Adipose Tissue-Derived Mesenchymal Stem Cells Extend the Lifespan and Enhance Liver Function in Hepatocyte Organoids
Abstract
:1. Introduction
2. Results
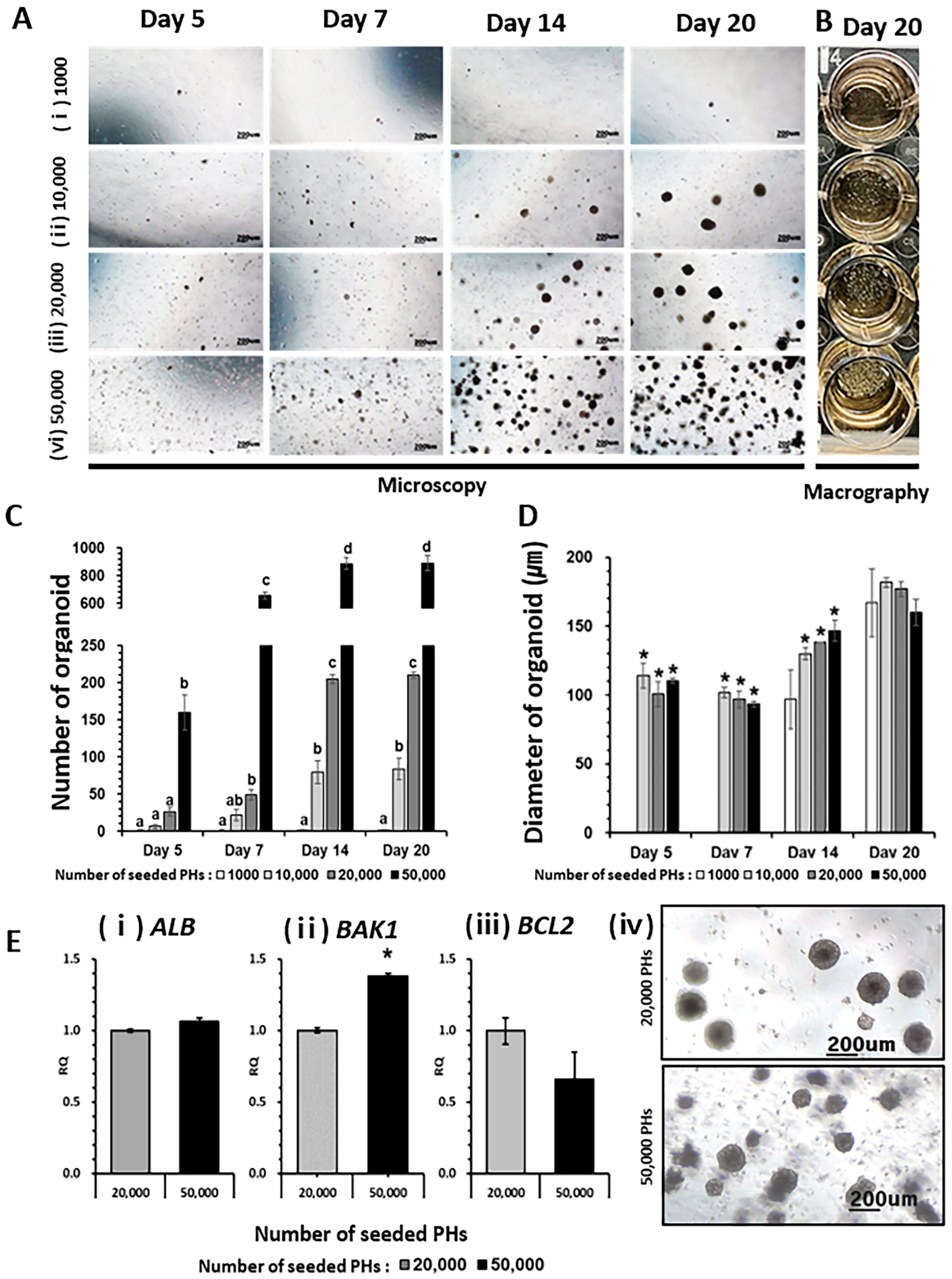
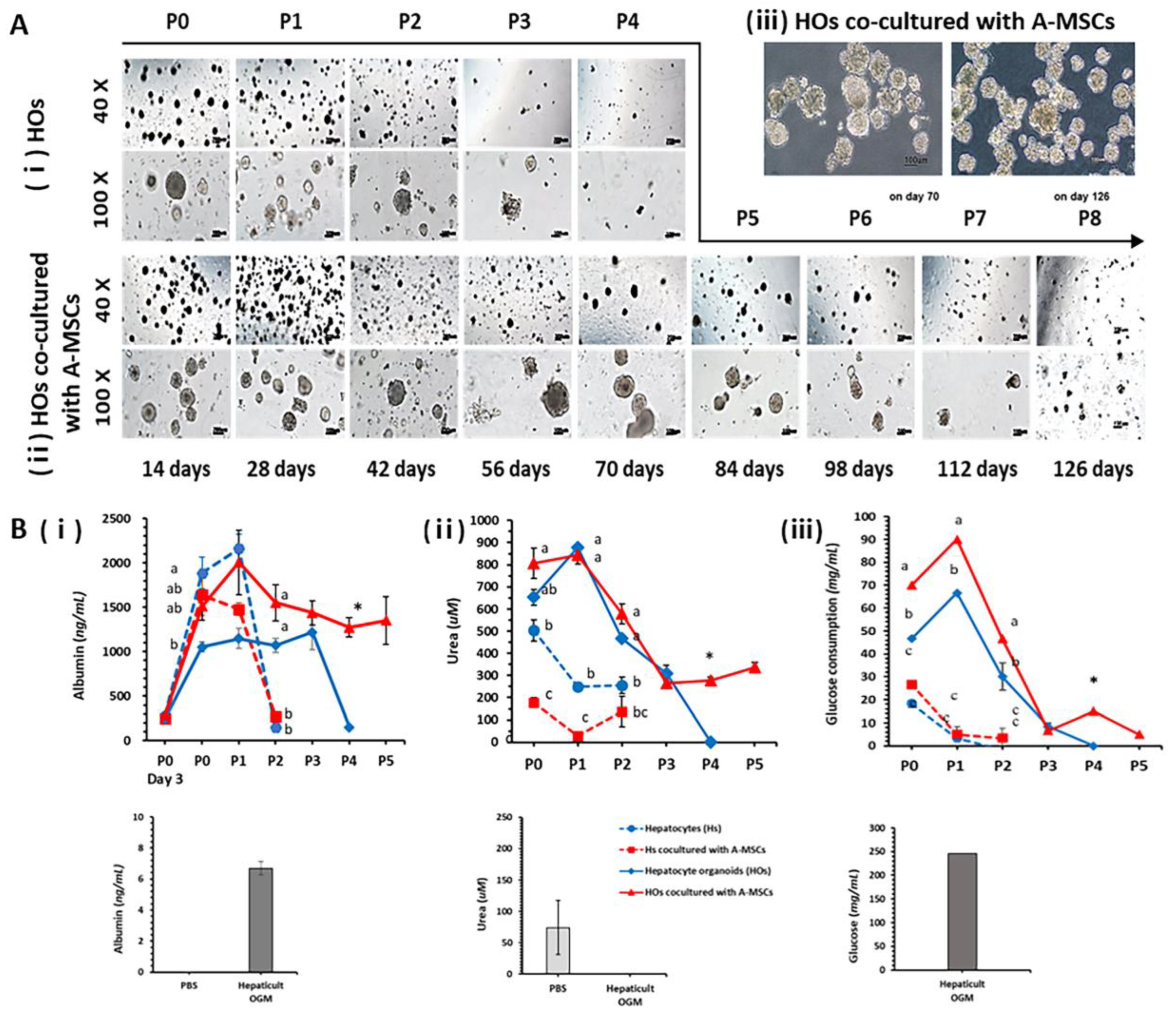
2.1. Generation of HO
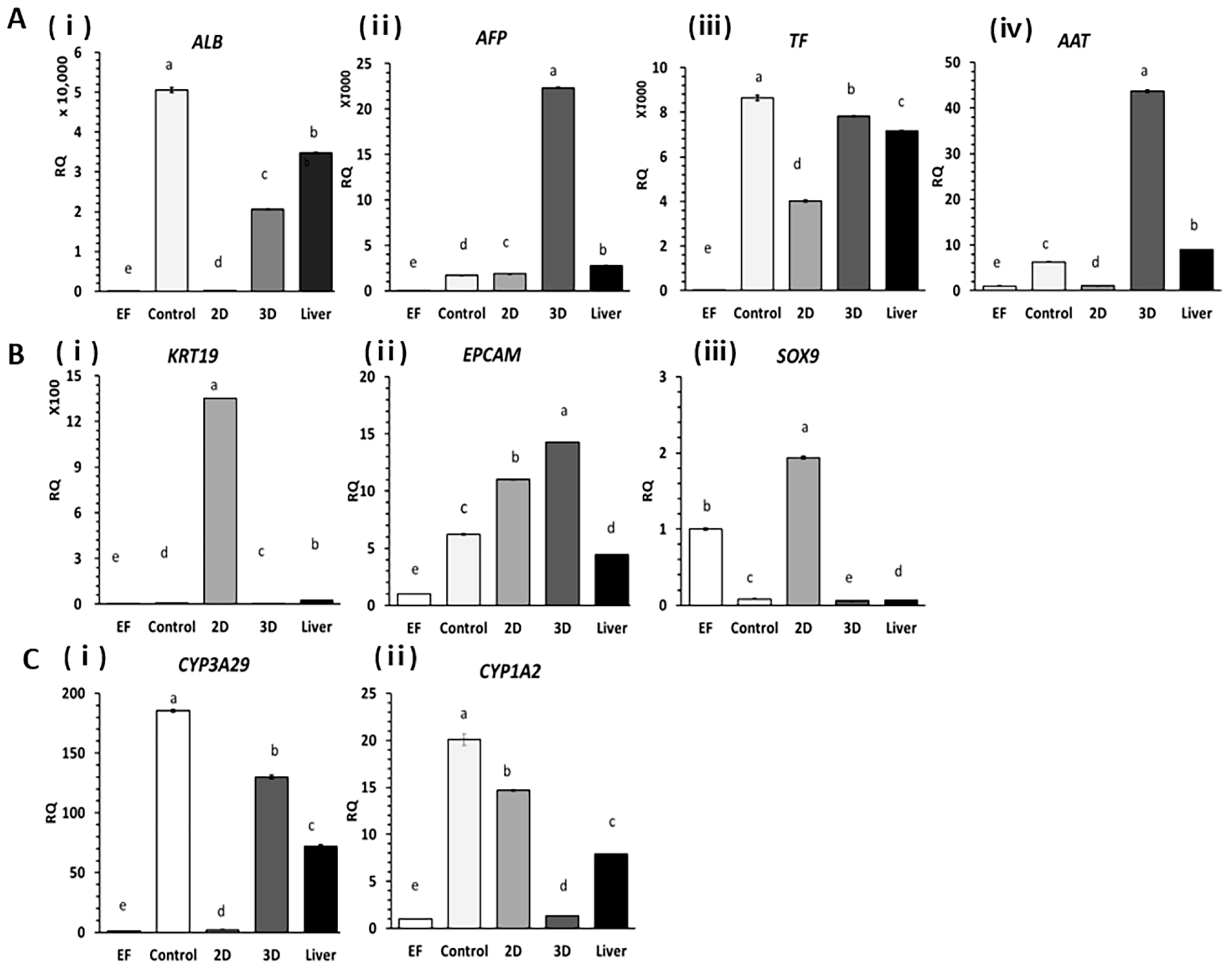
2.2. Expression of Liver-Specific Genes in HOs
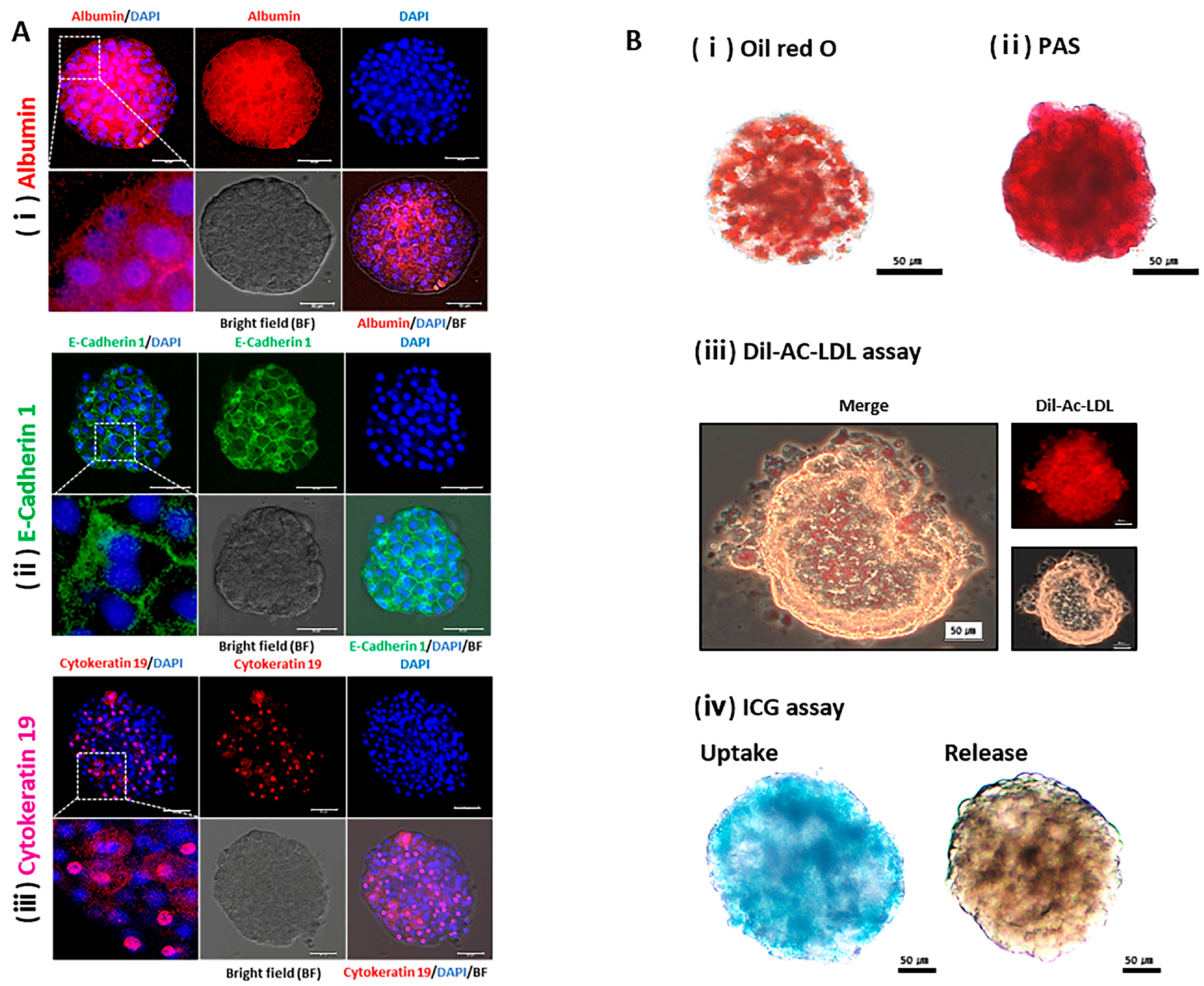
2.3. Physiological Characterization of the Liver using HOs
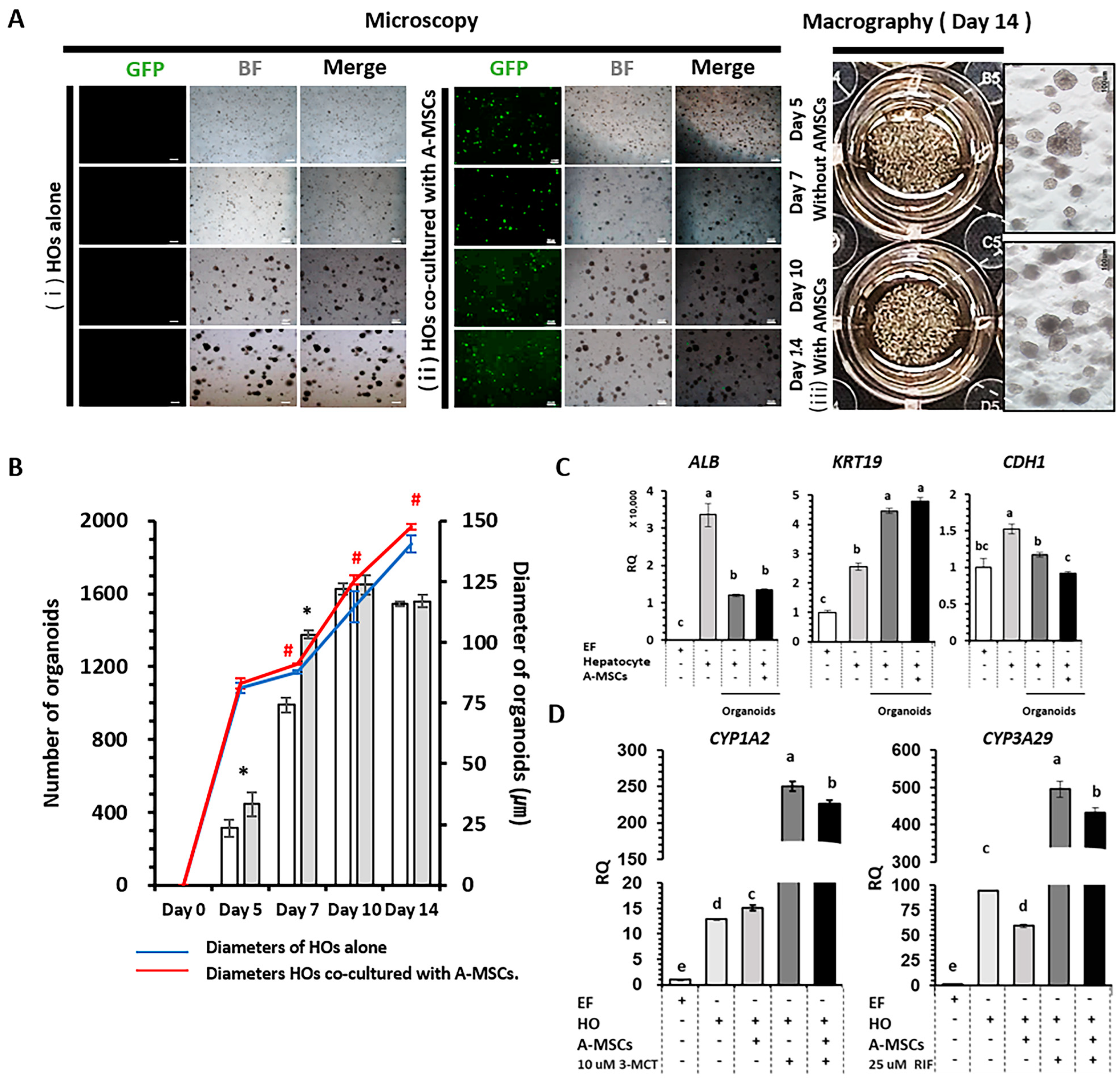
2.4. Co-Culture Effect of A-MSCs in Hepatocyte Cultures
2.5. Pharmacokinetics and Metabolic Capacity in HOs Co-Cultured with A-MSCs
2.6. Confirmation of In Vitro Expansion Ability in HOs
2.7. Analysis of Genes Related to Liver Function Maintenance and Lifespan during In Vitro Long-Term Culturing of HOs
3. Discussion
4. Materials and Methods
4.1. Chemicals and Medium
4.2. Experimental Samples and Ethics Statements
4.3. Generation of Hepatocyte Organoids
4.4. D and 3D Organoid Cultures of Hepatocytes for Analysis of Liver-Specific Genes
4.5. Immunofluorescence Staining
4.6. Liver Function Assay
4.7. Co-Culturing of HOs with A-MSCs
4.8. Assessment of Cytochrome P450 (CYP) Activity Using Inducers
4.9. Real-Time Reverse-Transcription Quantitative PCR (RT-qPCR)
4.10. Verification of Secretory Ability in Hepatocyte Organoids
