The Roles of the PSEUDO-RESPONSE REGULATORs in Circadian Clock and Flowering Time in Medicago truncatula
Abstract
:1. Introduction
2. Results
2.1. Identification of PRRs and Their Loss-of-Function Mutants in M. truncatula
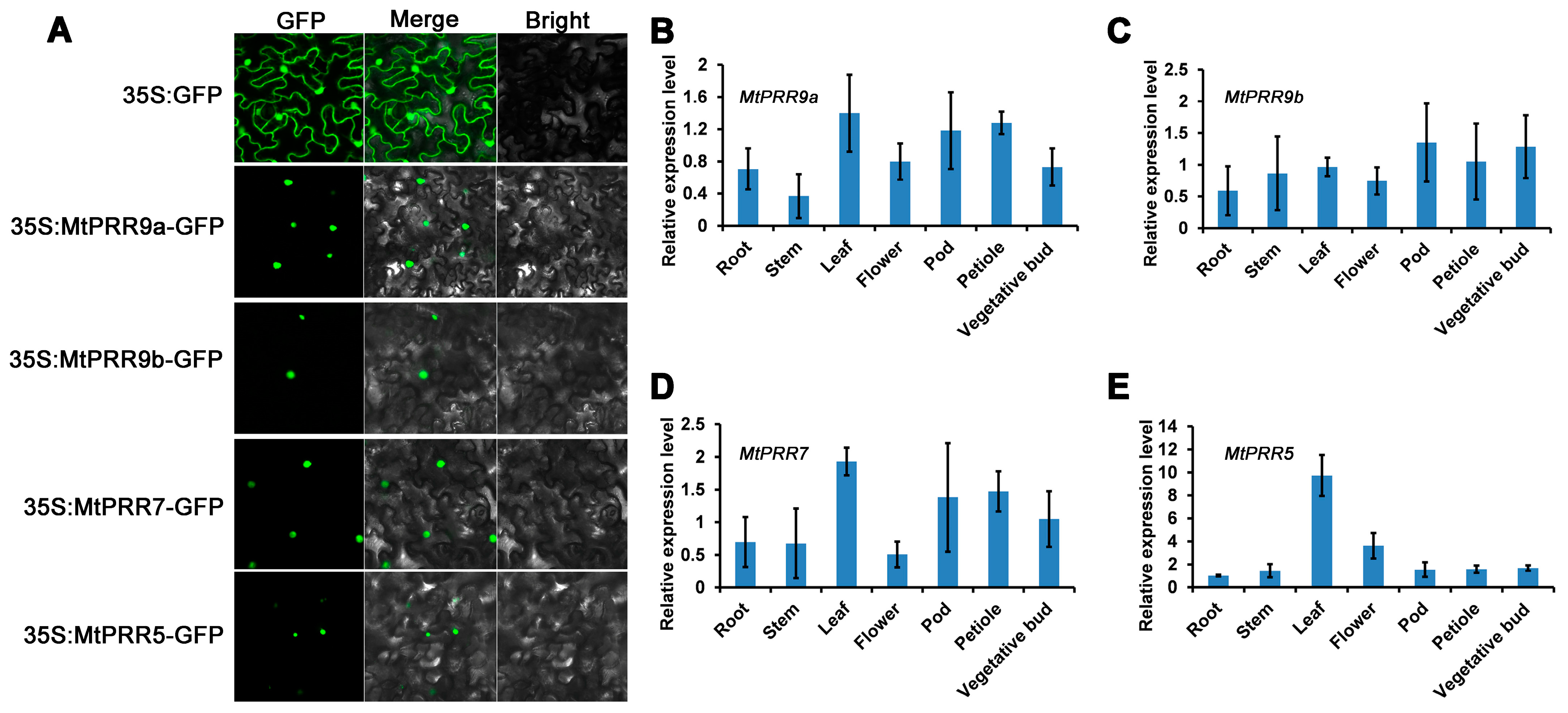
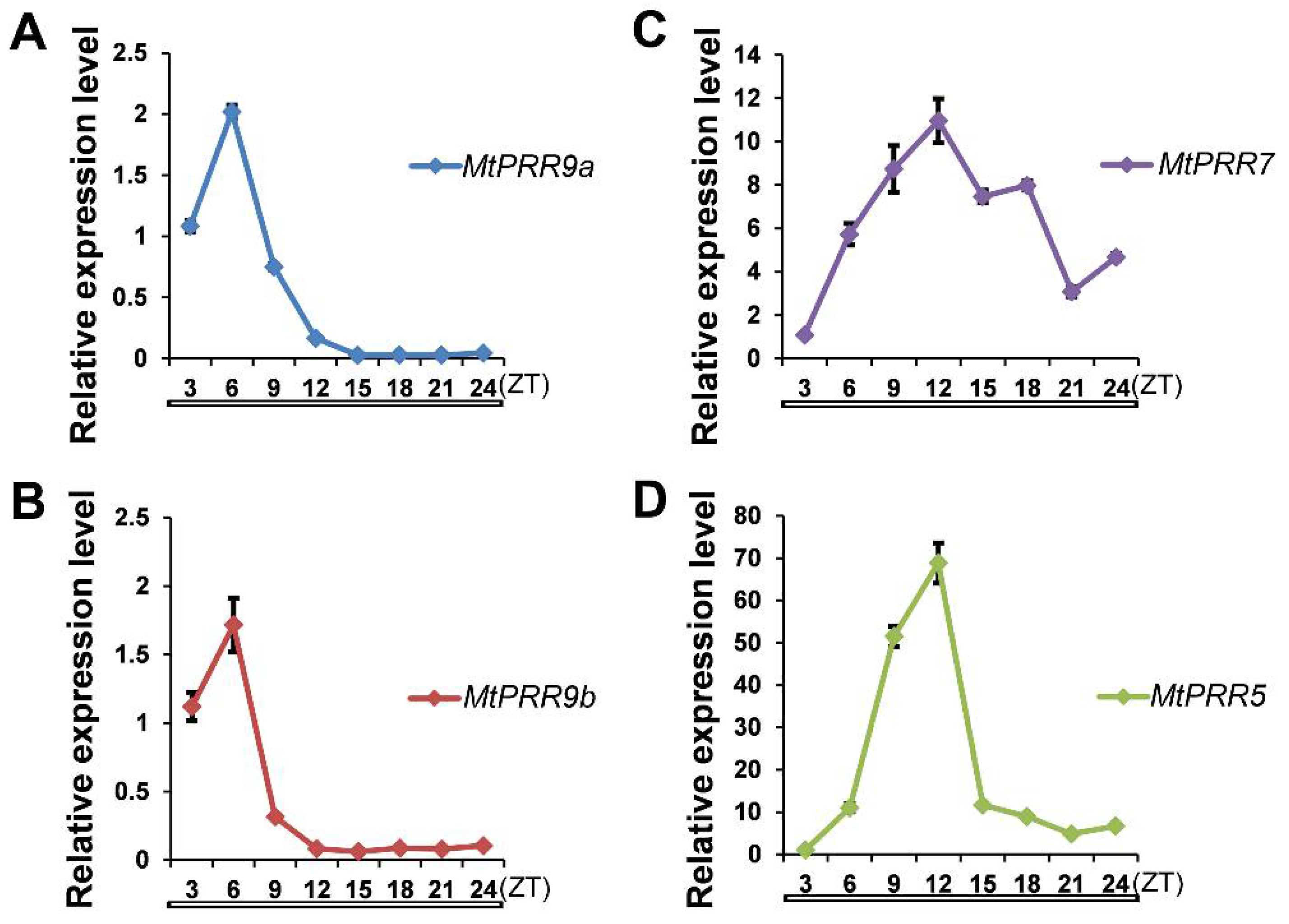
2.2. Subcellular Localizations of MtPRR9, 7, and 5 and Expression Patterns
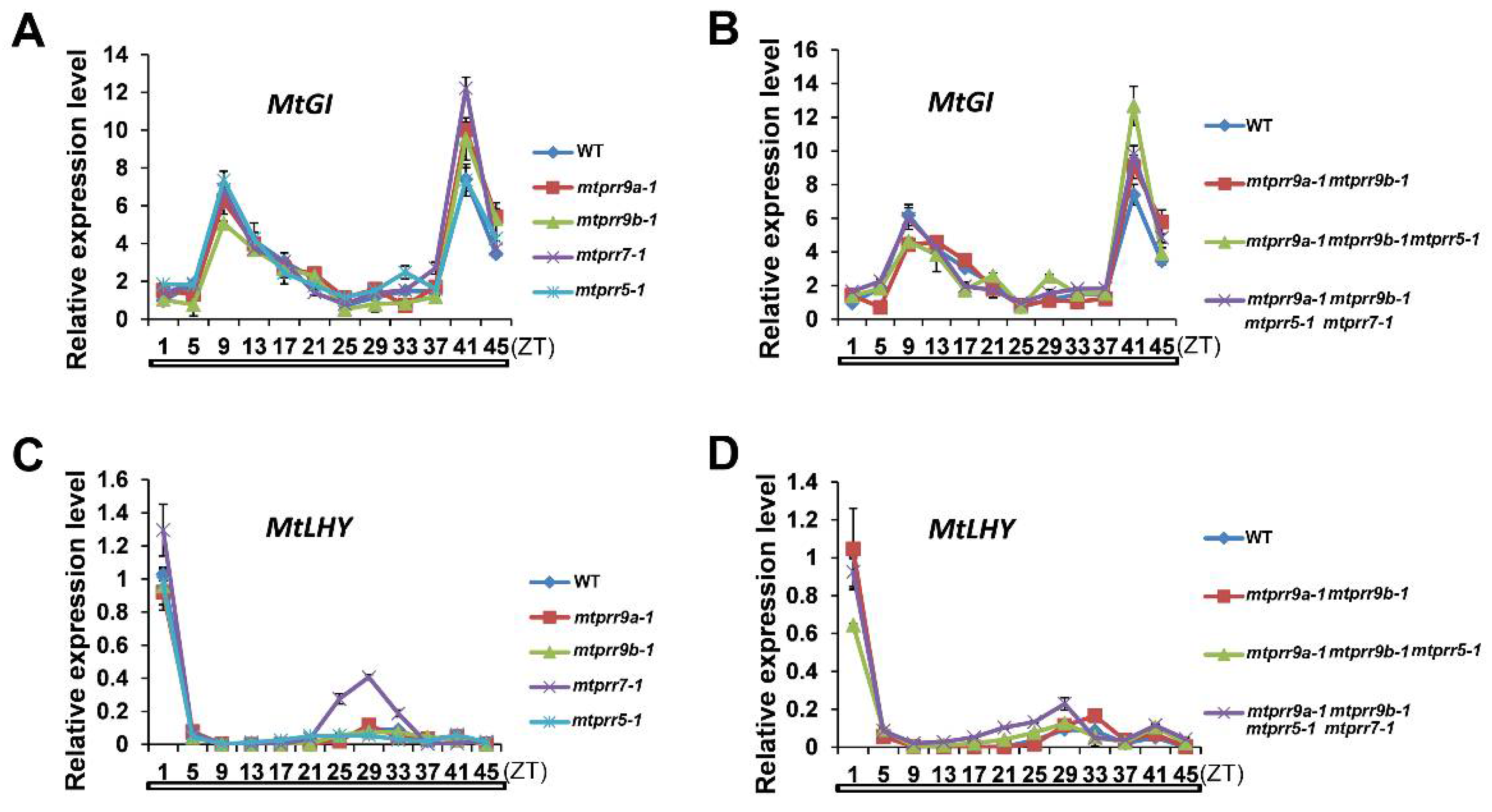
2.3. The Expression Patterns of Genes Associated with the Circadian Clock Are Altered in mtprr Mutants
2.4. MtPRR9, 7, and 5 Regulate Flowering Time
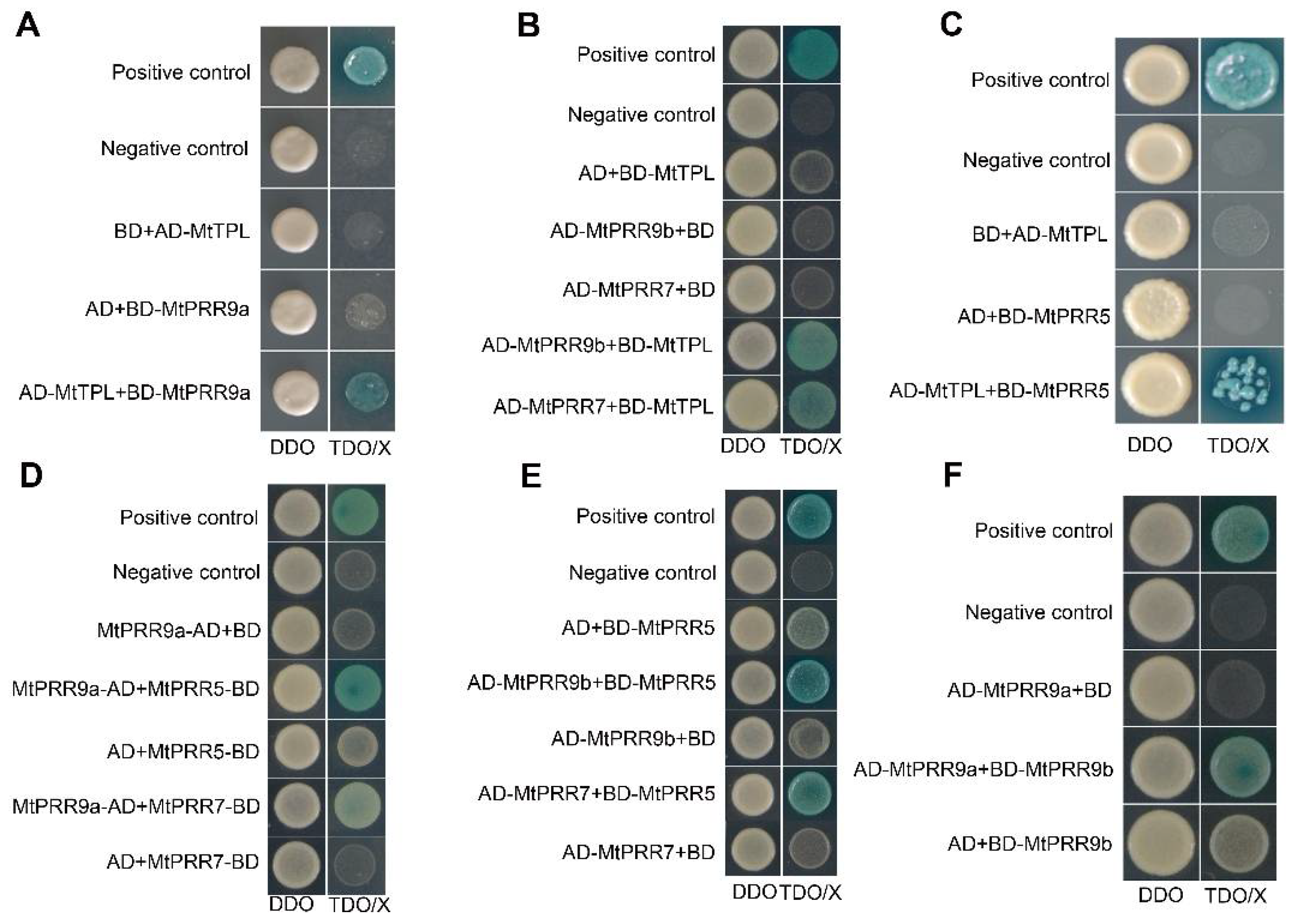
2.5. MtPRR9, 7, and 5 Physically Interact with MtTPL/MtTPR Proteins
2.6. MtPRR9, 7, and 5 Can Form Heterodimers
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Identification and Phylogenetic Analysis of MtPRRs
4.3. Conserved Domains Analysis
4.4. Subcellular Localization Analysis
4.5. RNA Extraction, RT-PCR, qRT-PCR, and Statistical Analysis
4.6. Double/Triple/Quadruple Mutant Generation
4.7. Y2H and BiFC Assays
4.8. Accession Numbers
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Harmer, S.L. The circadian system in higher plants. Annu. Rev. Plant Biol. 2009, 60, 357–377. [Google Scholar] [CrossRef] [PubMed]
- Gil, K.E.; Park, C.M. Thermal adaptation and plasticity of the plant circadian clock. New Phytol. 2019, 221, 1215–1229. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.W.; Rubio, V.; Lee, N.Y.; Bai, S.; Lee, S.Y.; Kim, S.S.; Liu, L.; Zhang, Y.; Irigoyen, M.L.; Sullivan, J.A.; et al. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell 2008, 32, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Goodspeed, D.; Chehab, E.W.; Min-Venditti, A.; Braam, J.; Covington, M.F. Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc. Natl. Acad. Sci. USA 2012, 109, 4674–4677. [Google Scholar] [CrossRef]
- Legnaioli, T.; Cuevas, J.; Mas, P. TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J. 2009, 28, 3745–3757. [Google Scholar] [CrossRef]
- Wang, W.; Barnaby, J.Y.; Tada, Y.; Li, H.; Tör, M.; Caldelari, D.; Lee, D.U.; Fu, X.D.; Dong, X. Timing of plant immune responses by a central circadian regulator. Nature 2011, 470, 110–114. [Google Scholar] [CrossRef]
