Time-Resolved X-ray Observation of Intracellular Crystallized Protein in Living Animal
Abstract
1. Introduction
2. Results
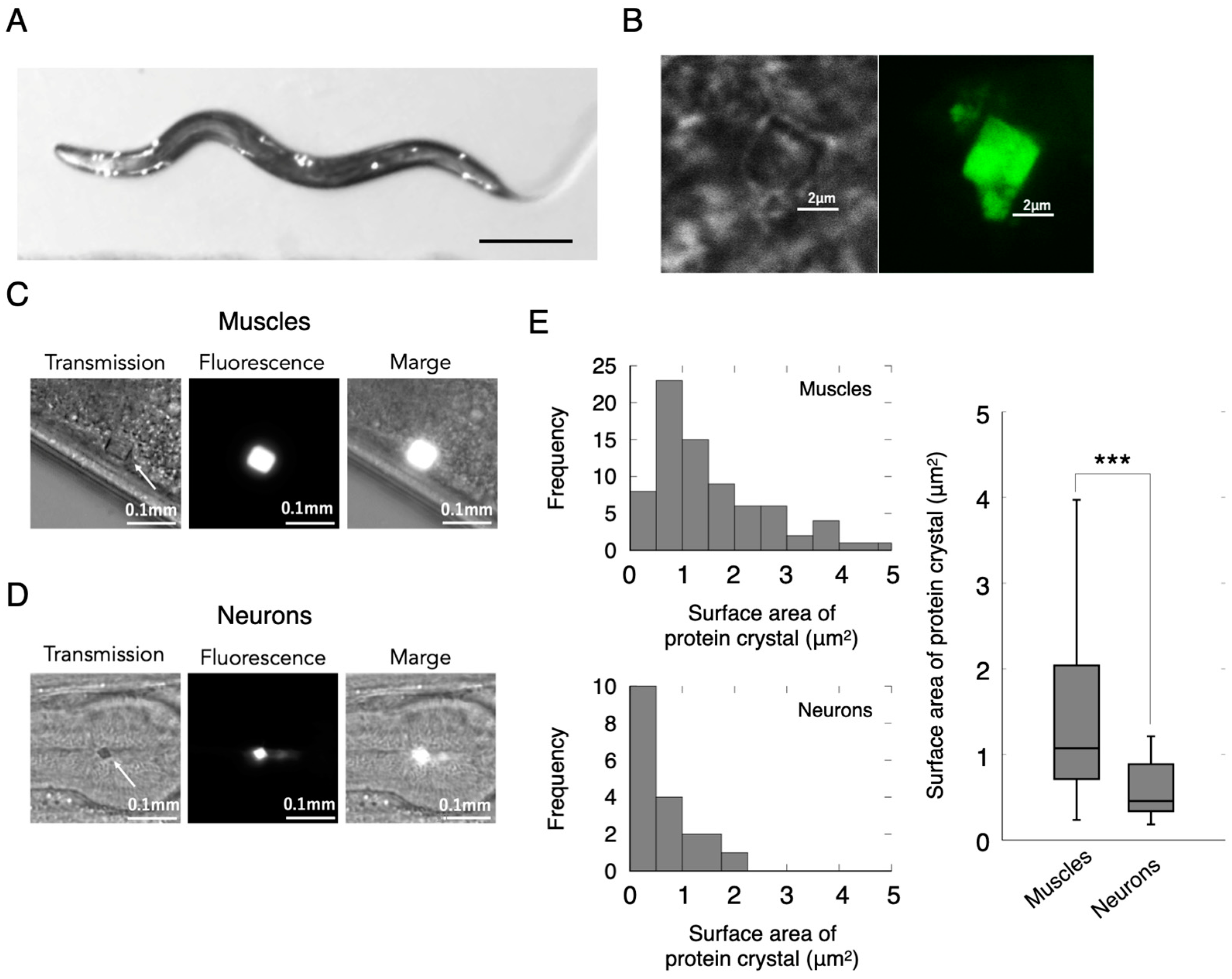
2.1. Observation of Crystallized Protein in the Living Animal Cells
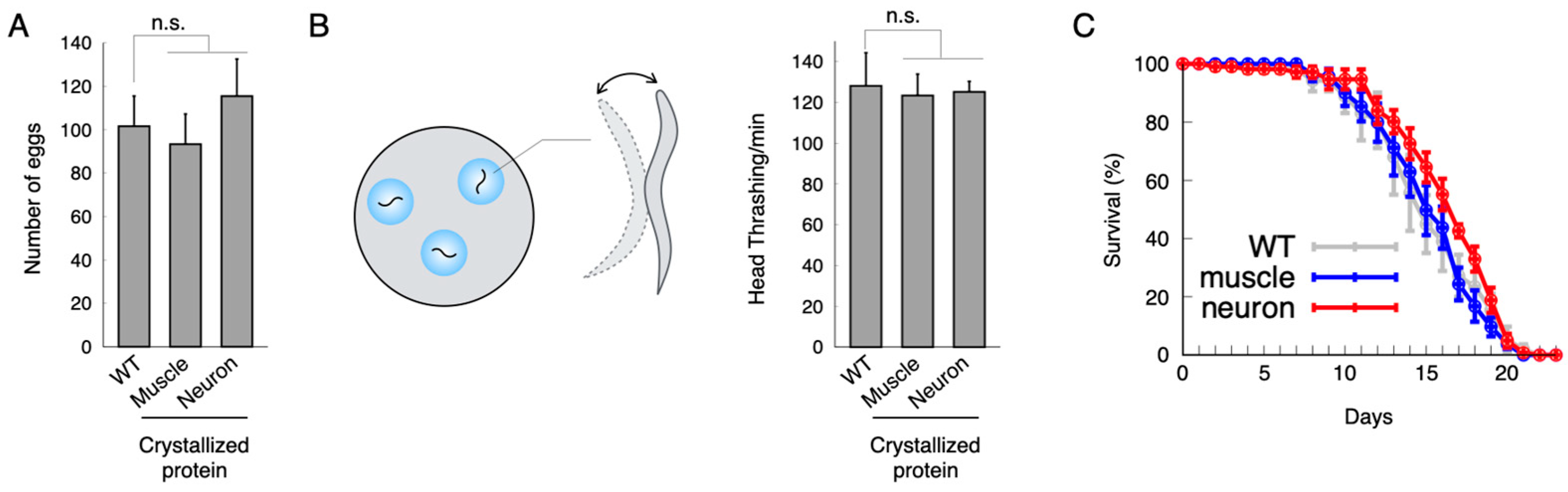
2.2. Toxicity Effect of the Crystallized Protein In Vivo
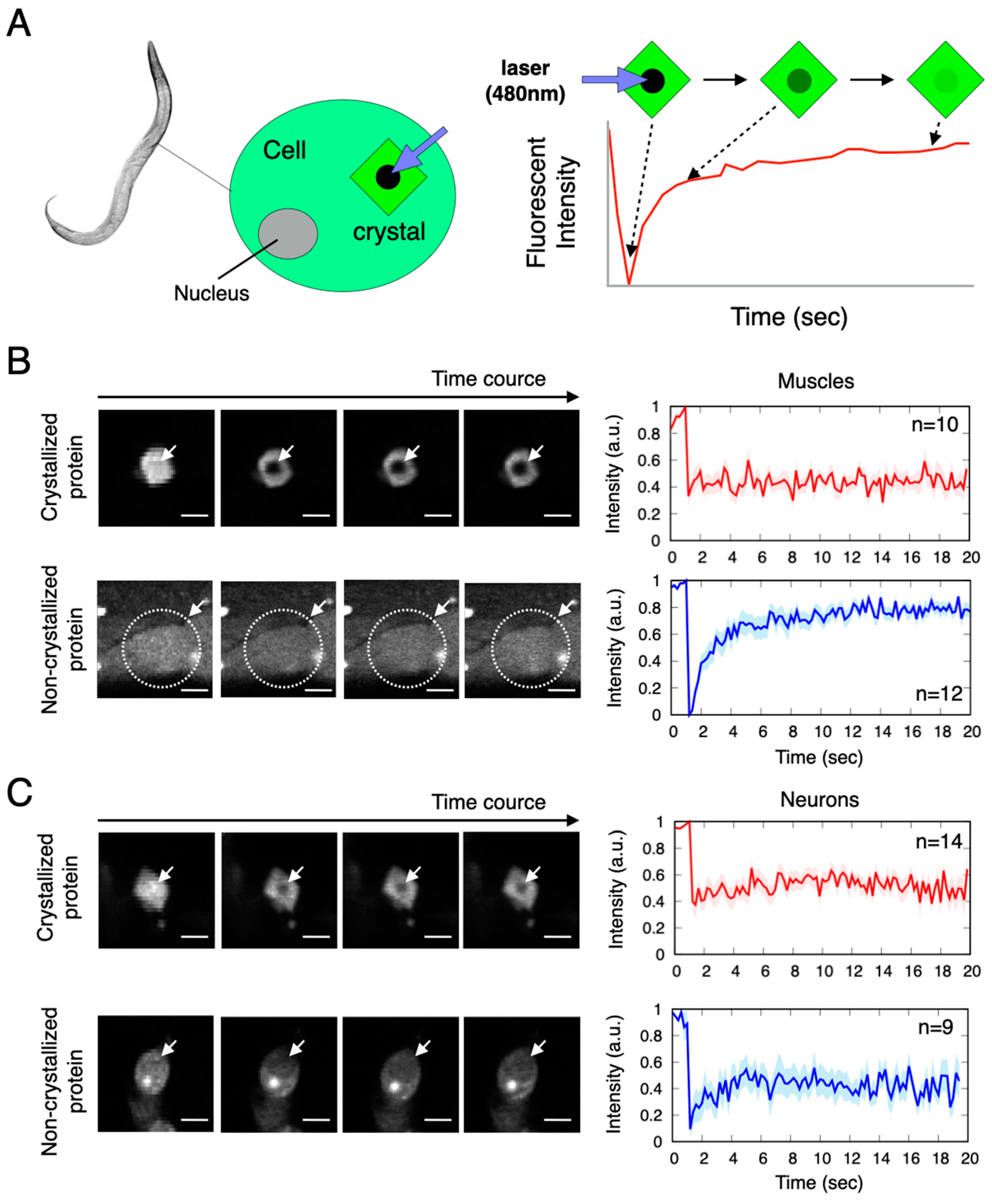
2.3. Translational Diffusion of Molecules in Aggregates Is Considerably Suppressed
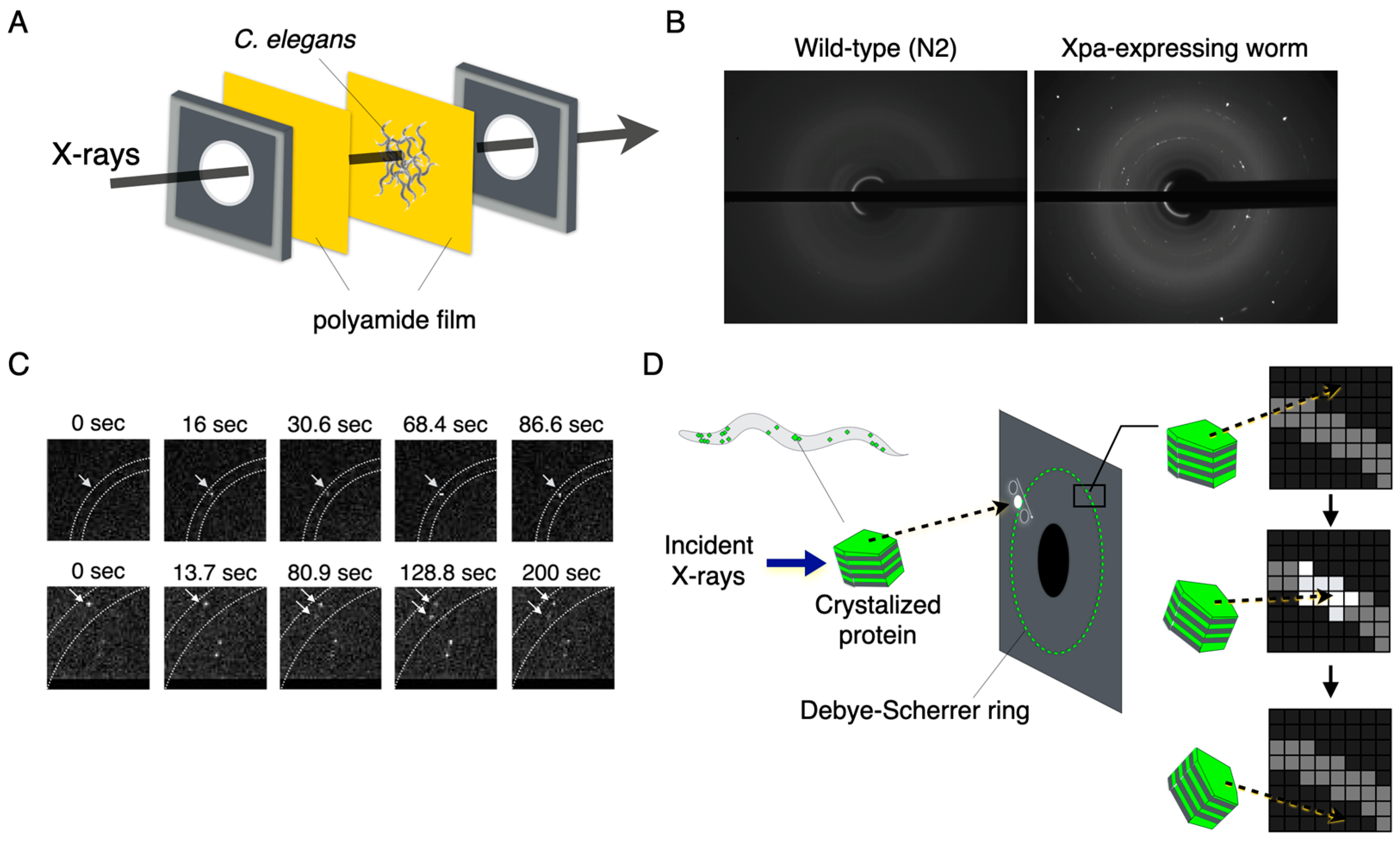
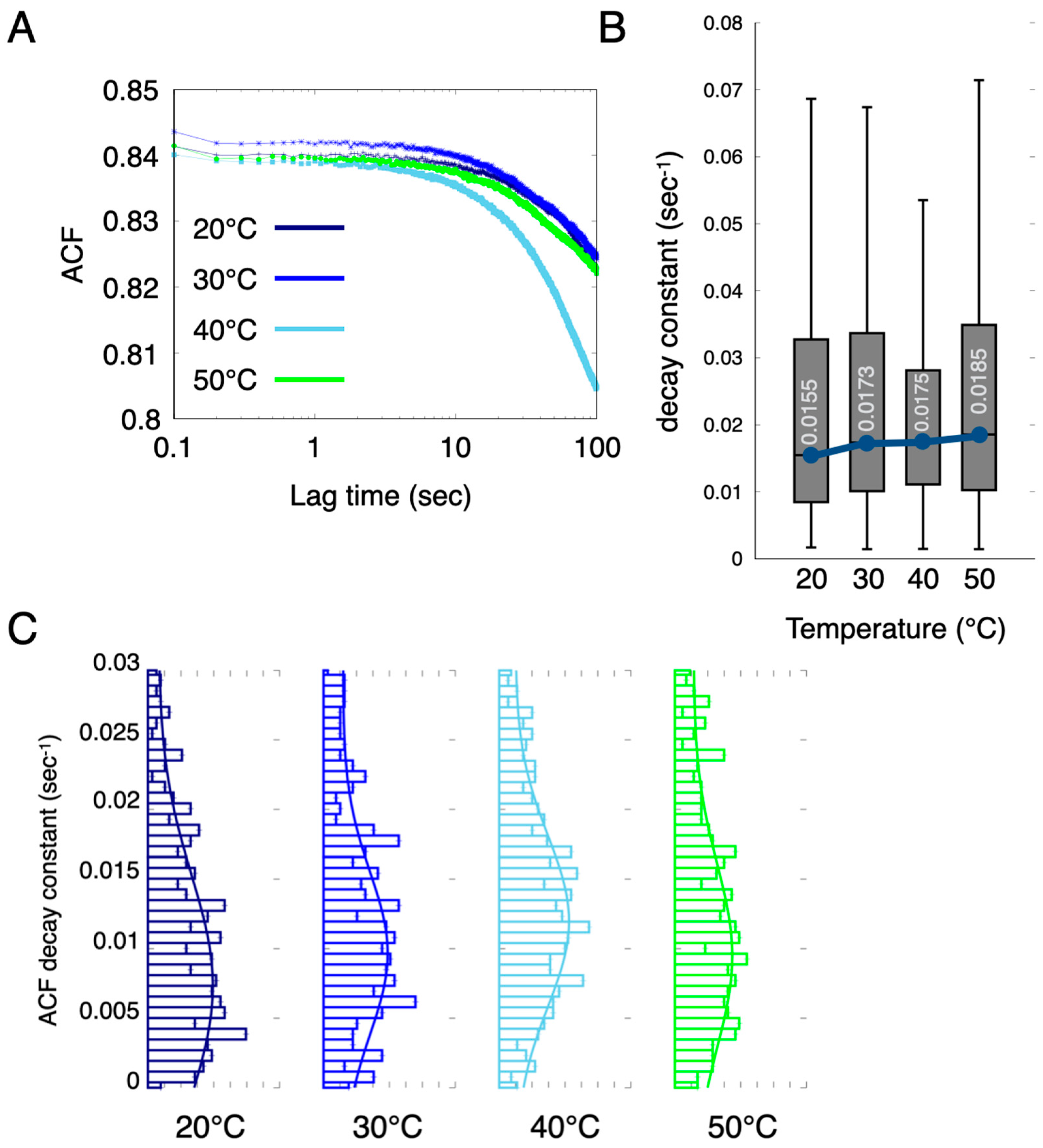
2.4. X-ray Blinking of the Crystallized Protein in Living Animal Cells
2.5. Diffracted X-ray Blinking of the Crystallized Proteins In Vivo
3. Discussion
4. Materials and Methods
4.1. Strains and Cultivation
4.2. Molecular Biology and Transgenic Animals
4.3. Microscopic Observations and FRAP
4.4. Toxicity Assessment
4.5. Sample Preparation for X-ray Diffraction Measurement
4.6. Diffracted X-ray Blinking
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ellis, R.J. Macromolecular Crowding: Obvious but Underappreciated. Trends Biochem. Sci. 2001, 26, 597–604. [Google Scholar] [CrossRef]
- Swaminathan, R.; Hoang, C.P.; Verkman, A.S. Photobleaching Recovery and Anisotropy Decay of Green Fluorescent Protein GFP-S65T in Solution and Cells: Cytoplasmic Viscosity Probed by Green Fluorescent Protein Translational and Rotational Diffusion. Biophys. J. 1997, 72, 1900–1907. [Google Scholar] [CrossRef] [PubMed]