4.11. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fisher, R.A.; Strom, S.C. Human hepatocyte transplantation: Worldwide results. Transplantation 2006, 82, 441–449. [Google Scholar] [CrossRef]
- Nguyen, D.G.; Funk, J.; Robbins, J.B.; Crogan-Grundy, C.; Presnell, S.C.; Singer, T.; Roth, A.B. Bioprinted 3D primary liver tissues allow assessment of organ-level response to clinical drug induced toxicity in vitro. PLoS ONE 2016, 11, e0158674. [Google Scholar] [CrossRef] [PubMed]
- Mun, S.J.; Ryu, J.S.; Lee, M.O.; Son, Y.S.; Oh, S.J.; Cho, H.S.; Son, M.Y.; Kim, D.S.; Kim, S.J.; Yoo, H.J.; et al. Generation of expandable human pluripotent stem cell-derived hepatocyte-like liver organoids. J. Hepatol. 2019, 71, 970–985. [Google Scholar] [CrossRef] [PubMed]
- Sampaziotis, F.; de Brito, M.C.; Madrigal, P.; Bertero, A.; Saeb-Parsy, K.; Soares, F.A.C.; Schrumpf, E.; Melum, E.; Karlsen, T.H.; Bradley, J.A.; et al. Cholangiocytes derived from human induced pluripotent stem cells for disease modeling and drug validation. Nat. Biotechnol. 2015, 33, 845–852. [Google Scholar] [CrossRef]
- Huang, P.; Zhang, L.; Gao, Y.; He, Z.; Yao, D.; Wu, Z.; Cen, J.; Chen, X.; Liu, C.; Hu, Y.; et al. Direct reprogramming of human fibroblasts to functional and expandable hepatocytes. Cell Stem Cell 2014, 14, 370–384. [Google Scholar] [CrossRef] [PubMed]
