Screening of Oral Potential Angiotensin-Converting Enzyme Inhibitory Peptides from Zizyphus jujuba Proteins Based on Gastrointestinal Digestion In Vivo
Abstract
:1. Introduction
2. Results and Discussion
2.1. Preparation and Components of Zizyphus jujuba Proteins
2.2. Peptidomics Analysis of Zizyphus jujuba Protein In Vivo
2.3. ACE-Inhibitory Activity of Jujube Protein after In Vitro and In Vivo Digestion
2.4. Setting 3D-QSAR Model
2.5. 3D-QSAR Model Prediction Results and Validation
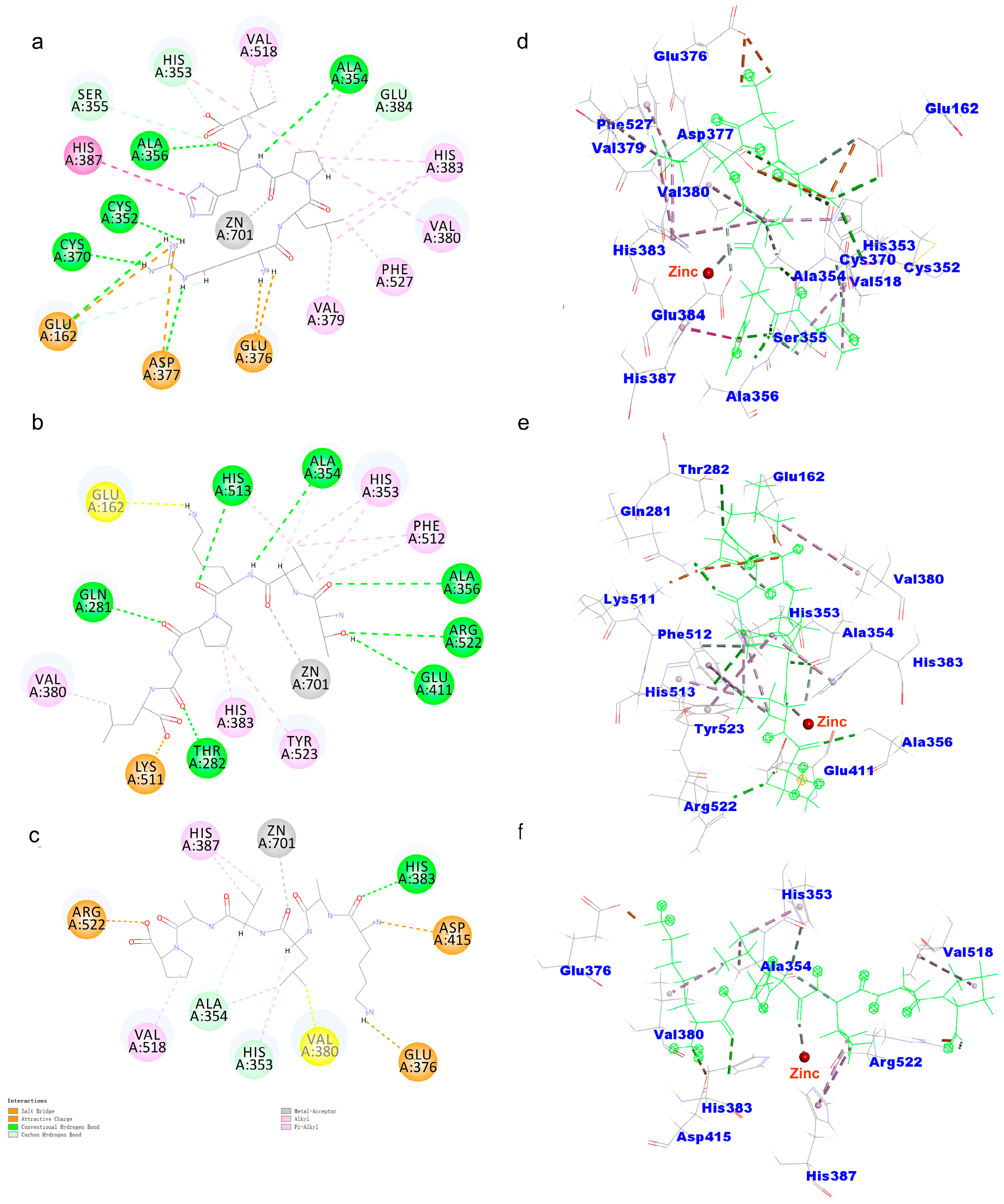
2.6. Molecular Docking
2.7. Stability of Peptides after Simulated Digestion In Vitro
3. Materials and Methods
3.1. Materials
3.2. Preparation of Zizyphus jujuba Protein
3.3. Prediction of the Activity of Zizyphus jujuba Proteins Based on In Silico Digestion
3.4. Peptidomics Analysis of Zizyphus jujuba Proteins after In Vivo Digestion
3.5. 3D-QSAR Model Setting and Screening for ACEI Peptides
3.6. ACE-Inhibitory Activity Assay In Vitro
3.7. Binding of Peptides to ACE via Molecular Docking
3.8. Stability In Vitro Digestion
3.9. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zou, M.; Chen, Y.L.; Sun-Waterhouse, D.; Zhang, Y.H.; Li, F. Immunomodulatory acidic polysaccharides from Zizyphus jujuba cv. Huizao: Insights into their chemical characteristics and modes of action. Food Chem. 2018, 258, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Naik, S.R.; Bhagat, S.; Shah, P.D.; Tare, A.A.; Ingawale, D.; Wadekar, R.R. Evaluation of anti-allergic and anti-anaphylactic activity of ethanolic extract of Zizyphus jujuba fruits in rodents. Rev. Bras. Farmacogn. 2013, 23, 811–818. [Google Scholar] [CrossRef]
- Al-Reza, S.M.; Rahman, A.; Lee, J.; Kang, S.C. Potential roles of essential oil and organic extracts of Zizyphus jujuba in inhibiting food-borne pathogens. Food Chem. 2010, 119, 981–986. [Google Scholar] [CrossRef]
- Park, H.J.; Jung, I.H.; Kwon, H.; Yu, J.; Jo, E.; Kim, H.; Park, S.J.; Lee, Y.C.; Kim, D.H.; Ryu, J.H. The ethanol extract of Zizyphus jujuba var. spinosa seeds ameliorates the memory deficits in Alzheimer’s disease model mice. J. Ethnopharmacol. 2019, 233, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Yang, M.; Huang, W.Z.; Wang, Y.; Yang, M.; Wang, Y.; Zhao, Z. Composition, antioxidant activities and hepatoprotective effects of the water extract of Ziziphus jujuba cv. Jinsixiaozao. RSC Adv. 2017, 7, 6511–6522. [Google Scholar] [CrossRef]
