The Development of New Methods to Stimulate the Production of Antimicrobial Peptides in the Larvae of the Black Soldier Fly Hermetia illucens
Abstract
:1. Introduction
2. Results
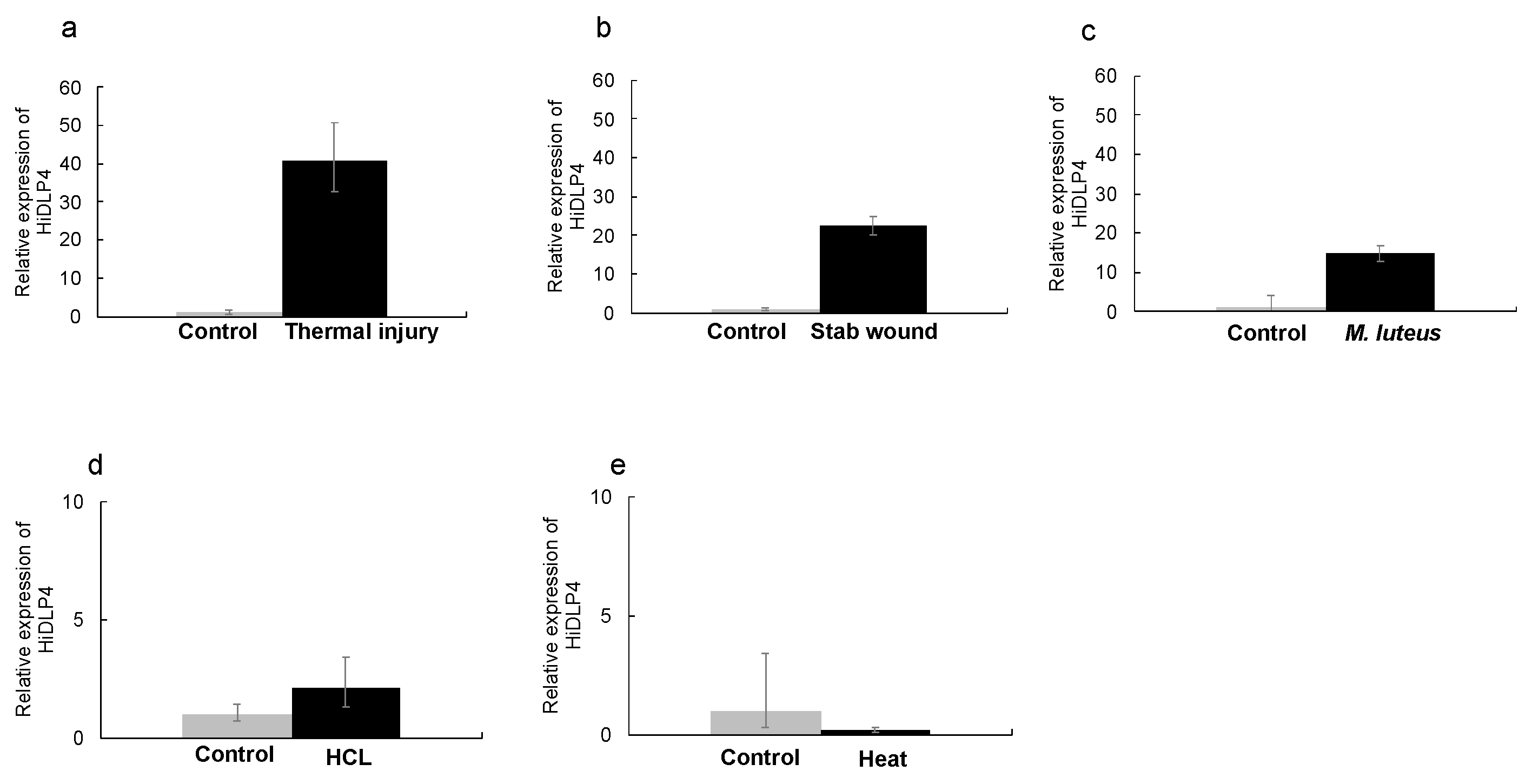
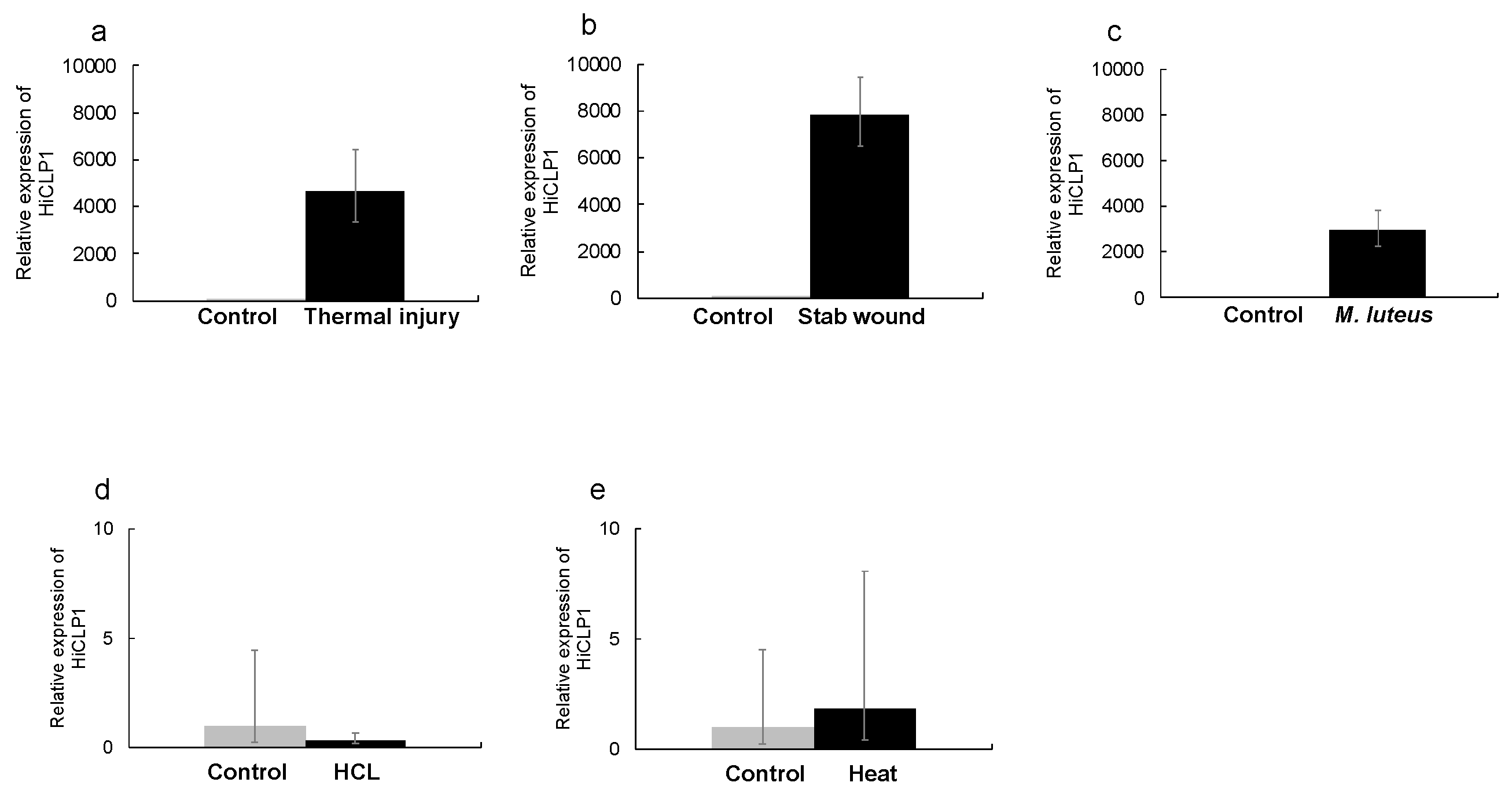
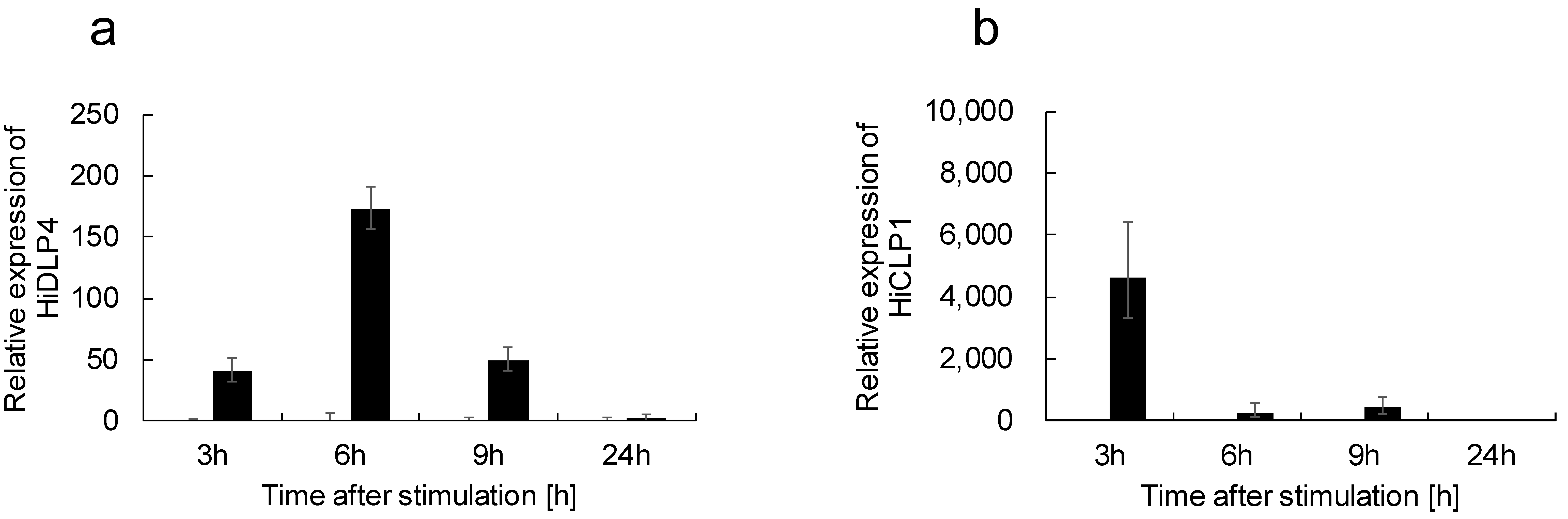
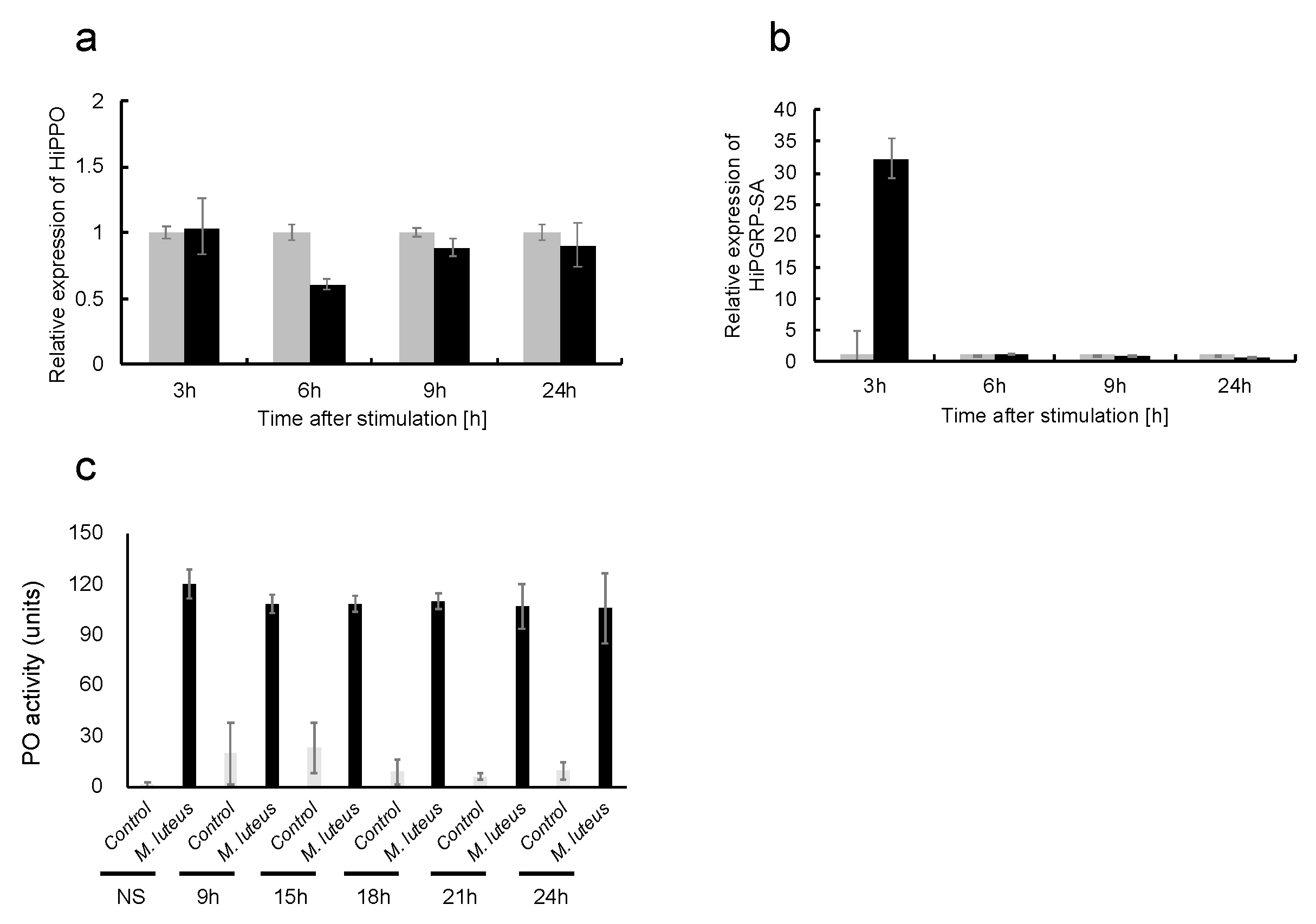
2.1. Thermal Injury Treatment Induces AMP Gene Expression