- Ullah, I.; Shin, Y.; Kim, Y.; Oh, K.B.; Hwang, S.; Kim, Y.I.; Lee, J.W.; Hur, T.Y.; Lee, S.; Ock, S.A. Effect of sex-specific differences on function of induced hepatocyte-like cells generated from male and female mouse embryonic fibroblasts. Stem Cell Res. Ther. 2021, 12, 79. [Google Scholar] [CrossRef]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, C.; Sachs, N.; Chiu, M.C.; Wong, B.H.; Chu, H.; Poon, V.K.; Wang, D.; Zhao, X.; Wen, L.; et al. Differentiated human airway organoids to assess infectivity of emerging influenza virus. Proc. Natl. Acad. Sci. USA 2018, 115, 6822–6827. [Google Scholar] [CrossRef]
- Huch, M.; Gehart, H.; van Boxtel, R.; Hamer, K.; Blokzijl, F.; Verstegen, M.M.; Ellis, E.; van Wenum, M.; Fuchs, S.A.; de Ligt, J.; et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 2015, 160, 299–312. [Google Scholar] [CrossRef]
- Hu, H.; Gehart, H.; Artegiani, B.; Löpez-Iglesias, C.; Dekkers, F.; Basak, O.; van Es, J.; Chuva de Sousa Lopes, S.M.; Begthel, H.; Korving, J.; et al. Long-term expansion of functional mouse and human hepatocytes as 3d organoids. Cell 2018, 175, 1591–1606.e19. [Google Scholar] [CrossRef]
- Hendriks, D.; Brouwers, J.F.; Hamer, K.; Geurts, M.H.; Luciana, L.; Massalini, S.; López-Iglesias, C.; Peters, P.J.; Rodríguez-Colman, M.J.; Chuva de Sousa Lopes, S.; et al. Engineered human hepatocyte organoids enable CRISPR-based target discovery and drug screening for steatosis. Nat. Biotechnol. 2023, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Khetani, S.R.; Bhatia, S.N. Microscale culture of human liver cells for drug development. Nat. Biotechnol. 2008, 26, 120–126. [Google Scholar] [CrossRef]