- Miao, W.; Sheng, L.; Yang, T.; Wu, G.; Zhang, M.; Sun, J.; Ainiwaer, A. The impact of flavonoids-rich Ziziphus jujuba Mill. Extract on Staphylococcus aureus biofilm formation. BMC Complement. Med. Ther. 2020, 20, 187. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, L.; Zhang, F.; Ma, H. Effects of multi-mode S-type ultrasound pretreatment on the preparation of ACE inhibitory peptide from rice protein. Food Chem. 2020, 331, 127216. [Google Scholar] [CrossRef] [PubMed]
- Sonklin, C.; Alashi, M.A.; Laohakunjit, N.; Kerdchoechuen, O.; Aluko, R.E. Identification of antihypertensive peptides from mung bean protein hydrolysate and their effects in spontaneously hypertensive rats. J. Funct. Foods 2020, 64, 103635. [Google Scholar] [CrossRef]
- Patil, S.P.; Goswami, A.; Kalia, K.; Kate, A.S. Plant-Derived Bioactive Peptides: A Treatment to Cure Diabetes. Int. J. Pept. Res. Ther. 2019, 26, 955–968. [Google Scholar] [CrossRef]
- Memarpoor-Yazdi, M.; Mahaki, H.; Zare-Zardini, H. Antioxidant activity of protein hydrolysates and purified peptides from Zizyphus jujuba fruits. J. Funct. Foods 2013, 5, 62–70. [Google Scholar] [CrossRef]
- Memarpoor-Yazdi, M.; Zare-Zardini, H.; Mogharrab, N.; Navapour, L. Purification, Characterization and Mechanistic Evaluation of Angiotensin Converting Enzyme Inhibitory Peptides Derived from Zizyphus Jujuba Fruit. Sci. Rep. 2020, 10, 3976. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Hua, Y.; Zhou, C.Y.; Xiong, Y.Z.; Pan, D.D.; Liu, Z.; Dang, Y.L. Umami peptides screened based on peptidomics and virtual screening from Ruditapes philippinarum and Mactra veneriformis clams. Food Chem. 2022, 394, 133504. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.Y.; Li, S.S.; Luo, Q.Y.; Shen, G.H.; Wu, H.J.; Li, M.L.; Liu, X.Y.; Chen, A.J.; Ye, M.; Zhang, Z.Q. Discovery and identification of antimicrobial peptides in Sichuan pepper (Zanthoxylum bungeanum Maxim) seeds by peptidomics and bioinformatics. Appl. Microbiol. Biotechnol. 2019, 103, 2217–2228. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.K.; Zhu, L.; Wu, G.C.; Liu, T.T.; Qi, X.G.; Zhang, H. Rapid Screening of Novel Dipeptidyl Peptidase-4 Inhibitory Peptides from Pea (Pisum sativum L.) Protein Using Peptidomics and Molecular Docking. J. Agric. Food Chem. 2022, 70, 10221–10228. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Chen, Y.; Zhao, W.; Zheng, F.; Ding, L.; Liu, J. Novel ACE inhibitory tripeptides from ovotransferrin using bioinformatics and peptidomics approaches. Sci. Rep. 2019, 9, 17434. [Google Scholar] [CrossRef] [PubMed]