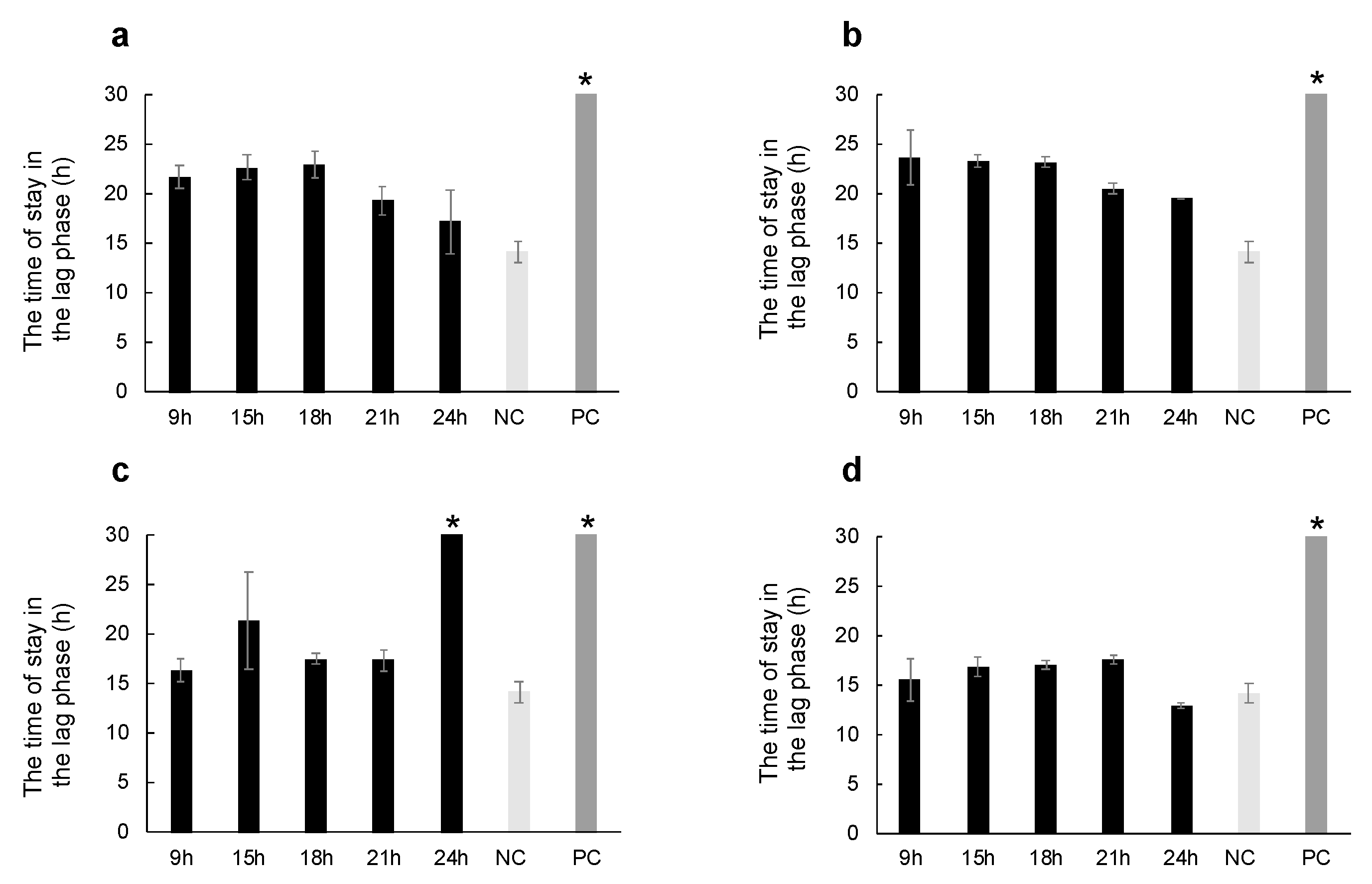
2.2. Thermal Injury Inhibits the Growth of Micrococcus luteus
2.3. Identification of Differentially Expressed Genes and Assignment of Drosophila melanogaster Homologs
2.4. Gene Enrichment Analysis of Differentially Expressed Genes
3. Discussion
4. Materials and Methods
4.1. Insects
4.2. HCl Treatment of the Larvae
4.3. Heat Treatment of the Larvae
