Novel Mouse Cell Lines and In Vivo Models for Human High-Grade Neuroendocrine Lung Carcinoma, Small Cell Lung Carcinoma (SCLC), and Large Cell Neuroendocrine Carcinoma (LCNEC)
Abstract
:1. Introduction
2. Results
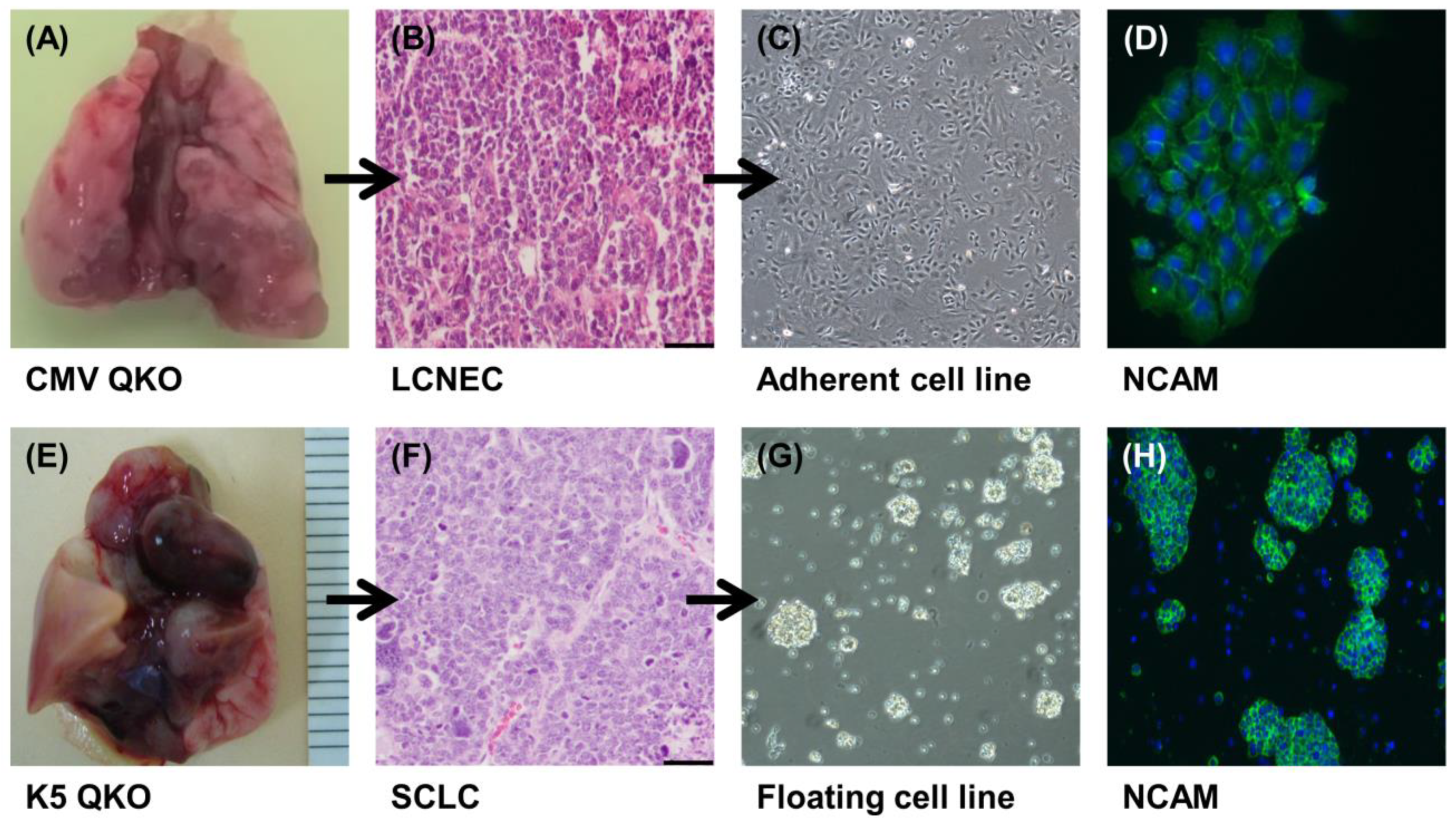
2.1. Generation of LCNEC and SCLC Cell Lines
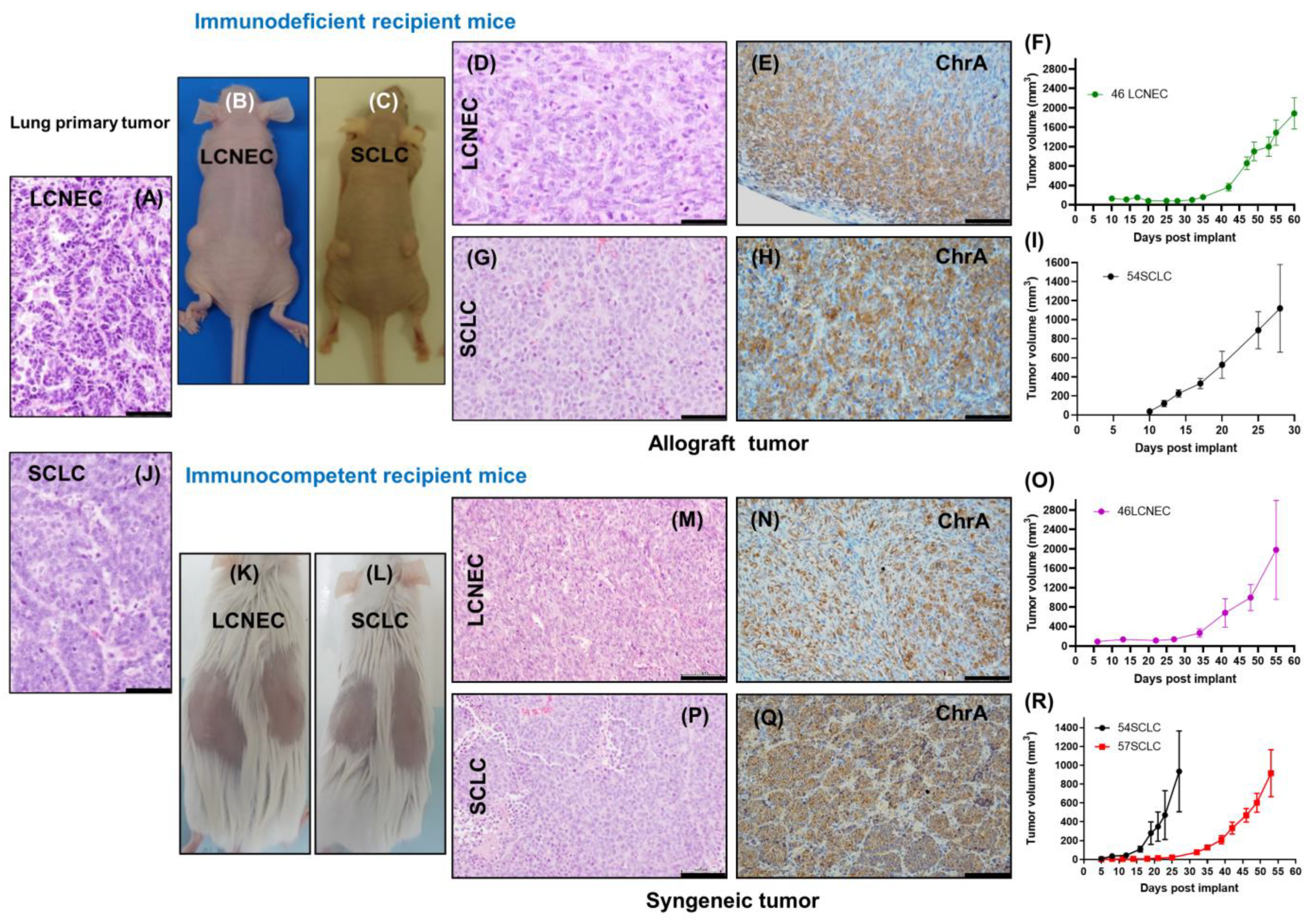
2.2. Evaluation of In Vivo Tumorigenic Potential of LCNEC and SCLC Cell Lines
2.3. The SCLC Syngeneic Tumors Showed the Ability to Form Metastasis
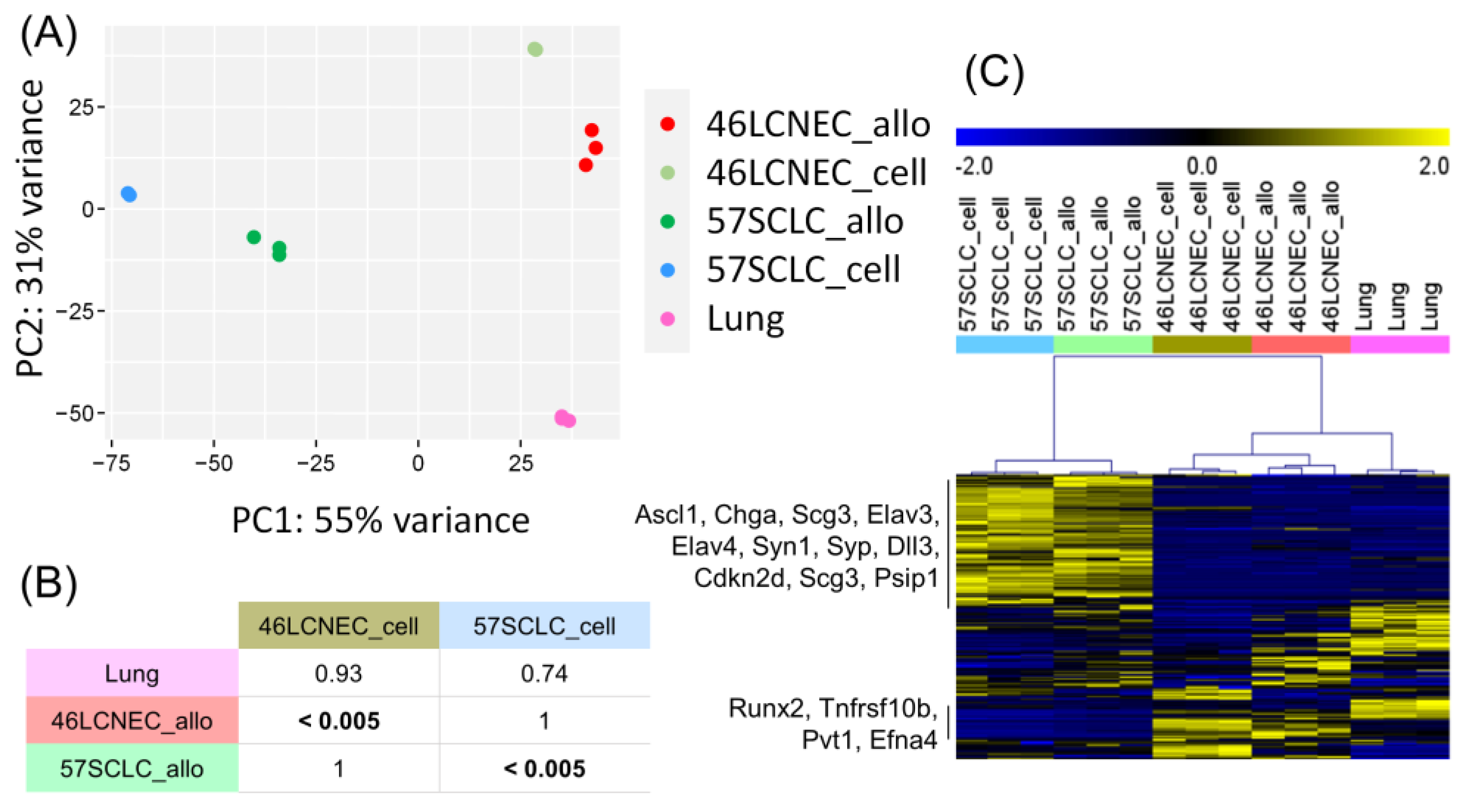
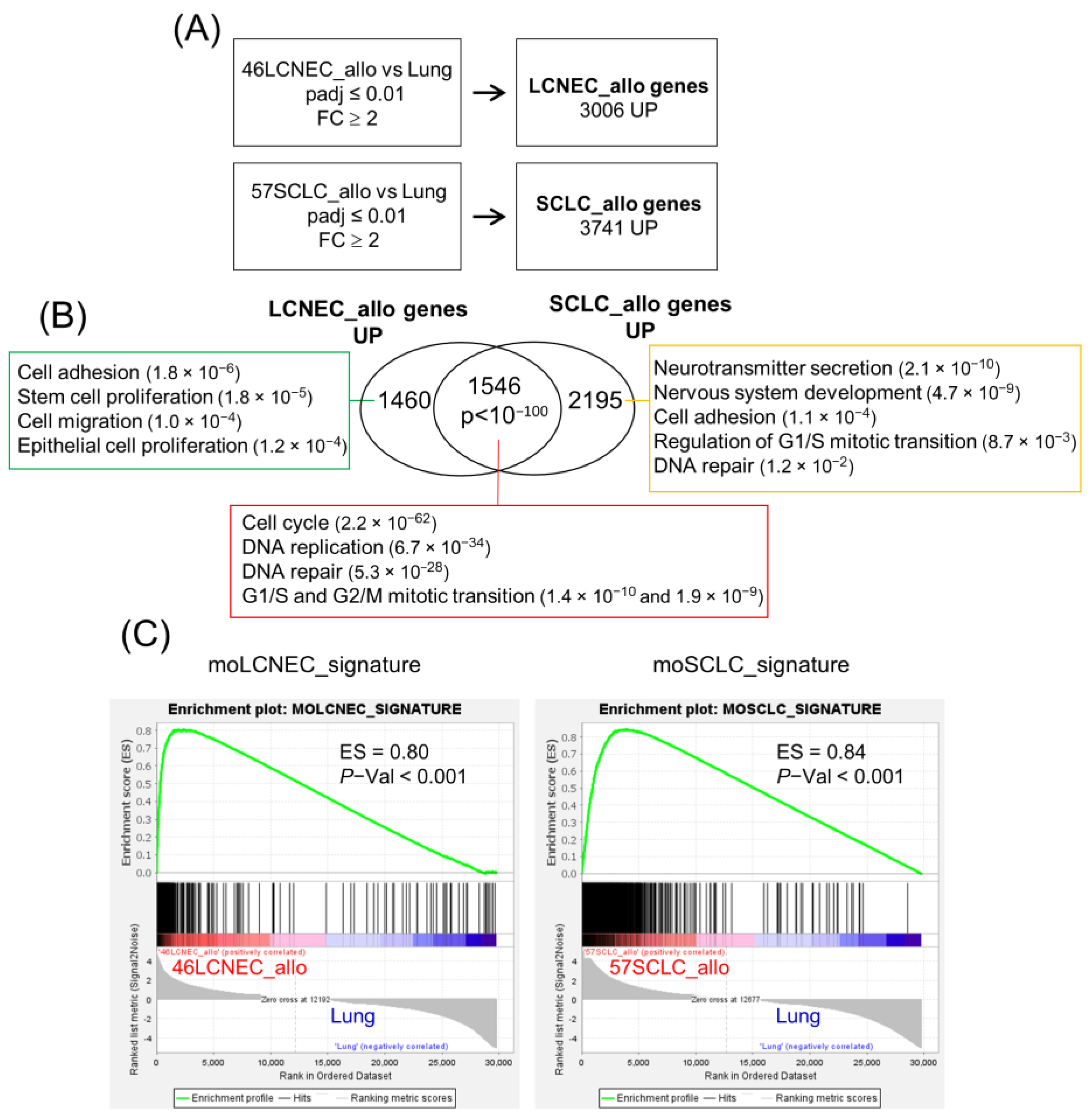
2.4. Transcriptional Profile of Mouse LCNEC and SCLC Cell Lines and Allografts
3. Discussion
4. Materials and Methods
4.1. Donor and Recipient Mice
4.2. Establishment of SCLC and LCNEC Murine Cell Lines and Cell Culture
4.3. Evaluation of Tumorigenic and Metastatic Potential
4.4. Genotyping
4.5. Immunofluorescence
4.6. Histology and Immunohistochemistry
4.7. RNA Isolation and Gene Expression Profiling
4.8. Gene Set Enrichment Analysis
4.9. Subclass Association
4.10. Human LCNEC/SCLC Classifier
4.11. Analysis of Biological Pathways
4.12. Data and Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rudin, C.M.; Brambilla, E.; Faivre-Finn, C.; Sage, J. Small-cell lung cancer. Nat. Rev. Dis. Primers. 2021, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Andrini, E.; Marchese, P.V.; De Biase, D.; Mosconi, C.; Siepe, G.; Panzuto, F.; Ardizzoni, A.; Campana, D.; Lamberti, G. Large Cell Neuroendocrine Carcinoma of the Lung: Current Understanding and Challenges. J. Clin. Med. 2022, 11, 1461. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, G.; Sonzogni, A.; Harari, S.; Albini, A.; Bresaola, E.; Marchio, C.; Massa, F.; Righi, L.; Gatti, G.; Papanikolaou, N.; et al. Classification of pulmonary neuroendocrine tumors: New insights. Transl. Lung Cancer Res. 2017, 6, 513–529. [Google Scholar] [CrossRef]
- Rudin, C.M.; Poirier, J.T.; Byers, L.A.; Dive, C.; Dowlati, A.; George, J.; Heymach, J.V.; Johnson, J.E.; Lehman, J.M.; MacPherson, D.; et al. Molecular subtypes of small cell lung cancer: A synthesis of human and mouse model data. Nat. Rev. 2019, 19, 289–297. [Google Scholar] [CrossRef]
- Gay, C.M.; Stewart, C.A.; Park, E.M.; Diao, L.; Groves, S.M.; Heeke, S.; Nabet, B.Y.; Fujimoto, J.; Solis, L.M.; Lu, W.; et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell 2021, 39, 346–360.e347. [Google Scholar] [CrossRef] [PubMed]
- George, J.; Walter, V.; Peifer, M.; Alexandrov, L.B.; Seidel, D.; Leenders, F.; Maas, L.; Muller, C.; Dahmen, I.; Delhomme, T.M.; et al. Integrative genomic profiling of large-cell neuroendocrine carcinomas reveals distinct subtypes of high-grade neuroendocrine lung tumors. Nat. Commun. 2018, 9, 1048. [Google Scholar] [CrossRef]
- Lantuejoul, S.; Fernandez-Cuesta, L.; Damiola, F.; Girard, N.; McLeer, A. New molecular classification of large cell neuroendocrine carcinoma and small cell lung carcinoma with potential therapeutic impacts. Transl. Lung Cancer Res. 2020, 9, 2233–2244. [Google Scholar] [CrossRef]