- Prabhakar, Y.S.; Gupta, S.P. Structure-Activity relationship study on angiotensin-converting enzyme inhibitors--investigation of hydrophobic interaction in inhibition mechanism. Indian J. Biochem. Biophys. 1985, 22, 318–320. [Google Scholar] [PubMed]
- Yan, W.L.; Lin, G.M.; Zhang, R.; Liang, Z.; Wu, L.; Wu, W. Studies on molecular mechanism between ACE and inhibitory peptides in different bioactivities by 3D-QSAR and MD simulations. J. Mol. Liq. 2020, 304, 112702. [Google Scholar] [CrossRef]
- Pan, D.D.; Cao, J.X.; Guo, H.Q.; Zhao, B. Studies on purification and the molecular mechanism of a novel ACE inhibitory peptide from whey protein hydrolysate. Food Chem. 2012, 130, 121–126. [Google Scholar] [CrossRef]
- Li, P.; Jia, J.; Fang, M.; Zhang, L.; Guo, M.; Xie, J.; Xia, Y.; Zhou, L.; Wei, D. In vitro and in vivo ACE inhibitory of pistachio hydrolysates and in silico mechanism of identified peptide binding with ACE. Process Biochem. 2014, 49, 898–904. [Google Scholar] [CrossRef]
- Liu, R.; Zhu, Y.; Chen, J.; Wu, H.; Shi, L.; Wang, X.; Wang, L. Characterization of ACE inhibitory peptides from Mactra veneriformis hydrolysate by nano-liquid chromatography electrospray ionization mass spectrometry (Nano-LC-ESI-MS) and molecular docking. Mar. Drugs 2014, 12, 3917–3928. [Google Scholar] [CrossRef]
- You, L.J.; Zhao, M.M.; Cui, C.; Zhao, H.F.; Yang, B. Effect of degree of hydrolysis on the antioxidant activity of loach (Misgurnus anguillicaudatus) protein hydrolysates. Innov. Food Sci. Emerg. 2009, 10, 235–240. [Google Scholar] [CrossRef]
- Xu, Q.; Hong, H.; Wu, J.; Yan, X. Bioavailability of bioactive peptides derived from food proteins across the intestinal epithelial membrane: A review. Trends Food Sci. Technol. 2019, 86, 399–411. [Google Scholar] [CrossRef]
- Miquel, E.; Ángel Gómez, J.; Alegría, A.; Barberá, R.; Farré, R.; Recio, I. Identification of casein phosphopeptides after simulated gastrointestinal digestion by tandem mass spectrometry. Eur. Food Res. Technol. 2006, 222, 48–53. [Google Scholar] [CrossRef]
