerJ 2019, 7, 6841. [Google Scholar] [CrossRef]
- Tsai, H.L.; Lue, W.L.; Lu, K.J.; Hsieh, M.H.; Wang, S.M.; Chen, J. Starch synthesis in Arabidopsis is achieved by spatial cotranscription of core starch metabolism genes. Plant Physiol. 2009, 151, 582–595. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.M.; Denyer, K.; Martin, C. The synthesis of the starch granule. Annu. Rev. Plant Biol. 1997, 48, 67–87. [Google Scholar] [CrossRef]
- Takaiwa, F.; Wakasa, Y.; Takagi, H.; Hiroi, T. Rice seed for delivery of vaccines to gut mucosal immune tissues. Plant Biotechnol. J. 2015, 13, 1041–1055. [Google Scholar] [CrossRef]
- Kim, Y.M.; Lee, J.Y.; Lee, T.; Lee, Y.H.; Kim, S.H.; Kang, S.H.; Yoon, U.H.; Ha, S.H.; Lim, S.H. The suppression of the glutelin storage protein gene in transgenic rice seeds results in a higher yield of recombinant protein. Plant Biotechnol. Rep. 2012, 6, 347–353. [Google Scholar] [CrossRef]
- Kim, H.J.; Lee, J.Y.; Yoon, U.H.; Lim, S.H.; Kim, Y.M. Effects of reduced prolamin on seed storage protein composition and the nutritional quality of rice. Int. J. Mol. Sci. 2013, 14, 73–84. [Google Scholar] [CrossRef]
- Lee, H.J.; Jo, Y.M.; Lee, J.Y.; Lim, S.H.; Kim, Y.M. Lack of Globulin Synthesis during Seed Development Alters Accumulation of Seed Storage Proteins in Rice. Int. J. Mol. Sci. 2015, 16, 17–36. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.; Lee, H.J.; Jo, Y.M.; Lim, S.H.; Rakwal, R.; Lee, J.Y.; Kim, Y.M. RNA Interference-Mediated Simultaneous Suppression of Seed Storage Proteins in Rice Grains. Front. Plant Sci. 2016, 31, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Du, H.; Tao, Y.; Xu, Y.; Wang, F.; Li, B.; Zhu, Q.H.; Niu, H.; Yang, J. Efficient breeding of low glutelin content rice germplasm by simultaneous editing multiple glutelin genes via CRISPR/Cas9. Plant Sci. 2022, 324, 111449. [Google Scholar] [CrossRef]
- Nagamine, A.; Matsusaka, H.; Ushijima, T.; Kawagoe, Y.; Ogawa, M.; Okita, T.W.; Kumamaru, T. A role for the cysteine-rich 10 kDa prolamin in protein body I formation in rice. Plant Cell Physiol. 2011, 52, 1003–1016. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Messing, J. RNA interference-mediated change in protein body morphology and seed opacity through loss of different zein proteins. Plant Physiol. 2010, 153, 337–347. [Google Scholar] [CrossRef]
- Kawakatsu, T.; Hirose, S.; Yasuda, H.; Takaiwa, F. Reducing rice seed storage protein accumulation leads to changes in nutrient quality and storage organelle formation. Plant Physiol. 2010, 154, 1842–1854. [Google Scholar] [CrossRef] [PubMed]
- Qian, D.; Chen, G.; Tian, L.; Qu, L.Q. OsDER1 Is an ER-Associated Protein Degradation Factor That Responds to ER Stress. Plant Physiol. 2018, 178, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Wakasa, Y.; Yasuda, H.; Oono, Y.; Kawakatsu, T.; Hirose, S.; Takahashi, H.; Hayashi, S.; Yang, L.; Takaiwa, F. Expression of ER quality control-related genes in response to changes in BiP1 levels in developing rice endosperm. Plant J. 2011, 65, 75–89. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Wang, L.; Lin, Q.; Yu, F. Rice seed storage proteins: Biosynthetic pathways and the effects of environmental factors. J. Integr. Plant Biol. 2021, 63, 1999–2019. [Google Scholar] [CrossRef] [PubMed]