- McClung, C.R. Wheels within wheels: New transcriptional feedback loops in the Arabidopsis circadian clock. F1000prime Rep. 2014, 6, 2. [Google Scholar] [CrossRef]
- Fogelmark, K.; Troein, C. Rethinking transcriptional activation in the Arabidopsis circadian clock. PLoS Comput. Biol. 2014, 10, e1003705. [Google Scholar] [CrossRef]
- Hsu, P.Y.; Harmer, S.L. Wheels within wheels: The plant circadian system. Trends Plant Sci. 2014, 19, 240–249. [Google Scholar] [CrossRef]
- Bendix, C.; Marshall, C.M.; Harmon, F.G. Circadian Clock Genes Universally Control Key Agricultural Traits. Mol. Plant 2015, 8, 1135–1152. [Google Scholar] [CrossRef]
- Kikis, E.A.; Khanna, R.; Quail, P.H. ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J. Cell Mol. Biol. 2005, 44, 300–313. [Google Scholar] [CrossRef] [PubMed]
- Hazen, S.P.; Schultz, T.F.; Pruneda-Paz, J.L.; Borevitz, J.O.; Ecker, J.R.; Kay, S.A. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. USA 2005, 102, 10387–10392. [Google Scholar] [CrossRef] [PubMed]
- Alabadí, D.; Oyama, T.; Yanovsky, M.J.; Harmon, F.G.; Más, P.; Kay, S.A. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 2001, 293, 880–883. [Google Scholar] [CrossRef] [PubMed]
- Farré, E.M.; Harmer, S.L.; Harmon, F.G.; Yanovsky, M.J.; Kay, S.A. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 2005, 15, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Nusinow, D.A. Into the Evening: Complex Interactions in the Arabidopsis Circadian Clock. Trends Genet. 2016, 32, 674–686. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kiba, T.; Kamioka, M.; Suzuki, T.; Yamashino, T.; Higashiyama, T.; Sakakibara, H.; Mizuno, T. Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl. Acad. Sci. USA 2012, 109, 17123–17128. [Google Scholar] [CrossRef]
- Matsushika, A.; Makino, S.; Kojima, M.; Mizuno, T. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: Insight into the plant circadian clock. Plant Cell Physiol. 2000, 41, 1002–1012. [Google Scholar] [CrossRef]