- Miquel, E.; Alegría, A.; Barberá, R.; Farré, R. Casein phosphopeptides released by simulated gastrointestinal digestion of infant formulas and their potential role in mineral binding. Int. Dairy J. 2006, 16, 992–1000. [Google Scholar] [CrossRef]
- Quirós, A.; del Mar Contreras, M.; Ramos, M.; Amigo, L.; Recio, I. Stability to gastrointestinal enzymes and structure-activity relationship of beta-casein-peptides with antihypertensive properties. Peptides 2009, 30, 1848–1853. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, L.; Jaworska, J.; Worth Andrew, P.; Cronin Mark, T.D.; McDowell Robert, M.; Gramatica, P. Methods for reliability and uncertainty assessment and for applicability evaluations of classification- and regression-based QSARs. Environ. Health Perspect. 2003, 111, 1361–1375. [Google Scholar] [CrossRef] [PubMed]
- Qi, C.; Lin, G.; Zhang, R.; Wu, W. Studies on the Bioactivities of ACE-inhibitory Peptides with Phenylalanine C-terminus Using 3D-QSAR, Molecular Docking and in vitro Evaluation. Mol. Inform. 2017, 36, 1600157. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Aluko, R.E.; Nakai, S. Structural Requirements of Angiotensin I-Converting Enzyme Inhibitory Peptides: Quantitative Structure−Activity Relationship Study of Di- and Tripeptides. J. Agric. Food Chem. 2006, 54, 732–738. [Google Scholar] [CrossRef] [PubMed]
- Natesh, R.; Schwager, S.L.U.; Sturrock, E.D.; Acharya, K.R. Crystal structure of the human angiotensin-converting enzyme–lisinopril complex. Nature 2003, 421, 551–554. [Google Scholar] [CrossRef]
- Kumada, Y.; Hashimoto, N.; Hasan, F.; Terashima, M.; Nakanishi, K.; Jungbauer, A.; Katoh, S. Screening of ACE-inhibitory peptides from a random peptide-displayed phage library using ACE-coupled liposomes. J. Biotechnol. 2007, 131, 144–149. [Google Scholar] [CrossRef]
- Auwal, S.M.; Zainal Abidin, N.; Zarei, M.; Tan, C.P.; Saari, N. Identification, structure-activity relationship and in silico molecular docking analyses of five novel angiotensin I-converting enzyme (ACE)-inhibitory peptides from stone fish (Actinopyga lecanora) hydrolysates. PLoS ONE 2019, 14, e0197644. [Google Scholar] [CrossRef] [PubMed]
- Murray, A.B.; FitzGerald, J.R. Angiotensin Converting Enzyme Inhibitory Peptides Derived from Food Proteins: Biochemistry, Bioactivity and Production. Curr. Pharm. Des. 2007, 13, 773–791. [Google Scholar] [CrossRef] [PubMed]