- Rivas, G.; Minton, A.P. Macromolecular Crowding In Vitro, In Vivo, and In Between. Trends Biochem. Sci. 2016, 41, 970–981. [Google Scholar] [CrossRef] [PubMed]
- Qing, J.; Chen, A.; Zhao, N. Quantifying the Protein–Protein Association Rate in Polymer Solutions: Crowding-Induced Diffusion and Energy Modifications. Phys. Chem. Chem. Phys. 2018, 20, 27937–27948. [Google Scholar] [CrossRef] [PubMed]
- Assunção, M.; Wong, C.W.; Richardson, J.J.; Tsang, R.; Beyer, S.; Raghunath, M.; Blocki, A. Macromolecular Dextran Sulfate Facilitates Extracellular Matrix Deposition by Electrostatic Interaction Independent from a Macromolecular Crowding Effect. Mater. Sci. Eng. C 2020, 106, 110280. [Google Scholar] [CrossRef] [PubMed]
- Nasreen, K.; Parray, Z.A.; Ahamad, S.; Ahmad, F.; Ahmed, A.; Freeh Alamery, S.; Hussain, T.; Hassan, M.I.; Islam, A. Interactions Under Crowding Milieu: Chemical-Induced Denaturation of Myoglobin Is Determined by the Extent of Heme Dissociation on Interaction with Crowders. Biomolecules 2020, 10, 490. [Google Scholar] [CrossRef] [PubMed]