- Murakami, M.; Ashikari, M.; Miura, K.; Yamashino, T.; Mizuno, T. The evolutionarily conserved OsPRR quintet: Rice pseudo-response regulators implicated in circadian rhythm. Plant Cell Physiol. 2003, 44, 1229–1236. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Sato, E.; Shimizu, T.; Nakamich, N.; Sato, S.; Kato, T.; Tabata, S.; Nagatani, A.; Yamashino, T.; Mizuno, T. Comparative genetic studies on the APRR5 and APRR7 genes belonging to the APRR1/TOC1 quintet implicated in circadian rhythm, control of flowering time, and early photomorphogenesis. Plant Cell Physiol. 2003, 44, 1119–1130. [Google Scholar] [CrossRef]
- Millar, A.J.; Carré, I.A.; Strayer, C.A.; Chua, N.H.; Kay, S.A. Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 1995, 267, 1161–1163. [Google Scholar] [CrossRef]
- Michael, T.P.; Salomé, P.A.; Yu, H.J.; Spencer, T.R.; Sharp, E.L.; McPeek, M.A.; Alonso, J.M.; Ecker, J.R.; McClung, C.R. Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 2003, 302, 1049–1053. [Google Scholar] [CrossRef] [PubMed]
- Nakamichi, N.; Kita, M.; Ito, S.; Yamashino, T.; Mizuno, T. PSEUDO-RESPONSE REGULATORS, PRR9, PRR7 and PRR5, together play essential roles close to the circadian clock of Arabidopsis thaliana. Plant Cell Physiol. 2005, 46, 686–698. [Google Scholar] [CrossRef] [PubMed]
- Kiba, T.; Henriques, R.; Sakakibara, H.; Chua, N.H. Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 2007, 19, 2516–2530. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, S.; Wang, L.; Han, L.; Suh, S.S.; Salomé, P.A.; McClung, C.R.; Somers, D.E. Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J. Biol. Chem. 2008, 283, 23073–23083. [Google Scholar] [CrossRef]
- Gendron, J.M.; Pruneda-Paz, J.L.; Doherty, C.J.; Gross, A.M.; Kang, S.E.; Kay, S.A. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 2012, 109, 3167–3172. [Google Scholar] [CrossRef]
- Jung, C.; Müller, A.E. Flowering time control and applications in plant breeding. Trends Plant Sci. 2009, 14, 563–573. [Google Scholar] [CrossRef]
- Pierre, J.B.; Huguet, T.; Barre, P.; Huyghe, C.; Julier, B. Detection of QTLs for flowering date in three mapping populations of the model legume species Medicago truncatula. Theor. Appl. Genet. 2008, 117, 609–620. [Google Scholar] [CrossRef]