- Lowczak, A.; Kolasinska-Cwikla, A.; Osowiecka, K.; Glinka, L.; Palucki, J.; Rzepko, R.; Doboszynska, A.; Cwikla, J.B. Outcomes of Patients with Pulmonary Large Cell Neuroendocrine Carcinoma in I-IV Stage. Medicina 2021, 57, 118. [Google Scholar] [CrossRef]
- Liang, J.; Guan, X.; Bao, G.; Yao, Y.; Zhong, X. Molecular subtyping of small cell lung cancer. Semin. Cancer Biol. 2022, 86, 450–462. [Google Scholar] [CrossRef]
- Drapkin, B.J.; Rudin, C.M. Advances in Small-Cell Lung Cancer (SCLC) Translational Research. Cold Spring Harb. Perspect. Med. 2021, 11, a038240. [Google Scholar] [CrossRef]
- Horn, L.; Mansfield, A.S.; Szczesna, A.; Havel, L.; Krzakowski, M.; Hochmair, M.J.; Huemer, F.; Losonczy, G.; Johnson, M.L.; Nishio, M.; et al. First-Line Atezolizumab plus Chemotherapy in Extensive-Stage Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 379, 2220–2229. [Google Scholar] [CrossRef] [PubMed]
- Paz-Ares, L.; Chen, Y.; Reinmuth, N.; Hotta, K.; Trukhin, D.; Statsenko, G.; Hochmair, M.J.; Ozguroglu, M.; Ji, J.H.; Garassino, M.C.; et al. Durvalumab, with or without tremelimumab, plus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer: 3-year overall survival update from CASPIAN. ESMO Open 2022, 7, 100408. [Google Scholar] [CrossRef]
- Ferone, G.; Lee, M.C.; Sage, J.; Berns, A. Cells of origin of lung cancers: Lessons from mouse studies. Genes Dev. 2020, 34, 1017–1032. [Google Scholar] [CrossRef] [PubMed]
- Lorz, C.; Oteo, M.; Santos, M. Neuroendocrine Lung Cancer Mouse Models: An Overview. Cancers 2020, 13, 14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Girard, L.; Zhang, Y.A.; Haruki, T.; Papari-Zareei, M.; Stastny, V.; Ghayee, H.K.; Pacak, K.; Oliver, T.G.; Minna, J.D.; et al. Small cell lung cancer tumors and preclinical models display heterogeneity of neuroendocrine phenotypes. Transl. Lung Cancer Res. 2018, 7, 32–49. [Google Scholar] [CrossRef] [PubMed]