- Takebe, T.; Enomura, M.; Yoshizawa, E.; Kimura, M.; Koike, H.; Ueno, Y.; Matsuzaki, T.; Yamazaki, T.; Toyohara, T.; Osafune, K.; et al. Vascularized and complex organ buds from diverse tissues via mesenchymal cell-driven condensation. Cell Stem Cell 2015, 16, 556–565. [Google Scholar] [CrossRef]
- He, Y.T.; Zhu, X.L.; Li, S.F.; Zhang, B.Q.; Li, Y.; Wu, Q.; Zhang, Y.L.; Zhou, Y.Y.; Li, L.; Qi, Y.N.; et al. Creating rat hepatocyte organoid as an in vitro model for drug testing. World. J. Stem Cells 2020, 12, 1184–1195. [Google Scholar] [CrossRef]
- Niu, Q.; Wang, T.; Wang, Z.; Wang, F.; Huang, D.; Sun, H.; Liu, H. Adipose-derived mesenchymal stem cell-secreted extracellular vesicles alleviate non-alcoholic fatty liver disease via delivering miR-223-3p. Adipocyte 2022, 11, 572–587. [Google Scholar] [CrossRef]
- Ock, S.A.; Lee, Y.M.; Park, J.S.; Shivakumar, S.B.; Moon, S.W.; Sung, N.J.; Lee, W.J.; Jang, S.J.; Park, J.M.; Lee, S.C. Evaluation of phenotypic, Functional and Molecular Characteristics of Porcine Mesenchymal stromal/stem cells Depending on Donor Age, Gender and Tissue Source. J. Vet. Med. Sci. 2016, 78, 987–995. [Google Scholar] [CrossRef]