- Burgarella, C.; Chantret, N.; Gay, L.; Prosperi, J.M.; Bonhomme, M.; Tiffin, P.; Young, N.D.; Ronfort, J. Adaptation to climate through flowering phenology: A case study in Medicago truncatula. Mol. Ecol. 2016, 25, 3397–3415. [Google Scholar] [CrossRef]
- Laurie, R.E.; Diwadkar, P.; Jaudal, M.; Zhang, L.; Hecht, V.; Wen, J.; Tadege, M.; Mysore, K.S.; Putterill, J.; Weller, J.L.; et al. The Medicago FLOWERING LOCUS T homolog, MtFTa1, is a key regulator of flowering time. Plant Physiol. 2011, 156, 2207–2224. [Google Scholar] [CrossRef]
- Zhang, X.; Zhai, H.; Wang, Y.; Tian, X.; Zhang, Y.; Wu, H.; Lü, S.; Yang, G.; Li, Y.; Wang, L.; et al. Functional conservation and diversification of the soybean maturity gene E1 and its homologs in legumes. Sci. Rep. 2016, 6, 29548. [Google Scholar] [CrossRef]
- Jaudal, M.; Zhang, L.; Che, C.; Li, G.; Tang, Y.; Wen, J.; Mysore, K.S.; Putterill, J. A SOC1-like gene MtSOC1a promotes flowering and primary stem elongation in Medicago. J. Exp. Bot. 2018, 69, 4867–4880. [Google Scholar] [CrossRef] [PubMed]
- Jaudal, M.; Zhang, L.; Che, C.; Putterill, J. Three Medicago MtFUL genes have distinct and overlapping expression patterns during vegetative and reproductive development and 35S:MtFULb accelerates flowering and causes a terminal flower phenotype in Arabidopsis. Front. Genet. 2015, 6, 50. [Google Scholar] [CrossRef] [PubMed]
- Jaudal, M.; Wen, J.; Mysore, K.S.; Putterill, J. Medicago PHYA promotes flowering, primary stem elongation and expression of flowering time genes in long days. BMC Plant Biol. 2020, 20, 329. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kita, M.; Niinuma, K.; Ito, S.; Yamashino, T.; Mizoguchi, T.; Mizuno, T. Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol. 2007, 48, 822–832. [Google Scholar] [CrossRef] [PubMed]
- Yanovsky, M.J.; Kay, S.A. Molecular basis of seasonal time measurement in Arabidopsis. Nature 2002, 419, 308–312. [Google Scholar] [CrossRef]
- Ito, S.; Niwa, Y.; Nakamichi, N.; Kawamura, H.; Yamashino, T.; Mizuno, T. Insight into missing genetic links between two evening-expressed pseudo-response regulator genes TOC1 and PRR5 in the circadian clock-controlled circuitry in Arabidopsis thaliana. Plant Cell Physiol. 2008, 49, 201–213. [Google Scholar] [CrossRef]