- Jahchan, N.S.; Lim, J.S.; Bola, B.; Morris, K.; Seitz, G.; Tran, K.Q.; Xu, L.; Trapani, F.; Morrow, C.J.; Cristea, S.; et al. Identification and Targeting of Long-Term Tumor-Propagating Cells in Small Cell Lung Cancer. Cell Rep. 2016, 16, 644–656. [Google Scholar] [CrossRef]
- Weiskopf, K.; Jahchan, N.S.; Schnorr, P.J.; Cristea, S.; Ring, A.M.; Maute, R.L.; Volkmer, A.K.; Volkmer, J.P.; Liu, J.; Lim, J.S.; et al. CD47-blocking immunotherapies stimulate macrophage-mediated destruction of small-cell lung cancer. J. Clin. Investig. 2016, 126, 2610–2620. [Google Scholar] [CrossRef]
- Shue, Y.T.; Lim, J.S.; Sage, J. Tumor heterogeneity in small cell lung cancer defined and investigated in pre-clinical mouse models. Transl. Lung Cancer Res. 2018, 7, 21–31. [Google Scholar] [CrossRef]
- Lallo, A.; Gulati, S.; Schenk, M.W.; Khandelwal, G.; Berglund, U.W.; Pateras, I.S.; Chester, C.P.E.; Pham, T.M.; Kalderen, C.; Frese, K.K.; et al. Ex vivo culture of cells derived from circulating tumour cell xenograft to support small cell lung cancer research and experimental therapeutics. Br. J. Pharmacol. 2019, 176, 436–450. [Google Scholar] [CrossRef]
- Santos, M. New models of large-cell neuroendocrine carcinoma and small-cell lung carcinoma. Mol. Cell. Oncol. 2020, 7, 1702413. [Google Scholar] [CrossRef]