- Ellis, R.J. Macromolecular Crowding: An Important but Neglected Aspect of the Intracellular Environment. Curr. Opin. Struct. Biol. 2001, 11, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wang, Y.; Pielak, G.J. Translational and Rotational Diffusion of a Small Globular Protein under Crowded Conditions. J. Phys. Chem. B 2009, 113, 13390–13392. [Google Scholar] [CrossRef]
- Wang, Y.; Li, C.; Pielak, G.J. Effects of Proteins on Protein Diffusion. J. Am. Chem. Soc. 2010, 132, 9392–9397. [Google Scholar] [CrossRef]
- Lee, H.B.; Cong, A.; Leopold, H.; Currie, M.; Boersma, A.J.; Sheets, E.D.; Heikal, A.A. Rotational and Translational Diffusion of Size-Dependent Fluorescent Probes in Homogeneous and Heterogeneous Environments. Phys. Chem. Chem. Phys. 2018, 20, 24045–24057. [Google Scholar] [CrossRef]
- Yamamoto, J.; Oura, M.; Yamashita, T.; Miki, S.; Jin, T.; Haraguchi, T.; Hiraoka, Y.; Terai, H.; Kinjo, M. Rotational Diffusion Measurements Using Polarization-Dependent Fluorescence Correlation Spectroscopy Based on Superconducting Nanowire Single-Photon Detector. Opt. Express 2015, 23, 32633. [Google Scholar] [CrossRef]
- Yamamoto, J.; Matsui, A.; Gan, F.; Oura, M.; Ando, R.; Matsuda, T.; Gong, J.P.; Kinjo, M. Quantitative Evaluation of Macromolecular Crowding Environment Based on Translational and Rotational Diffusion Using Polarization Dependent Fluorescence Correlation Spectroscopy. Sci. Rep. 2021, 11, 10594. [Google Scholar] [CrossRef]