- Chen, H.S.; Joo, D.J.; Shaheen, M.; Li, Y.; Wang, Y.; Yang, J.; Nicolas, C.T.; Predmore, K.; Amiot, B.; Michalak, G.; et al. Randomized trial of spheroid reservoir bioartificial liver in porcine model of posthepatectomy liver failure. Hepatology 2019, 69, 329–342. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, J.; Ren, H.; Liu, Y.; Xiang, C.; Li, C.; Lu, S.; Shi, Y.; Deng, H.; Shi, X. Hepatic spheroids derived from human induced pluripotent stem cells in bio-artificial liver rescue porcine acute liver failure. Cell Res. 2020, 30, 95–97. [Google Scholar] [CrossRef] [PubMed]
- Hochleitner, B.; Hengster, P.; Duo, L.; Bucher, H.; Klima, G.; Margreiter, R. A novel bioartificial liver with culture of porcine hepatocyte aggregates under simulated microgravity. Artif. Organs 2005, 29, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wu, Q.; Wang, Y.; Weng, C.; He, Y.; Gao, M.; Yang, G.; Li, L.; Chen, F.; Shi, Y.; et al. Novel spheroid reservoir bioartificial liver improves survival of nonhuman primates in a toxin-induced model of acute liver failure. Theranostics 2018, 8, 5562–5574. [Google Scholar] [CrossRef]
- Krüger, M.; Samsom, R.A.; Oosterhoff, L.A.; van Wolferen, M.E.; Kooistra, H.S.; Geijsen, N.; Penning, L.C.; Kock, L.M.; Sainz-Arnal, P.; Baptista, P.M.; et al. High level of polarized engraftment of porcine intrahepatic cholangiocyte organoids in decellularized liver scaffolds. J. Cell. Mol. Med. 2022, 26, 4949–4958. [Google Scholar] [CrossRef] [PubMed]