- Ohta, M.; Takaiwa, F. OsHrd3 is necessary for maintaining the quality of endoplasmic reticulum-derived protein bodies in rice endosperm. J. Exp. Bot. 2015, 66, 4585–4593. [Google Scholar] [CrossRef][Green Version]
- Ohta, M.; Wakasa, Y.; Takahashi, H.; Hayashi, S.; Kudo, K.; Takaiwa, F. Analysis of rice ER-resident J-proteins reveals diversity and functional differentiation of the ER-resident Hsp70 system in plants. J. Exp. Bot. 2013, 64, 29–41. [Google Scholar] [CrossRef]
- Ohta, M.; Takaiwa, F. OsERdj7 is an ER-resident J-protein involved in ER quality control in rice endosperm. J. Plant Physiol. 2020, 245, 153109. [Google Scholar] [CrossRef]
- Howell, S.H. Endoplasmic reticulum stress responses in plants. Annu. Rev. Plant Biol. 2013, 64, 477–499. [Google Scholar] [CrossRef]
- Muench, D.G.; Wu, Y.; Zhang, Y.; Li, X.; Boston, R.S.; Okita, T.W. Molecular cloning, expression and subcellular localization of a BiP homolog from rice endosperm tissue. Plant Cell Physiol. 1997, 38, 404–412. [Google Scholar] [CrossRef]
- Takemoto, Y.; Coughlan, S.J.; Okita, T.W.; Satoh, H.; Ogawa, M.; Kumamaru, T. The rice mutant esp2 greatly accumulates the glutelin precursor and deletes the protein disulfide isomerase. Plant Physiol. 2002, 128, 1212–1222. [Google Scholar] [CrossRef]
- Onda, Y.; Nagamine, A.; Sakurai, M.; Kumamaru, T.; Ogawa, M.; Kawagoe, Y. Distinct roles of protein disulfide isomerase and P5 sulfhydryl oxidoreductases in multiple pathways for oxidation of structurally diverse storage proteins in rice. Plant Cell 2011, 23, 210–223. [Google Scholar] [CrossRef]
- Tian, L.; Dai, L.L.; Yin, Z.J.; Fukuda, M.; Kumamaru, T.; Dong, X.B.; Xu, X.P.; Qu, L.Q. Small GTPase Sar1 is crucial for proglutelin and α-globulin export from the endoplasmic reticulum in rice endosperm. J. Exp. Bot. 2013, 64, 2831–2845. [Google Scholar] [CrossRef]
- Hanton, S.L.; Chatre, L.; Matheson, L.A.; Rossi, M.; Held, M.A.; Brandizzi, F. Plant Sar1 isoforms with near-identical protein sequences exhibit different localisations and effects on secretion. Plant Mol. Biol. 2008, 67, 283–294. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, S.; Liu, S.; Jiang, L.; Chen, L.; Ren, Y.; Han, X.; Liu, F.; Ji, S.; Liu, X.; et al. The vacuolar processing enzyme OsVPE1 is required for efficient glutelin processing in rice. Plant J. 2009, 58, 606–617. [Google Scholar] [CrossRef]
- Kumamaru, T.; Uemura, Y.; Inoue, Y.; Takemoto, Y.; Siddiqui, S.U.; Ogawa, M.; Hara-Nishimura, I.; Satoh, H. Vacuolar processing enzyme plays an essential role in the crystalline structure of glutelin in rice seed. Plant Cell Physiol. 2010, 51, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E.; Lykke-Andersen, J. mRNA surveillance: The perfect persist. J. Cell Sci. 2002, 115, 3033–3038. [Google Scholar] [CrossRef] [PubMed]