- Lazaro, S.; Perez-Crespo, M.; Lorz, C.; Bernardini, A.; Oteo, M.; Enguita, A.B.; Romero, E.; Hernandez, P.; Tomas, L.; Morcillo, M.A.; et al. Differential development of large-cell neuroendocrine or small-cell lung carcinoma upon inactivation of 4 tumor suppressor genes. Proc. Natl. Acad. Sci. USA 2019, 116, 22300–22306. [Google Scholar] [CrossRef]
- Lazaro, S.; Lorz, C.; Enguita, A.B.; Seller, I.; Paramio, J.M.; Santos, M. Pten and p53 Loss in the Mouse Lung Causes Adenocarcinoma and Sarcomatoid Carcinoma. Cancers 2022, 14, 3671. [Google Scholar] [CrossRef]
- Ko, J.; Winslow, M.M.; Sage, J. Mechanisms of small cell lung cancer metastasis. EMBO Mol. Med. 2021, 13, e13122. [Google Scholar] [CrossRef] [PubMed]
- Reich, M.; Liefeld, T.; Gould, J.; Lerner, J.; Tamayo, P.; Mesirov, J.P. GenePattern 2.0. Nat. Genet. 2006, 38, 500–501. [Google Scholar] [CrossRef] [PubMed]
- Hoshida, Y.; Brunet, J.P.; Tamayo, P.; Golub, T.R.; Mesirov, J.P. Subclass mapping: Identifying common subtypes in independent disease data sets. PLoS ONE 2007, 2, e1195. [Google Scholar] [CrossRef] [PubMed]
- Tlemsani, C.; Pongor, L.; Elloumi, F.; Girard, L.; Huffman, K.E.; Roper, N.; Varma, S.; Luna, A.; Rajapakse, V.N.; Sebastian, R.; et al. SCLC-CellMiner: A Resource for Small Cell Lung Cancer Cell Line Genomics and Pharmacology Based on Genomic Signatures. Cell Rep. 2020, 33, 108296. [Google Scholar] [CrossRef]
- Gazdar, A.F.; Savage, T.K.; Johnson, J.E.; Berns, A.; Sage, J.; Linnoila, R.I.; MacPherson, D.; McFadden, D.G.; Farago, A.; Jacks, T.; et al. The comparative pathology of genetically engineered mouse models for neuroendocrine carcinomas of the lung. J. Thorac. Oncol. 2015, 10, 553–564. [Google Scholar] [CrossRef]