- Yanger, K.; Knigin, D.; Zong, Y.; Maggs, L.; Gu, G.; Akiyama, H.; Pikarsky, E.; Stanger, B.Z. Adult hepatocytes are generated by self-duplication rather than stem cell differentiation. Cell Stem Cell 2014, 15, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.Y.; Lu, R.M.; Liao, M.Y.; Yu, J.; Chung, C.H.; Kao, C.F.; Wu, H.C. Epithelial cell adhesion molecule regulation is associated with the maintenance of the undifferentiated phenotype of human embryonic stem cells. J. Biol. Chem. 2010, 285, 8719–8732. [Google Scholar] [CrossRef] [PubMed]
- Ng, V.Y.; Ang, S.N.; Chan, J.X.; Choo, A.B. Characterization of epithelial cell adhesion molecule as a surface marker on undifferentiated human embryonic stem cells. Stem Cells 2010, 28, 29–35. [Google Scholar] [CrossRef]
- Gires, O. EpCAM in hepatocytes and their progenitors. J. Hepatol. 2012, 56, 490–492. [Google Scholar] [CrossRef]
- Aini, W.; Miyagawa-Hayashino, A.; Ozeki, M.; Adeeb, S.; Hirata, M.; Tamaki, K.; Uemoto, S.; Haga, H. Accelerated telomere reduction and hepatocyte senescence in tolerated human liver allografts. Transpl. Immunol. 2014, 31, 55–59. [Google Scholar] [CrossRef]
- Levy, G.; Bomze, D.; Heinz, S.; Ramachandran, S.D.; Noerenberg, A.; Cohen, M.; Shibolet, O.; Sklan, E.; Braspenning, J.; Nahmias, Y. Long-term culture and expansion of primary human hepatocytes. Nat. Biotechnol. 2015, 33, 1264–1271. [Google Scholar] [CrossRef] [PubMed]