- Hoof, A.V.; Wagner, E.J. A brief survey of mRNA surveillance. Trends Biochem. Sci. 2011, 36, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Smith, D.F. Hop as an adaptor in the heat shock protein 70 (Hsp70) and hsp90 chaperone machinery. J. Biol. Chem. 1998, 273, 35194–35200. [Google Scholar] [CrossRef]
- Taipale, M.; Jarosz, D.F.; Lindquist, S. HSP90 at the hub of protein homeostasis: Emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 2010, 7, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Lu, Y.H.; Song, M.; Wang, Y.; Xu, W.; Wu, L.; Wang, H.; Ma, Z. Overexpression of a Triticum aestivum Calreticulin gene (TaCRT1) Improves Salinity Tolerance in Tobacco. PLoS ONE 2015, 10, e0140591. [Google Scholar] [CrossRef] [PubMed]
- Christensen, A.; Svensson, K.; Persson, S.; Jung, J.; Michalak, M.; Widell, S.; Sommarin, M. Functional characterization of Arabidopsis calreticulin1a: A key alleviator of endoplasmic reticulum stress. Plant Cell Physiol. 2008, 49, 912–924. [Google Scholar] [CrossRef] [PubMed]
- Saijo, Y.; Tintor, N.; Lu, X.; Rauf, P.; Pajerowska-Mukhtar, K.; Häweker, H.; Dong, X.; Robatzek, S.; Schulze-Lefert, P. Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 2009, 28, 3439–3449. [Google Scholar] [CrossRef] [PubMed]
- Christensen, A.; Svensson, K.; Thelin, L.; Zhang, W.; Tintor, N.; Prins, D.; Funke, N.; Michalak, M.; Schulze-Lefert, P.; Saijo, Y.; et al. Higher plant calreticulins have acquired specialized functions in Arabidopsis. PLoS ONE 2010, 5, e11342. [Google Scholar] [CrossRef] [PubMed]
- Shergalis, A.G.; Hu, S.; Bankhead, A., 3rd; Neamati, N. Role of the ERO1-PDI interaction in oxidative protein folding and disease. Pharmacol. Ther. 2020, 210, 107525. [Google Scholar] [CrossRef]
- Christianson, J.C.; Shaler, T.A.; Tyler, R.E.; Kopito, R.R. OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat. Cell Biol. 2008, 10, 272–282. [Google Scholar] [CrossRef]
- Okita, T.W.; Nakata, P.A.; Anderson, J.M.; Sowokinos, J.; Morell, M.; Preiss, J. The Subunit Structure of Potato Tuber ADPglucose Pyrophosphorylase. Plant Physiol. 1990, 93, 785–790. [Google Scholar] [CrossRef]
- Smith-White, B.J.; Preiss, J. Comparison of proteins of ADP-glucose pyrophosphorylase from diverse sources. J. Mol. Evol. 1992, 34, 449–464. [Google Scholar] [CrossRef]
- Villand, P.; Olsen, O.A.; Kleczkowski, L.A. Molecular characterization of multiple cDNA clones for ADP-glucose pyrophosphorylase from Arabidopsis thaliana. Plant Mol. Biol. 1993, 23, 1279–1284. [Google Scholar] [CrossRef]
- Kawagoe, Y.; Kubo, A.; Satoh, H.; Takaiwa, F.; Nakamura, Y. Roles of isoamylase and ADP-glucose pyrophosphorylase in starch granule synthesis in rice endosperm. Plant J. 2005, 42, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Zheng, F.Q.; Shen, G.Z.; Gao, J.P.; Snustad, D.P.; Li, M.G.; Zhang, J.L.; Hong, M.M. The amylose content in rice endosperm is related to the post-transcriptional regulation of the waxy gene. Plant J. 1995, 7, 613–622. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Huang, B.; Zhang, M.; Zhang, X.; Rivenbark, J.; Lappe, R.L.; James, M.G.; Myers, A.M.; Hennen-Bierwagen, T.A. Functional interactions between starch synthase III and isoamylase-type starch-debranching enzyme in maize endosperm. Plant Physiol. 2012, 158, 679–692. [Google Scholar] [CrossRef]