- Shimizu, H.; Iwamoto, M.; Konno, T.; Nihei, A.; Sasaki, Y.C.; Oiki, S. Global twisting motion of single molecular KcsA potassium channel upon gating. Cell 2008, 132, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Sekiguchi, H.; Suzuki, Y.; Nishino, Y.; Kobayashi, S.; Shimoyama, Y.; Cai, W.; Nagata, K.; Okada, M.; Ichiyanagi, K.; Ohta, N.; et al. Real Time Ligand-Induced Motion Mappings of AChBP and nAChR Using X-ray Single Molecule Tracking. Sci. Rep. 2014, 4, 6384. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, S.; Mio, K.; Kuramochi, M.; Sekiguchi, H.; Ikezaki, K.; Mio, M.; Hengphasatporn, K.; Shigeta, Y.; Kubo, T.; Sasaki, Y.C. Agonist and Antagonist-Diverted Twisting Motions of a Single TRPV1 Channel. J. Phys. Chem. B 2020, 124, 11617–11624. [Google Scholar] [CrossRef]
- Sekiguchi, H.; Kuramochi, M.; Ikezaki, K.; Okamura, Y.; Yoshimura, K.; Matsubara, K.; Chang, J.-W.; Ohta, N.; Kubo, T.; Mio, K.; et al. Diffracted X-ray Blinking Tracks Single Protein Motions. Sci. Rep. 2018, 8, 17090. [Google Scholar] [CrossRef]
- Schönherr, R.; Rudolph, J.M.; Redecke, L. Protein Crystallization in Living Cells. Biol. Chem. 2018, 399, 751–772. [Google Scholar] [CrossRef] [PubMed]
- Katsube-Tanaka, T. Analysis and Improvement of Seed Storage Proteins. Jpn. J. Crop Sci. 2010, 79, 76–80. [Google Scholar] [CrossRef][Green Version]