- Matsushika, A.; Murakami, M.; Ito, S.; Nakamichi, N.; Yamashino, T.; Mizuno, T. Characterization of Circadian-associated pseudo-response regulators: I. Comparative studies on a series of transgenic lines misexpressing five distinctive PRR Genes in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2007, 71, 527–534. [Google Scholar] [CrossRef]
- Turner, A.; Beales, J.; Faure, S.; Dunford, R.P.; Laurie, D.A. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 2005, 310, 1031–1034. [Google Scholar] [CrossRef]
- Murphy, R.L.; Klein, R.R.; Morishige, D.T.; Brady, J.A.; Rooney, W.L.; Miller, F.R.; Dugas, D.V.; Klein, P.E.; Mullet, J.E. Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc. Natl. Acad. Sci. USA 2011, 108, 16469–16474. [Google Scholar] [CrossRef]
- Murakami, M.; Matsushika, A.; Ashikari, M.; Yamashino, T.; Mizuno, T. Circadian-associated rice pseudo response regulators (OsPRRs): Insight into the control of flowering time. Biosci. Biotechnol. Biochem. 2005, 69, 410–414. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kudo, T.; Makita, N.; Kiba, T.; Kinoshita, T.; Sakakibara, H. Flowering time control in rice by introducing Arabidopsis clock-associated PSEUDO-RESPONSE REGULATOR 5. Biosci. Biotechnol. Biochem. 2020, 84, 970–979. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Dong, L.; Fang, C.; Liu, S.; Kong, L.; Cheng, Q.; Chen, L.; Su, T.; Nan, H.; Zhang, D.; et al. Stepwise selection on homeologous PRR genes controlling flowering and maturity during soybean domestication. Nat. Genet. 2020, 52, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, S.; Wu, T.; Liu, L.; Sun, X.; Cai, Y.; Li, J.; Jia, H.; Yuan, S.; Chen, L.; et al. Natural variation and CRISPR/Cas9-mediated mutation in GmPRR37 affect photoperiodic flowering and contribute to regional adaptation of soybean. Plant Biotechnol. J. 2020, 18, 1869–1881. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, Y.H.; Li, Y.; Lu, H.; Hong, H.; Tian, Y.; Li, H.; Zhao, T.; Zhou, X.; Liu, J.; et al. A Domestication-Associated Gene GmPRR3b Regulates the Circadian Clock and Flowering Time in Soybean. Mol. Plant 2020, 13, 745–759. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Liu, H.; Kong, Y.; Wen, L.; Zhao, Y.; Zhou, C.; Han, L. Late Elongated Hypocotyl Positively Regulates Salt Stress Tolerance in Medicago truncatula. Int. J. Mol. Sci. 2023, 24, 9948. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Han, L.; Liu, X.; Wang, H.; Wen, L.; Yu, X.; Xu, X.; Kong, F.; Fu, C.; Mysore, K.S.; et al. The nodulation and nyctinastic leaf movement is orchestrated by clock gene LHY in Medicago truncatula. J. Integr. Plant Biol. 2020, 62, 1880–1895. [Google Scholar] [CrossRef]
- Kong, Y.; Zhang, Y.; Liu, X.; Meng, Z.; Yu, X.; Zhou, C.; Han, L. The Conserved and Specific Roles of the LUX ARRHYTHMO in Circadian Clock and Nodulation. Int. J. Mol. Sci. 2022, 23, 3473. [Google Scholar] [CrossRef]
- Cheng, X.; Wang, M.; Lee, H.K.; Tadege, M.; Ratet, P.; Udvardi, M.; Mysore, K.S.; Wen, J. An efficient reverse genetics platform in the model legume Medicago truncatula. New Phytol. 2014, 201, 1065–1076. [Google Scholar] [CrossRef]
- Wang, L.; Kim, J.; Somers, D.E. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. USA 2013, 110, 761–766. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kiba, T.; Henriques, R.; Mizuno, T.; Chua, N.H.; Sakakibara, H. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 2010, 22, 594–605. [Google Scholar] [CrossRef]
- Wang, H.; Xu, Y.; Hong, L.; Zhang, X.; Wang, X.; Zhang, J.; Ding, Z.; Meng, Z.; Wang, Z.Y.; Long, R.; et al. HEADLESS Regulates Auxin Response and Compound Leaf Morphogenesis in Medicago truncatula. Front. Plant Sci. 2019, 10, 1024. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Fujiwara, S.; Somers, D.E. PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J. 2010, 29, 1903–1915. [Google Scholar] [CrossRef]
- Para, A.; Farré, E.M.; Imaizumi, T.; Pruneda-Paz, J.L.; Harmon, F.G.; Kay, S.A. PRR3 Is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell 2007, 19, 3462–3473. [Google Scholar] [CrossRef] [PubMed]
- Jalal, A.; Sun, J.; Chen, Y.; Fan, C.; Liu, J.; Wang, C. Evolutionary Analysis and Functional Identification of Clock-Associated PSEUDO-RESPONSE REGULATOR (PRRs) Genes in the Flowering Regulation of Roses. Int. J. Mol. Sci. 2022, 23, 7335. [Google Scholar] [CrossRef]