- Yamanouchi, H.; Nakamura, Y. Organ Specificity of Isoforms of Starch Branching Enzyme (Q-Enzyme) in Rice. Plant Cell Physiol. 1992, 33, 985–991. [Google Scholar] [CrossRef]
- Nakamura, Y.; Umemoto, T.; Ogata, N.; Kuboki, Y.; Yano, M.; Sasaki, T. Starch debranching enzyme (R-enzyme or pullulanase) from developing rice endosperm: Purification, cDNA and chromosomal localization of the gene. Planta 1996, 199, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Fujita, N.; Yoshida, M.; Kondo, T.; Saito, K.; Utsumi, Y.; Tokunaga, T.; Nishi, A.; Satoh, H.; Park, J.H.; Jane, J.L.; et al. Characterization of SSIIIa-deficient mutants of rice: The function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiol. 2007, 144, 2009–2023. [Google Scholar] [CrossRef] [PubMed]
- She, K.C.; Kusano, H.; Koizumi, K.; Yamakawa, H.; Hakata, M.; Imamura, T.; Fukuda, M.; Naito, N.; Tsurumaki, Y.; Yaeshima, M.; et al. A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant Cell 2010, 22, 3280–3294. [Google Scholar] [CrossRef]
- Feng, T.; Wang, L.; Li, L.; Liu, Y.; Chong, K.; Theißen, G.; Meng, Z. OsMADS14 and NF-YB1 cooperate in the direct activation of OsAGPL2 and Waxy during starch synthesis in rice endosperm. New Phytol. 2022, 234, 77–92. [Google Scholar] [CrossRef]
- Bai, A.N.; Lu, X.D.; Li, D.Q.; Liu, J.X.; Liu, C.M. NF-YB1-regulated expression of sucrose transporters in aleurone facilitates sugar loading to rice endosperm. Cell Res. 2016, 26, 384–388. [Google Scholar] [CrossRef]
- Xu, J.J.; Zhang, X.F.; Xue, H.W. Rice aleurone layer specific OsNF-YB1 regulates grain filling and endosperm development by interacting with an ERF transcription factor. J. Exp. Bot. 2016, 67, 6399–6411. [Google Scholar] [CrossRef] [PubMed]
- Bello, B.K.; Hou, Y.; Zhao, J.; Jiao, G.; Wu, Y.; Li, Z.; Wang, Y.; Tong, X.; Wang, W.; Yuan, W.; et al. NF-YB1-YC12-bHLH144 complex directly activates Wx to regulate grain quality in rice (Oryza sativa L.). Plant Biotechnol. J. 2019, 17, 1222–1235. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Ren, Y.; Li, W.; Wu, F.; Yang, W.; Huang, X.; Yao, J. NF-YC12 is a key multi-functional regulator of accumulation of seed storage substances in rice. J. Exp. Bot. 2019, 70, 3765–3780. [Google Scholar] [CrossRef] [PubMed]
- Long, W.; Wang, Y.; Zhu, S.; Jing, W.; Wang, Y.; Ren, Y.; Tian, Y.; Liu, S.; Liu, X.; Chen, L.; et al. FLOURY SHRUNKEN ENDOSPERM1 Connects Phospholipid Metabolism and Amyloplast Development in Rice. Plant Physiol. 2018, 177, 698–712. [Google Scholar] [CrossRef]
- Furbank, T.R.; Scofield, N.G.; Tatsuro, H.; Xin-Ding, W.; John, P.W.; Christina, O.E. Cellular localisation and function of a sucrose transporter OsSUT1 in developing rice grains. Func. Plant Biol. 2001, 28, 1187–1196. [Google Scholar] [CrossRef]