- Alzebdeh, D.A.; Matthew, H.W. Metabolic oscillations in co-cultures of hepatocytes and mesenchymal stem cells: Effects of seeding arrangement and culture mixing. J. Cell. Biochem. 2017, 118, 3003–3015. [Google Scholar] [CrossRef]
- Ma, J.; Li, Y.; Wu, M.; Li, X. Oxidative stress-mediated p53/p21WAF1/CIP1 pathway may be involved in microcystin-LR-induced cytotoxicity in HepG2 cells. Chemosphere 2018, 194, 773–783. [Google Scholar] [CrossRef]
- Knobeloch, D.; Ehnert, S.; Schyschka, L.; Büchler, P.; Schoenberg, M.; Kleeff, J.; Thasler, W.E.; Nussler, N.C.; Godoy, P.; Hengstler, J.; et al. Human hepatocytes: Isolation, culture, and quality procedures. Methods Mol. Biol. 2012, 806, 99–120. [Google Scholar] [CrossRef]
- Seglen, P.O. Preparation of isolated rat liver cells. Methods Cell Biol. 1976, 13, 29–83. [Google Scholar] [CrossRef] [PubMed]
- Lecluyse, E.L.; Alexandre, E. Isolation and culture of primary hepatocytes from resected human liver tissue. Methods Mol. Biol. 2010, 640, 57–82. [Google Scholar] [CrossRef]
- Martignoni, M.; Groothuis, G.M.; de-de Kanter, R. Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Expert Opin. Drug Metab. Toxicol. 2006, 2, 875–894. [Google Scholar] [CrossRef]
- Kakuni, M.; Yamasaki, C.; Tachibana, A.; Yoshizane, Y.; Ishida, Y.; Tateno, C. Chimeric mice with humanized livers: A unique tool for in vivo and in vitro enzyme induction studies. Int. J. Mol. Sci. 2013, 15, 58–74. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.T.; Lee, S.C.; Gao, Y.; Kim, K.P.; Song, G.; An, S.Y.; Adachi, K.; Jang, Y.J.; Kim, J.; Oh, K.J.; et al. Small Molecules Facilitate Single Factor-Mediated Hepatic Reprogramming. Cell Rep. 2016, 15, 814–829. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ock, S.A.; Kim, S.-Y.; Ju, W.S.; Kim, Y.-I.; Wi, H.-Y.; Lee, P. Adipose Tissue-Derived Mesenchymal Stem Cells Extend the Lifespan and Enhance Liver Function in Hepatocyte Organoids. Int. J. Mol. Sci. 2023, 24, 15429. https://doi.org/10.3390/ijms242015429
Ock SA, Kim S-Y, Ju WS, Kim Y-I, Wi H-Y, Lee P. Adipose Tissue-Derived Mesenchymal Stem Cells Extend the Lifespan and Enhance Liver Function in Hepatocyte Organoids. International Journal of Molecular Sciences. 2023; 24(20):15429. https://doi.org/10.3390/ijms242015429
Chicago/Turabian StyleOck, Sun A, Seo-Yeon Kim, Won Seok Ju, Young-Im Kim, Ha-Yeon Wi, and Poongyeon Lee. 2023. "Adipose Tissue-Derived Mesenchymal Stem Cells Extend the Lifespan and Enhance Liver Function in Hepatocyte Organoids" International Journal of Molecular Sciences 24, no. 20: 15429. https://doi.org/10.3390/ijms242015429
APA StyleOck, S. A., Kim, S.-Y., Ju, W. S., Kim, Y.-I., Wi, H.-Y., & Lee, P. (2023). Adipose Tissue-Derived Mesenchymal Stem Cells Extend the Lifespan and Enhance Liver Function in Hepatocyte Organoids. International Journal of Molecular Sciences, 24(20), 15429. https://doi.org/10.3390/ijms242015429