- George, J.; Lim, J.S.; Jang, S.J.; Cun, Y.; Ozretic, L.; Kong, G.; Leenders, F.; Lu, X.; Fernandez-Cuesta, L.; Bosco, G.; et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015, 524, 47–53. [Google Scholar] [CrossRef]
- Gazdar, A.F.; Girard, L.; Lockwood, W.W.; Lam, W.L.; Minna, J.D. Lung cancer cell lines as tools for biomedical discovery and research. J. Natl. Cancer Inst. 2010, 102, 1310–1321. [Google Scholar] [CrossRef]
- Calbo, J.; van Montfort, E.; Proost, N.; van Drunen, E.; Beverloo, H.B.; Meuwissen, R.; Berns, A. A functional role for tumor cell heterogeneity in a mouse model of small cell lung cancer. Cancer Cell 2011, 19, 244–256. [Google Scholar] [CrossRef]
- Kwon, M.C.; Proost, N.; Song, J.Y.; Sutherland, K.D.; Zevenhoven, J.; Berns, A. Paracrine signaling between tumor subclones of mouse SCLC: A critical role of ETS transcription factor Pea3 in facilitating metastasis. Genes Dev. 2015, 29, 1587–1592. [Google Scholar] [CrossRef] [PubMed]
- Denny, S.K.; Yang, D.; Chuang, C.H.; Brady, J.J.; Lim, J.S.; Gruner, B.M.; Chiou, S.H.; Schep, A.N.; Baral, J.; Hamard, C.; et al. Nfib Promotes Metastasis through a Widespread Increase in Chromatin Accessibility. Cell 2016, 166, 328–342. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Bioll. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, P3. [Google Scholar] [CrossRef]