- Jiang, H.W.; Dian, W.M.; Liu, F.Y.; Wu, P. Cloning and characterization of a glucose 6-phosphate/phosphate translocator from Oryza sativa. J. Zhejiang Univ. Sci. 2003, 4, 331–335. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wei, X.; Ren, Y.; Qiu, J.; Jiao, G.; Guo, X.; Tang, S.; Wan, J.; Hu, P. OsBT1 encodes an ADP-glucose transporter involved in starch synthesis and compound granule formation in rice endosperm. Sci. Rep. 2017, 7, 40124. [Google Scholar] [CrossRef]
- Belhaj, K.; Chaparro-Garcia, A.; Kamoun, S.; Nekrasov, V. Plant genome editing made easy: Targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 2013, 9, 39. [Google Scholar] [CrossRef]
- Li, Z.; Trick, H.N. Rapid method for high-quality RNA isolation from seed endosperm containing high levels of starch. BioTechniques 2005, 38, 872–876. [Google Scholar] [CrossRef]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinforma Biomath. 2013, 3, 71–85. [Google Scholar]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Erratum: Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 888. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinforma 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]


| Bin Code a | Annotation | 1a-1-2 vs. Ilmi | 5b-5-3 vs. Ilmi | 1c-2-3 vs. Ilmi | Total Number of Non-Redundant Genes | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | ||||||||||
| No | % | No | % | No | % | No | % | No | % | No | % | No | % | ||
| 1 | Photosyntheisis | 18 | 1.86% | 0 | 0.00% | 31 | 1.39% | 0 | 0.00% | 17 | 2.21% | 0 | 0.00% | 34 | 1.01% |
| 2/3/4/5/6/8 b | Carbohydrate metabolism | 7 | 0.72% | 4 | 2.07% | 4 | 0.18% | 8 | 2.07% | 1 | 0.13% | 1 | 1.14% | 20 | 0.59% |
| 9. | Mitochondrial electron transport/ATP synthesis | 5 | 0.52% | 0 | 0.00% | 20 | 0.90% | 2 | 0.52% | 10 | 1.30% | 0 | 0.00% | 23 | 0.68% |
| 10 | Cell wall | 6 | 0.62% | 4 | 2.07% | 13 | 0.58% | 5 | 1.30% | 1 | 0.13% | 0 | 0.00% | 27 | 0.80% |
| 11 | Lipid metabolism | 1 | 0.10% | 5 | 2.59% | 8 | 0.36% | 10 | 2.59% | 1 | 0.13% | 0 | 0.00% | 20 | 0.59% |
| 13 | Amino acid metabolism | 5 | 0.52% | 0 | 0.00% | 7 | 0.31% | 2 | 0.52% | 0 | 0.00% | 0 | 0.00% | 14 | 0.42% |
| 15/16 | Metal handling & secondary metabolism | 4 | 0.41% | 1 | 0.52% | 7 | 0.31% | 3 | 0.78% | 1 | 0.13% | 0 | 0.00% | 13 | 0.39% |
| 17 | Hormone metabolism | 13 | 1.34% | 2 | 1.04% | 19 | 0.85% | 4 | 1.04% | 2 | 0.26% | 0 | 0.00% | 38 | 1.13% |
| 18/19 | Co-factor/vitamine metabolism and tetrapyrrole synthesis | 2 | 0.21% | 0 | 0.00% | 4 | 0.18% | 1 | 0.26% | 0 | 0.00% | 0 | 0.00% | 6 | 0.18% |
| 20 | Stress | 24 | 2.48% | 6 | 3.11% | 29 | 1.30% | 13 | 3.37% | 3 | 0.39% | 0 | 0.00% | 65 | 1.93% |
| 21 | Redox | 4 | 0.41% | 0 | 0.00% | 6 | 0.27% | 3 | 0.78% | 0 | 0.00% | 1 | 1.14% | 14 | 0.42% |
| 23/24/25 c | Other metabolism-related BINs | 3 | 0.31% | 1 | 0.52% | 2 | 0.09% | 6 | 1.55% | 0 | 0.00% | 0 | 0.00% | 11 | 0.33% |
| 26 | Miscellaneous | 16 | 1.65% | 11 | 5.70% | 35 | 1.57% | 17 | 4.40% | 7 | 0.91% | 3 | 3.41% | 70 | 2.08% |
| 27.1 | RNA.processing | 110 | 11.35% | 35 | 18.13% | 145 | 6.51% | 36 | 9.33% | 127 | 16.49% | 32 | 36.36% | 258 | 7.67% |
| 27.3 | RNA.regulation of transcription | 15 | 1.55% | 6 | 3.11% | 43 | 1.93% | 13 | 3.37% | 7 | 0.91% | 0 | 0.00% | 73 | 2.17% |
| 27.2/4 | RNA.transcription and RNA binding | 5 | 0.52% | 0 | 0.00% | 8 | 0.36% | 3 | 0.78% | 1 | 0.13% | 0 | 0.00% | 14 | 0.42% |