- Kim, J.A.; Kim, J.S.; Hong, J.K.; Lee, Y.H.; Choi, B.S.; Seol, Y.J.; Jeon, C.H. Comparative mapping, genomic structure, and expression analysis of eight pseudo-response regulator genes in Brassica rapa. Mol. Genet. Genom. 2012, 287, 373–388. [Google Scholar] [CrossRef] [PubMed]
- Hotta, C.T. The evolution and function of the PSEUDO RESPONSE REGULATOR gene family in the plant circadian clock. Genet. Mol. Biol. 2022, 45 (Suppl. S1), e20220137. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Yu, Y.; Liu, M.; Song, Y.; Li, H.; Sun, J.; Wang, Q.; Xie, Q.; Wang, L.; Xu, X. BBX19 fine-tunes the circadian rhythm by interacting with PSEUDO-RESPONSE REGULATOR proteins to facilitate their repressive effect on morning-phased clock genes. Plant Cell 2021, 33, 2602–2617. [Google Scholar] [CrossRef]
- Maeda, A.E.; Nakamichi, N. Plant clock modifications for adapting flowering time to local environments. Plant Physiol. 2022, 190, 952–967. [Google Scholar] [CrossRef]
- Hayama, R.; Sarid-Krebs, L.; Richter, R.; Fernández, V.; Jang, S.; Coupland, G. PSEUDO RESPONSE REGULATORs stabilize CONSTANS protein to promote flowering in response to day length. EMBO J. 2017, 36, 904–918. [Google Scholar] [CrossRef]
- Kwon, C.T.; Koo, B.H.; Kim, D.; Yoo, S.C.; Paek, N.C. Casein kinases I and 2α phosphorylate oryza sativa pseudo-response regulator 37 (OsPRR37) in photoperiodic flowering in rice. Mol. Cells 2015, 38, 81–88. [Google Scholar]
- Wong, A.C.; Hecht, V.F.; Picard, K.; Diwadkar, P.; Laurie, R.E.; Wen, J.; Mysore, K.; Macknight, R.C.; Weller, J.L. Isolation and functional analysis of CONSTANS-LIKE genes suggests that a central role for CONSTANS in flowering time control is not evolutionarily conserved in Medicago truncatula. Front. Plant Sci. 2014, 5, 486. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jiang, A.; Thomson, G.; Kerr-Phillips, M.; Phan, C.; Krueger, T.; Jaudal, M.; Wen, J.; Mysore, K.S.; Putterill, J. Overexpression of Medicago MtCDFd1_1 Causes Delayed Flowering in Medicago via Repression of MtFTa1 but Not MtCO-Like Genes. Front. Plant Sci. 2019, 10, 1148. [Google Scholar] [CrossRef] [PubMed]