- FitzGerald, R.J.; Meisel, H. Milk protein-derived peptide inhibitors of angiotensin-I-converting enzyme. Br. J. Nutr. 2000, 84 (Suppl. S1), 33–37. [Google Scholar] [CrossRef] [PubMed]
- Afrin, S.; Rakib, M.A.; Kim, B.H.; Kim, J.O.; Ha, Y.L. Eritadenine from Edible Mushrooms Inhibits Activity of Angiotensin Converting Enzyme in Vitro. J. Agric. Food Chem. 2016, 64, 2263–2268. [Google Scholar] [CrossRef] [PubMed]
- Henda, Y.B.; Labidi, A.; Arnaudin, I.; Bridiau, N.; Delatouche, R.; Maugard, T.; Piot, J.-M.; Sannier, F.; Thiéry, V.; Bordenave-Juchereau, S. Measuring Angiotensin-I Converting Enzyme Inhibitory Activity by Micro Plate Assays: Comparison Using Marine Cryptides and Tentative Threshold Determinations with Captopril and Losartan. J. Agric. Food Chem. 2013, 61, 10685–10690. [Google Scholar] [CrossRef] [PubMed]
- Pina, A.S.; Roque, A.C.A. Studies on the molecular recognition between bioactive peptides and angiotensin-converting enzyme. J. Mol. Recognit. 2009, 22, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Lai, J.; He, P.; Pan, L.; Zhang, Y.; Zhang, M.; Wu, H. Screening, ACE-inhibitory mechanism and structure-activity relationship of a novel ACE-inhibitory peptide from Lepidium meyenii (Maca) protein hydrolysate. Food Biosci. 2023, 52, 102374. [Google Scholar] [CrossRef]
- Soleymanzadeh, N.; Mirdamadi, S.; Mirzaei, M.; Kianirad, M. Novel β-casein derived antioxidant and ACE-inhibitory active peptide from camel milk fermented by Leuconostoc lactis PTCC1899: Identification and molecular docking. Int. Dairy J. 2019, 97, 201–208. [Google Scholar] [CrossRef]
- Li, W.; Chen, W.; Ma, H.; Wu, D.; Zhang, Z.; Yang, Y. Structural characterization and angiotensin-converting enzyme (ACE) inhibitory mechanism of Stropharia rugosoannulata mushroom peptides prepared by ultrasound. Ultrason. Sonochem. 2022, 88, 106074. [Google Scholar] [CrossRef]
- Li, J.; Huo, X.; Zheng, Y.; Guo, Y.; Feng, C. ACE-Inhibitory Peptides Identified from Quinoa Bran Glutelin-2 Hydrolysates: In Silico Screening and Characterization, Inhibition Mechanisms of ACE, Coordination with Zinc Ions, and Stability. Plant Foods Hum. Nutr. 2023, 78, 419–425. [Google Scholar] [CrossRef]