4.4. Stab Wound to the Larvae
4.5. Thermal Injury to the Larvae
4.6. Injection of Micrococcus luteus
4.7. Total RNA Purification and cDNA Synthesis from Fat Body Samples and RT-qPCR Analysis
4.8. cDNA Cloning
4.9. Hemolymph Collection for Phenoloxidase Activity
4.10. Measurement of Phenoloxidase Activity
4.11. RNA-Seq Analysis
4.12. Gene Enrichment and Molecular Interaction Analyses
4.13. Hemolymph Collection for Antimicrobial Activity Assay
4.14. Antimicrobial Activity Assay
4.15. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Diener, S.; Zurbrügg, C.; Tockner, K. Conversion of organic material by black soldier fly larvae: Establishing optimal feeding rates. Waste Manag. Res. 2009, 27, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Diener, S.; Zurbrügg, C.; Gutiérrez, F.R.; Nguyen, D.H.; Morel, A.; Koottatep, T.; Tockner, K. Black soldier fly larvae for organic waste treatment-prospects and constraints. Proc. WasteSafe 2011, 2, 13–15. [Google Scholar]
- United Nations. World Urbanization Prospects. The 2018 Version. 9 (United Nations, 2019). Available online: https://population.un.org/wup/publications/Files/WUP2018-Report.pdf (accessed on 15 January 2023).