- Mori, H.; Nakazawa, H.; Ikeda, K. Entomovirus. Cytoplasmic polyhedrosis virus and polyhedrin. Uirusu 1998, 48, 81–88. [Google Scholar] [CrossRef]
- Tsutsui, H.; Jinno, Y.; Shoda, K.; Tomita, A.; Matsuda, M.; Yamashita, E.; Katayama, H.; Nakagawa, A.; Miyawaki, A. A Diffraction-Quality Protein Crystal Processed as an Autophagic Cargo. Mol. Cell 2015, 58, 186–193. [Google Scholar] [CrossRef]
- Axelrod, D.; Koppel, D.E.; Schlessinger, J.; Elson, E.; Webb, W.W. Mobility Measurement by Analysis of Fluorescence Photobleaching Recovery Kinetics. Biophys. J. 1976, 16, 1055–1069. [Google Scholar] [CrossRef]
- Rayan, G.; Guet, J.E.; Taulier, N.; Pincet, F.; Urbach, W. Recent Applications of Fluorescence Recovery after Photobleaching (FRAP) to Membrane Bio-Macromolecules. Sensors 2010, 10, 5927–5948. [Google Scholar] [CrossRef] [PubMed]
- Kuramochi, M.; Omata, H.; Ishihara, M.; Hanslin, S.Ø.; Mizumaki, M.; Kawamura, N.; Osawa, H.; Suzuki, M.; Mio, K.; Sekiguchi, H.; et al. Tilting and Rotational Motions of Silver Halide Crystal with Diffracted X-ray Blinking. Sci. Rep. 2021, 11, 4097. [Google Scholar] [CrossRef] [PubMed]
- Coe, L.; Evan, A.; Worcester, E. Kidney stone disease. J. Clin. Investig. 2005, 115, 2598–2608. [Google Scholar] [CrossRef] [PubMed]