- Jiang, Q.; Chen, Q.; Zhang, T.; Liu, M.; Duan, S.; Sun, X. The Antihypertensive Effects and Potential Molecular Mechanism of Microalgal Angiotensin I-Converting Enzyme Inhibitor-Like Peptides: A Mini Review. Int. J. Mol. Sci. 2021, 22, 4068. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Yang, W.; Zhu, Q.; Zhang, G.; Zhang, X.; Liu, L.; Li, X.; Hussain, M.; Ni, C.; Jiang, X. Proteolysis and ACE-inhibitory peptide profile of Cheddar cheese: Effect of digestion treatment and different probiotics. LWT 2021, 145, 111295. [Google Scholar] [CrossRef]
- Ahmed, T.; Sun, X.; Udenigwe, C.C. Role of structural properties of bioactive peptides in their stability during simulated gastrointestinal digestion: A systematic review. Trends Food Sci. Technol. 2022, 120, 265–273. [Google Scholar] [CrossRef]
- Pei, J.; Gao, X.; Pan, D.; Hua, Y.; He, J.; Liu, Z.; Dang, Y. Advances in the stability challenges of bioactive peptides and improvement strategies. Curr. Res. Food Sci. 2022, 5, 2162–2170. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Jiang, Y.; Zhou, X.; Bi, H.; Yang, B. Structure identification of soybean peptides and their immunomodulatory activity. Food Chem. 2021, 359, 129970. [Google Scholar] [CrossRef] [PubMed]
- Sangsawad, P.; Roytrakul, S.; Yongsawatdigul, J. Angiotensin converting enzyme (ACE) inhibitory peptides derived from the simulated in vitro gastrointestinal digestion of cooked chicken breast. J. Funct. Foods 2017, 29, 77–83. [Google Scholar] [CrossRef]
- Pekkoh, J.; Phinyo, K.; Thurakit, T.; Lomakool, S.; Duangjan, K.; Ruangrit, K.; Pumas, C.; Jiranusornkul, S.; Yooin, W.; Cheirsilp, B.; et al. Lipid Profile, Antioxidant and Antihypertensive Activity, and Computational Molecular Docking of Diatom Fatty Acids as ACE Inhibitors. Antioxidants 2022, 11, 186. [Google Scholar] [CrossRef]
- Pei, J.Y.; Liu, Z.; Pan, D.D.; Zhao, Y.F.; Dang, Y.L.; Gao, X.C. Transport, Stability, and In Vivo Hypoglycemic Effect of a Broccoli-Derived DPP-IV Inhibitory Peptide VPLVM. J. Agric. Food Chem. 2022, 70, 4934–4941. [Google Scholar] [CrossRef]
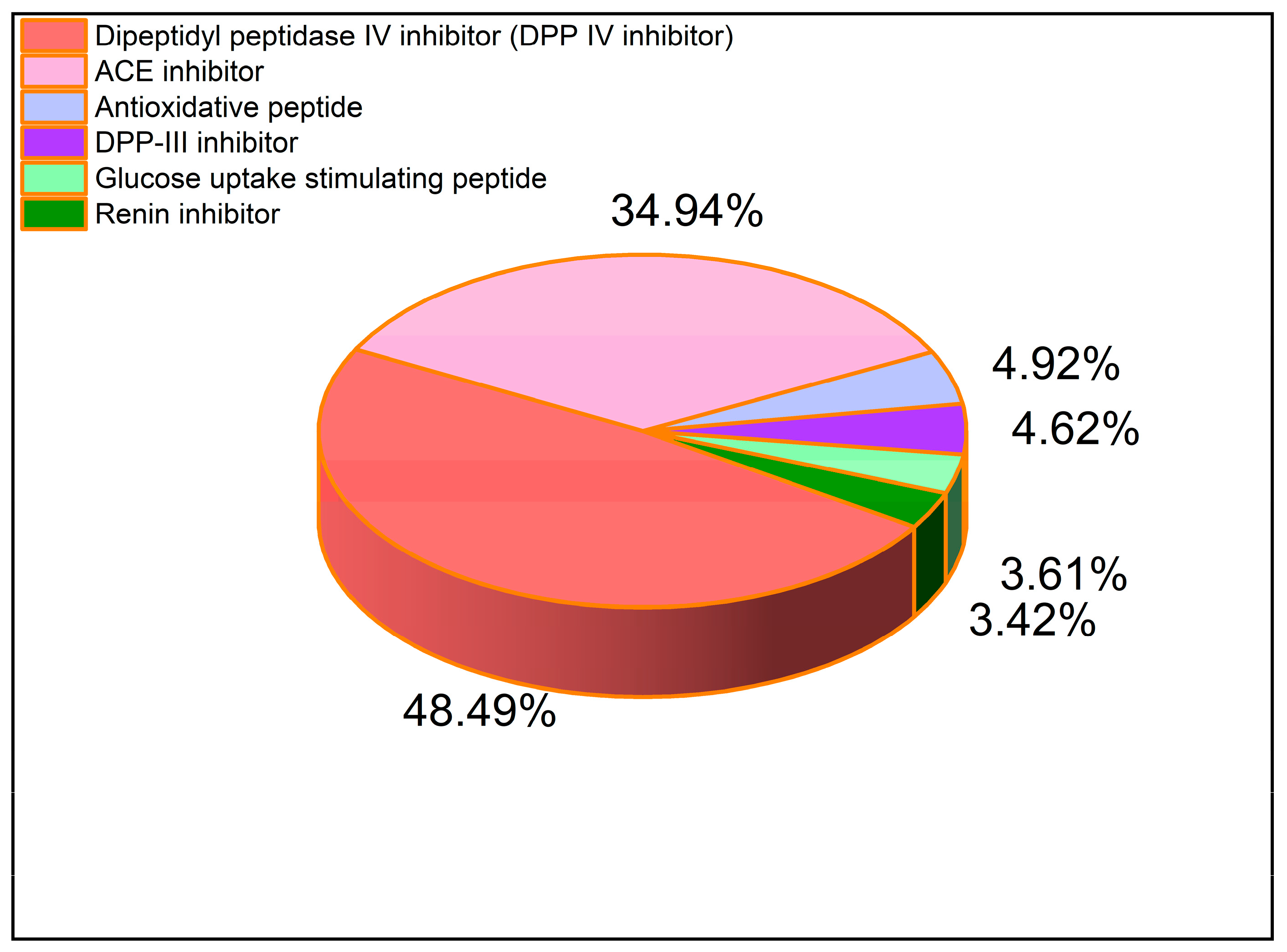

| Sample Name | ACE-Inhibitory IC50 (mg/mL) |
|---|---|
| Simulate gastrointestinal digestion of jujube protein | 0.34 |
| Jujube protein hydrolysates in mice intestinal tract | 0.21 |
| No | Sequence | Peptide | Exact Mass | ACE Inhibition Rate (%) (2 mg/mL) | ACE IC50 (µM) |