| 28 | DNA.synthesis and repair | 4 | 0.41% | 2 | 1.04% | 16 | 0.72% | 4 | 1.04% | 5 | 0.65% | 1 | 1.14% | 28 | 0.83% |
| 29.1/3/6/7/8 d | Protein others | 7 | 0.72% | 1 | 0.52% | 9 | 0.40% | 4 | 1.04% | 2 | 0.26% | 2 | 2.27% | 20 | 0.59% |
| 29.2.1/2/3/4 | Protein synthesis-ribosome, initiation, & elongation | 49 | 5.06% | 0 | 0.00% | 44 | 1.98% | 4 | 1.04% | 21 | 2.73% | 7 | 7.95% | 83 | 2.47% |
| 29.2.6 | Protein synthesis-ribosomal RNA | 12 | 1.24% | 1 | 0.52% | 102 | 4.58% | 2 | 0.52% | 80 | 10.39% | 1 | 1.14% | 113 | 3.36% |
| 29.2.7 | Protein synthesis_transfer RNA | 56 | 5.78% | 2 | 1.04% | 129 | 5.80% | 2 | 0.52% | 89 | 11.56% | 3 | 3.41% | 145 | 4.31% |
| 29.4 | Protein postranslational modification | 5 | 0.52% | 6 | 3.11% | 21 | 0.94% | 13 | 3.37% | 1 | 0.13% | 0 | 0.00% | 43 | 1.28% |
| 29.5 | Protein.degradation | 7 | 0.72% | 9 | 4.66% | 27 | 1.21% | 21 | 5.44% | 2 | 0.26% | 1 | 1.14% | 59 | 1.75% |
| 30 | Signalling | 16 | 1.65% | 2 | 1.04% | 22 | 0.99% | 13 | 3.37% | 4 | 0.52% | 0 | 0.00% | 47 | 1.40% |
| 31 | Cell organization | 8 | 0.83% | 1 | 0.52% | 15 | 0.67% | 3 | 0.78% | 1 | 0.13% | 2 | 2.27% | 26 | 0.77% |
| 33 | Development | 7 | 0.72% | 3 | 1.55% | 15 | 0.67% | 20 | 5.18% | 0 | 0.00% | 1 | 1.14% | 43 | 1.28% |
| 34 | Transport | 14 | 1.44% | 7 | 3.63% | 19 | 0.85% | 14 | 3.63% | 1 | 0.13% | 2 | 2.27% | 52 | 1.55% |
| 35 | Unknown | 546 | 56.35% | 84 | 43.52% | 1426 | 64.06% | 160 | 41.45% | 386 | 50.13% | 31 | 35.23% | 2009 | 59.72% |
| Total | 969 | 100.00% | 193 | 100.00% | 2226 | 100.00% | 386 | 100.00% | 770 | 100.00% | 88 | 100.00% | 3364 | 100.00% | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chandra, D.; Cho, K.; Pham, H.A.; Lee, J.-Y.; Han, O. Down-Regulation of Rice Glutelin by CRISPR-Cas9 Gene Editing Decreases Carbohydrate Content and Grain Weight and Modulates Synthesis of Seed Storage Proteins during Seed Maturation. Int. J. Mol. Sci. 2023, 24, 16941. https://doi.org/10.3390/ijms242316941
Chandra D, Cho K, Pham HA, Lee J-Y, Han O. Down-Regulation of Rice Glutelin by CRISPR-Cas9 Gene Editing Decreases Carbohydrate Content and Grain Weight and Modulates Synthesis of Seed Storage Proteins during Seed Maturation. International Journal of Molecular Sciences. 2023; 24(23):16941. https://doi.org/10.3390/ijms242316941
Chicago/Turabian StyleChandra, Deepanwita, Kyoungwon Cho, Hue Anh Pham, Jong-Yeol Lee, and Oksoo Han. 2023. "Down-Regulation of Rice Glutelin by CRISPR-Cas9 Gene Editing Decreases Carbohydrate Content and Grain Weight and Modulates Synthesis of Seed Storage Proteins during Seed Maturation" International Journal of Molecular Sciences 24, no. 23: 16941. https://doi.org/10.3390/ijms242316941
APA StyleChandra, D., Cho, K., Pham, H. A., Lee, J.-Y., & Han, O. (2023). Down-Regulation of Rice Glutelin by CRISPR-Cas9 Gene Editing Decreases Carbohydrate Content and Grain Weight and Modulates Synthesis of Seed Storage Proteins during Seed Maturation. International Journal of Molecular Sciences, 24(23), 16941. https://doi.org/10.3390/ijms242316941