- Carey, M.C. Pathogenesis of gallstones. Am. J. Surg. 1993, 165, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Martillo, M.A.; Nazzal, L.; Crittenden, D.B. The crystallization of monosodium urate. Curr. Rheumatol. Rep. 2014, 16, 400. [Google Scholar] [CrossRef]
- Kerr, R. Imaging the Activity of Neurons and Muscles. WormBook 2006, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chronis, N. Worm Chips: Microtools for C. elegans Biology. Lab. Chip 2010, 10, 432–437. [Google Scholar] [CrossRef]
- Tracy, M.A.; Pecora, R. Diffusion in Rod/Sphere Composite Liquids. In Application of Scattering Methods to the Dynamics of Polymer Systems; Ewen, B., Fischer, E.W., Fytas, G., Eds.; Progress in Colloid & Polymer Science; Steinkopff: Darmstadt, Germany, 1993; Volume 91, pp. 165–170. ISBN 978-3-7985-0952-8. [Google Scholar]
- Alam, S.; Mukhopadhyay, A. Translational Anisotropy and Rotational Diffusion of Gold Nanorods in Colloidal Sphere Solutions. Langmuir 2015, 31, 8780–8785. [Google Scholar] [CrossRef]
- Mello, C.C.; Kramer, J.M.; Stinchcomb, D.; Ambros, V. Efficient Gene Transfer in C. elegans: Extrachromosomal Maintenance and Integration of Transforming Sequences. EMBO J. 1991, 10, 3959–3970. [Google Scholar] [CrossRef]
- Mazenko, G.; Banavar, J.R.; Gomer, R. Diffusion Coefficients and the Time Auto-Correlation Function of Density Fluctuations. Surf. Sci. 1981, 107, 459–468. [Google Scholar] [CrossRef]