- Animal Products Safety Division, Food Safety and Consumer Affairs Bureau, Ministry of Agriculture, Forestry and Fisheries. Chikusan Shiryo. 2023. Available online: https://www.maff.go.jp/j/chikusan/sinko/lin/l_siryo/attach/pdf/index-936.pdf (accessed on 9 January 2023).
- Van Huis, A.; Van Itterbeeck, J.; Klunder, H.; Mertens, E.; Halloran, A.; Muir, G.; Vantomme, P. Edible Insects, Future Prospects for Food and Feed Security; Food and Agriculture Organization of the United Nations: Rome, Italy, 2013; p. 171. [Google Scholar]
- Chia, S.Y.; Tanga, C.M.; Osuga, I.M.; Alaru, A.O.; Mwangi, D.M.; Githinji, M.; Subramanian, S.; Fiaboe, K.K.M.; Ekesi, S.; van Loon, J.J.A.; et al. Effect of dietary replacement of fishmeal by insect meal on growth performance, blood profiles and economics of growing pigs in Kenya. Animals 2019, 9, 705. [Google Scholar] [CrossRef] [PubMed]
- Riddick, E.W. Insect Protein as a Partial Replacement for Fishmeal in the Diets of Juvenile Fish and Crustaceans in Mass Production of Beneficial Organisms; Morales-Ramos, J.A., Rojas, M.G., Shapiro-Ilan, D.I., Eds.; Academic Press: Cambridge, MA, USA, 2014; pp. 565–582. [Google Scholar]
- Butaye, P.; Devriese, L.A.; Haesebrouck, F. Antimicrobial growth promoters used in animal feed, effects of less well known antibiotics on gram-positive bacteria. Clin. Microbiol. Rev. 2003, 16, 175–188. [Google Scholar] [CrossRef] [PubMed]
- López-Gálvez, G.; López-Alonso, M.; Pechova, A.; Mayo, B.; Dierick, N.; Gropp, J. Alternatives to antibiotics and trace elements (copper and zinc) to improve gut health and zootechnical parameters in piglets: A review. Anim. Feed Sci. Technol. 2021, 271, 114727. [Google Scholar] [CrossRef]
- Gadde, U.; Kim, W.H.; Oh, S.T.; Lillehoj, H.S. Alternatives to antibiotics for maximizing growth performance and feed efficiency in poultry: A review. Anim. Health Res. Rev. 2017, 18, 26–45. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Brower, C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.; Robinson, T.P.; Teillant, A.; Laxminarayan, R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef]
- Nhung, N.T.; Cuong, N.V.; Thwaites, G.; Carrique-Mas, J. Antimicrobial usage and antimicrobial resistance in animal production in Southeast Asia, a review. Antibiotics 2016, 5, 37. [Google Scholar] [CrossRef]
- Manniello, M.D.; Moretta, A.; Salvia, R.; Scieuzo, C.; Lucchetti, D.; Vogel, H.; Sgambato, A.; Falabella, P. Insect antimicrobial peptides, Potential weapons to counteract the antibiotic resistance. Cell. Mol. Life Sci. 2021, 78, 4259–4282. [Google Scholar] [CrossRef]
- Wu, Q.; Patočka, J.; Kuča, K. Insect antimicrobial peptides, a mini review. Toxins 2018, 10, 461. [Google Scholar] [CrossRef]
- Ferrandon, D.; Imler, J.L.; Hetru, C.; Hoffmann, J.A. The Drosophila systemic immune response, sensing and signalling during bacterial and fungal infections. Nat. Rev. Immunol. 2007, 7, 862–874. [Google Scholar] [CrossRef] [PubMed]
- Zdybicka-Barabas, A.; Bulak, P.; Polakowski, C.; Bieganowski, A.; Waśko, A.; Cytryńska, M. Immune response in the larvae of the black soldier fly Hermetia illucens. Invertebr. Surviv. J. 2017, 14, 9–17. [Google Scholar]
- Park, S.I.; Yoe, S.M. A novel cecropin-like peptide from black soldier fly, Hermetia illucens: Isolation, structural and functional characterization. Entomol. Res. 2017, 47, 115–124. [Google Scholar] [CrossRef]