- Crepy, M.; Yanovsky, M.J.; Casal, J.J. Blue Rhythms between GIGANTEA and Phytochromes. Plant Signal. Behav. 2007, 2, 530–532. [Google Scholar] [CrossRef] [PubMed]
- Park, M.J.; Kwon, Y.J.; Gil, K.E.; Park, C.M. LATE ELONGATED HYPOCOTYL regulates photoperiodic flowering via the circadian clock in Arabidopsis. BMC Plant Biol. 2016, 16, 114. [Google Scholar] [CrossRef]
- Tadege, M.; Wen, J.; He, J.; Tu, H.; Kwak, Y.; Eschstruth, A.; Cayrel, A.; Endre, G.; Zhao, P.X.; Chabaud, M.; et al. Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. Cell Mol. Biol. 2008, 54, 335–347. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, J.; Zhang, J.; Zhou, C.; Han, L. Genome-wide characterization of AINTEGUMENTA-LIKE family in Medicago truncatula reveals the significant roles of AINTEGUMENTAs in leaf growth. Front. Plant Sci. 2022, 13, 1050462. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, J.; Xie, Y.; Liu, X.; Wen, L.; Wang, H.; Zhang, J.; Li, J.; Han, L.; Yu, X.; et al. LATE MERISTEM IDENTITY1 regulates leaf margin development via the auxin transporter gene SMOOTH LEAF MARGIN1. Plant Physiol. 2021, 187, 218–235. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Zhang, J.; Liu, X.; Kong, Y.; Han, L. The Roles of the PSEUDO-RESPONSE REGULATORs in Circadian Clock and Flowering Time in Medicago truncatula. Int. J. Mol. Sci. 2023, 24, 16834. https://doi.org/10.3390/ijms242316834
Wang X, Zhang J, Liu X, Kong Y, Han L. The Roles of the PSEUDO-RESPONSE REGULATORs in Circadian Clock and Flowering Time in Medicago truncatula. International Journal of Molecular Sciences. 2023; 24(23):16834. https://doi.org/10.3390/ijms242316834
Chicago/Turabian StyleWang, Xiao, Juanjuan Zhang, Xiu Liu, Yiming Kong, and Lu Han. 2023. "The Roles of the PSEUDO-RESPONSE REGULATORs in Circadian Clock and Flowering Time in Medicago truncatula" International Journal of Molecular Sciences 24, no. 23: 16834. https://doi.org/10.3390/ijms242316834
APA StyleWang, X., Zhang, J., Liu, X., Kong, Y., & Han, L. (2023). The Roles of the PSEUDO-RESPONSE REGULATORs in Circadian Clock and Flowering Time in Medicago truncatula. International Journal of Molecular Sciences, 24(23), 16834. https://doi.org/10.3390/ijms242316834