- Hunter, G.L.; Edmond, K.V.; Elsesser, M.T.; Weeks, E.R. Tracking Rotational Diffusion of Colloidal Clusters. Opt. Express 2011, 19, 17189. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuramochi, M.; Sugawara, I.; Shinkai, Y.; Mio, K.; Sasaki, Y.C. Time-Resolved X-ray Observation of Intracellular Crystallized Protein in Living Animal. Int. J. Mol. Sci. 2023, 24, 16914. https://doi.org/10.3390/ijms242316914
Kuramochi M, Sugawara I, Shinkai Y, Mio K, Sasaki YC. Time-Resolved X-ray Observation of Intracellular Crystallized Protein in Living Animal. International Journal of Molecular Sciences. 2023; 24(23):16914. https://doi.org/10.3390/ijms242316914
Chicago/Turabian StyleKuramochi, Masahiro, Ibuki Sugawara, Yoichi Shinkai, Kazuhiro Mio, and Yuji C. Sasaki. 2023. "Time-Resolved X-ray Observation of Intracellular Crystallized Protein in Living Animal" International Journal of Molecular Sciences 24, no. 23: 16914. https://doi.org/10.3390/ijms242316914
APA StyleKuramochi, M., Sugawara, I., Shinkai, Y., Mio, K., & Sasaki, Y. C. (2023). Time-Resolved X-ray Observation of Intracellular Crystallized Protein in Living Animal. International Journal of Molecular Sciences, 24(23), 16914. https://doi.org/10.3390/ijms242316914