- Sultana, A.; Luo, H.; Ramakrishna, S. Harvesting of antimicrobial peptides from insect (Hermetia illucens) and its applications in the food packaging. Appl. Sci. 2021, 11, 6991. [Google Scholar] [CrossRef]
- Vogel, H.; Müller, A.; Heckel, D.G.; Gutzeit, H.; Vilcinskas, A. Nutritional immunology: Diversification and diet-dependent expression of antimicrobial peptides in the black soldier fly Hermetia illucens. Dev. Comp. Immunol. 2018, 78, 141–148. [Google Scholar] [CrossRef]
- Park, S.I.; Kim, J.W.; Yoe, S.M. Purification and characterization of a novel antibacterial peptide from black soldier fly (Hermetia illucens) larvae. Dev. Comp. Immunol. 2015, 52, 98–106. [Google Scholar] [CrossRef]
- Patyra, E.; Kwiatek, K. Insect Meals and Insect Antimicrobial Peptides as an Alternative for Antibiotics and Growth Promoters in Livestock Production. Pathogens 2023, 12, 854. [Google Scholar] [CrossRef]
- Lee, K.S.; Yun, E.Y.; Goo, T.W. Evaluation of the antimicrobial activity of an extract of Lactobacillus casei-infected Hermetia illucens larvae produced using an automatic injection system. Animals 2020, 10, 2121. [Google Scholar] [CrossRef]
- Tang, Z.; Xu, L.; Shi, B.; Deng, H.; Lai, X.; Liu, J.; Sun, Z. Oral administration of synthetic porcine beta-defensin-2 improves growth performance and cecal microbial flora and down-regulates the expression of intestinal toll-like receptor-4 and inflammatory cytokines in weaned piglets challenged with enterotoxigenic Escherichia coli. Anim. Sci. J. 2016, 87, 1258–1266. [Google Scholar]
- Peng, Z.; Wang, A.; Xie, L.; Song, W.; Wang, J.; Yin, Z.; Zhou, D.; Li, F. Use of recombinant porcine β-defensin 2 as a medicated feed additive for weaned piglets. Sci. Rep. 2016, 6, 26790. [Google Scholar] [CrossRef]
- Wen, L.F.; He, J.G. Dose-response effects of an antimicrobial peptide, a cecropin hybrid, on growth performance, nutrient utilisation, bacterial counts in the digesta and intestinal morphology in broilers. Br. J. Nutr. 2012, 108, 1756–1763. [Google Scholar] [CrossRef] [PubMed]
- Mwangi, J.; Hao, X.; Lai, R.; Zhang, Z.Y. Antimicrobial peptides, new hope in the war against multidrug resistance. Zool. Res. 2019, 40, 488–505. [Google Scholar] [CrossRef] [PubMed]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef] [PubMed]
- Gorman, M.J.; An, C.; Kanost, M.R. Characterization of tyrosine hydroxylase from Manduca sexta. Insect Biochem. Mol. Biol. 2017, 37, 1327–1337. [Google Scholar] [CrossRef]
- Kurata, S.; Ariki, S.; Kawabata, S. Recognition of pathogens and activation of immune responses in Drosophila and horseshoe crab innate immunity. Immunobiology 2006, 211, 237–249. [Google Scholar] [CrossRef]
- Kurata, S. Recognition and elimination of diversified pathogens in insect defense systems. Mol. Divers. 2006, 10, 599–605. [Google Scholar] [CrossRef]
- Tzou, P.; De Gregorio, E.; Lemaitre, B. How Drosophila combats microbial infection: A model to study innate immunity and host-pathogen interactions. Curr. Opin. Microbiol. 2002, 5, 102–110. [Google Scholar] [CrossRef]
- Kanost, M.R.; Gorman, M.J. Phenoloxidases in Insect Immunity in Insect Immunology; Beckage, N., Ed.; Academic Press: Cambridge, MA, USA; Elsevier: Amsterdam, The Netherlands, 2008; pp. 69–86. [Google Scholar]