|---|---|---|---|---|---|
| 1 | Leu-Glu-Lys-Pro-Leu-Leu | LEKPLL | 711.45 | 24.2 | ________ |
| 2 | Leu-Glu-Lys-Leu-Val-Thr | LEKLVT | 701.43 | 30.5 | ________ |
| 3 | Arg-Leu-Pro-His-Val | RLPHV | 620.38 | 100.0 | 6.01 |
| 4 | Thr-Val-Lys-Pro-Gly-Leu | TVKPGL | 613.38 | 100.0 | 3.81 |
| 5 | Tyr-Leu-His-Leu | YLHL | 544.30 | 65.4 | ________ |
| 6 | Arg-Phe-Pro-Arg | RFPR | 574.33 | 22.9 | ________ |
| 7 | Lys-Ala-Leu-Val-Ala-Pro | KALVAP | 597.38 | 100.0 | 17.06 |
| 8 | Lys-Val-Lys-Pro-Leu | KVKPL | 583.41 | −14.2 | ________ |
| 9 | Pro-Arg-Pro-Lys-Pro-Pro-Pro | PRPKPPP | 787.47 | 74.8 | ________ |
| 10 | Pro-Glu-Arg-Lys | PERK | 528.30 | 4.5 | ________ |
| * Positive control | Captopril | 5 ng/mL | 63.4 |
| Name | CDOCKER Energy (kcal/mol) | CDOCKER Interaction Energy (kcal/mol) | Simulated Gastrointestinal Digestion Stability % |
|---|---|---|---|
| RLPHV | −106.075 | −101.716 | 82 |
| TVKPGL | −104.036 | −90.9375 | 90 |
| KALVAP | −102.312 | −110.661 | 78 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, X.; Zhang, C.; Wang, N.; Lin, J.-M.; Dang, Y.; Zhao, Y. Screening of Oral Potential Angiotensin-Converting Enzyme Inhibitory Peptides from Zizyphus jujuba Proteins Based on Gastrointestinal Digestion In Vivo. Int. J. Mol. Sci. 2023, 24, 15848. https://doi.org/10.3390/ijms242115848
Gao X, Zhang C, Wang N, Lin J-M, Dang Y, Zhao Y. Screening of Oral Potential Angiotensin-Converting Enzyme Inhibitory Peptides from Zizyphus jujuba Proteins Based on Gastrointestinal Digestion In Vivo. International Journal of Molecular Sciences. 2023; 24(21):15848. https://doi.org/10.3390/ijms242115848
Chicago/Turabian StyleGao, Xinchang, Chaoying Zhang, Ning Wang, Jin-Ming Lin, Yali Dang, and Yufen Zhao. 2023. "Screening of Oral Potential Angiotensin-Converting Enzyme Inhibitory Peptides from Zizyphus jujuba Proteins Based on Gastrointestinal Digestion In Vivo" International Journal of Molecular Sciences 24, no. 21: 15848. https://doi.org/10.3390/ijms242115848
APA StyleGao, X., Zhang, C., Wang, N., Lin, J.-M., Dang, Y., & Zhao, Y. (2023). Screening of Oral Potential Angiotensin-Converting Enzyme Inhibitory Peptides from Zizyphus jujuba Proteins Based on Gastrointestinal Digestion In Vivo. International Journal of Molecular Sciences, 24(21), 15848. https://doi.org/10.3390/ijms242115848