- Hosack, D.A.; Dennis, G., Jr.; Sherman, B.T.; Lane, H.C.; Lempicki, R.A. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003, 4, R70. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Recuero, E.; Lázaro, S.; Lorz, C.; Enguita, A.B.; Garcia-Escudero, R.; Santos, M. Novel Mouse Cell Lines and In Vivo Models for Human High-Grade Neuroendocrine Lung Carcinoma, Small Cell Lung Carcinoma (SCLC), and Large Cell Neuroendocrine Carcinoma (LCNEC). Int. J. Mol. Sci. 2023, 24, 15284. https://doi.org/10.3390/ijms242015284
Recuero E, Lázaro S, Lorz C, Enguita AB, Garcia-Escudero R, Santos M. Novel Mouse Cell Lines and In Vivo Models for Human High-Grade Neuroendocrine Lung Carcinoma, Small Cell Lung Carcinoma (SCLC), and Large Cell Neuroendocrine Carcinoma (LCNEC). International Journal of Molecular Sciences. 2023; 24(20):15284. https://doi.org/10.3390/ijms242015284
Chicago/Turabian StyleRecuero, Enrique, Sara Lázaro, Corina Lorz, Ana Belén Enguita, Ramón Garcia-Escudero, and Mirentxu Santos. 2023. "Novel Mouse Cell Lines and In Vivo Models for Human High-Grade Neuroendocrine Lung Carcinoma, Small Cell Lung Carcinoma (SCLC), and Large Cell Neuroendocrine Carcinoma (LCNEC)" International Journal of Molecular Sciences 24, no. 20: 15284. https://doi.org/10.3390/ijms242015284
APA StyleRecuero, E., Lázaro, S., Lorz, C., Enguita, A. B., Garcia-Escudero, R., & Santos, M. (2023). Novel Mouse Cell Lines and In Vivo Models for Human High-Grade Neuroendocrine Lung Carcinoma, Small Cell Lung Carcinoma (SCLC), and Large Cell Neuroendocrine Carcinoma (LCNEC). International Journal of Molecular Sciences, 24(20), 15284. https://doi.org/10.3390/ijms242015284