- Cossins, A. Temperature Biology of Animals in Body Temperature in Tachymetabolic Animals; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012; pp. 98–154. [Google Scholar]
- Takano, Y.; Sakamoto, T.; Tabunoki, H.; Yoshimura, J.; Iwabuchi, K. Integrated effects of thermal acclimation and challenge temperature on cellular immunity in the plusiine moth larvae Chrysodeixis eriosoma (Lepidoptera: Noctuidae). Physiol. Entomol. 2021, 46, 52–59. [Google Scholar] [CrossRef]
- Stramer, B.; Winfield, M.; Shaw, T.; Millard, T.H.; Woolner, S.; Martin, P. Gene induction following wounding of wild-type versus macrophage-deficient Drosophila embryos. EMBO Rep. 2008, 9, 465–471. [Google Scholar] [CrossRef]
- Fehlbaum, P.; Bulet, P.; Michaut, L.; Lagueux, M.; Broekaert, W.F.; Hetru, C.; Hoffmann, J.A. Insect immunity. Septic injury of Drosophila induces the synthesis of a potent antifungal peptide with sequence homology to plant antifungal peptides. J. Biol. Chem. 1994, 269, 33159–33163. [Google Scholar] [CrossRef]
- Hoffmann, J.A.; Hetru, C.; Reichhart, J.M. The humoral antibacterial response of Drosophila. FEBS Lett. 1993, 325, 63–66. [Google Scholar] [CrossRef] [PubMed]
- Shaukat, Z.; Liu, D.; Gregory, S. Sterile inflammation in Drosophila. Mediat. Inflamm. 2015, 2015, 369286. [Google Scholar] [CrossRef] [PubMed]
- Buchon, N.; Silverman, N.; Cherry, S. Immunity in Drosophila melanogaster-From microbial recognition to whole-organism physiology. Nat. Rev. Immunol. 2014, 14, 796–810. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Sun, J.; Nishiyama, T.; Shimizu, K.; Kadota, K. TCC: An R package for comparing tag count data with robust normalization strategies. BMC Bioinform. 2013, 14, 219. [Google Scholar] [CrossRef]
- Orchard, S.; Ammari, M.; Aranda, B.; Breuza, L.; Briganti, L.; Broackes-Carter, F.; Campbell, N.H.; Chavali, G.; Chen, C.; del-Toro, N.; et al. The MIntAct project-IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014, 42, D358–D363. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakagawa, A.; Sakamoto, T.; Kanost, M.R.; Tabunoki, H. The Development of New Methods to Stimulate the Production of Antimicrobial Peptides in the Larvae of the Black Soldier Fly Hermetia illucens. Int. J. Mol. Sci. 2023, 24, 15765. https://doi.org/10.3390/ijms242115765
Nakagawa A, Sakamoto T, Kanost MR, Tabunoki H. The Development of New Methods to Stimulate the Production of Antimicrobial Peptides in the Larvae of the Black Soldier Fly Hermetia illucens. International Journal of Molecular Sciences. 2023; 24(21):15765. https://doi.org/10.3390/ijms242115765
Chicago/Turabian StyleNakagawa, Atsuyoshi, Takuma Sakamoto, Michael R. Kanost, and Hiroko Tabunoki. 2023. "The Development of New Methods to Stimulate the Production of Antimicrobial Peptides in the Larvae of the Black Soldier Fly Hermetia illucens" International Journal of Molecular Sciences 24, no. 21: 15765. https://doi.org/10.3390/ijms242115765
APA StyleNakagawa, A., Sakamoto, T., Kanost, M. R., & Tabunoki, H. (2023). The Development of New Methods to Stimulate the Production of Antimicrobial Peptides in the Larvae of the Black Soldier Fly Hermetia illucens. International Journal of Molecular Sciences, 24(21), 15765. https://doi.org/10.3390/ijms242115765





