Exosomal miR-205-5p Improves Endometrial Receptivity by Upregulating E-Cadherin Expression through ZEB1 Inhibition
Abstract
:1. Introduction
2. Results
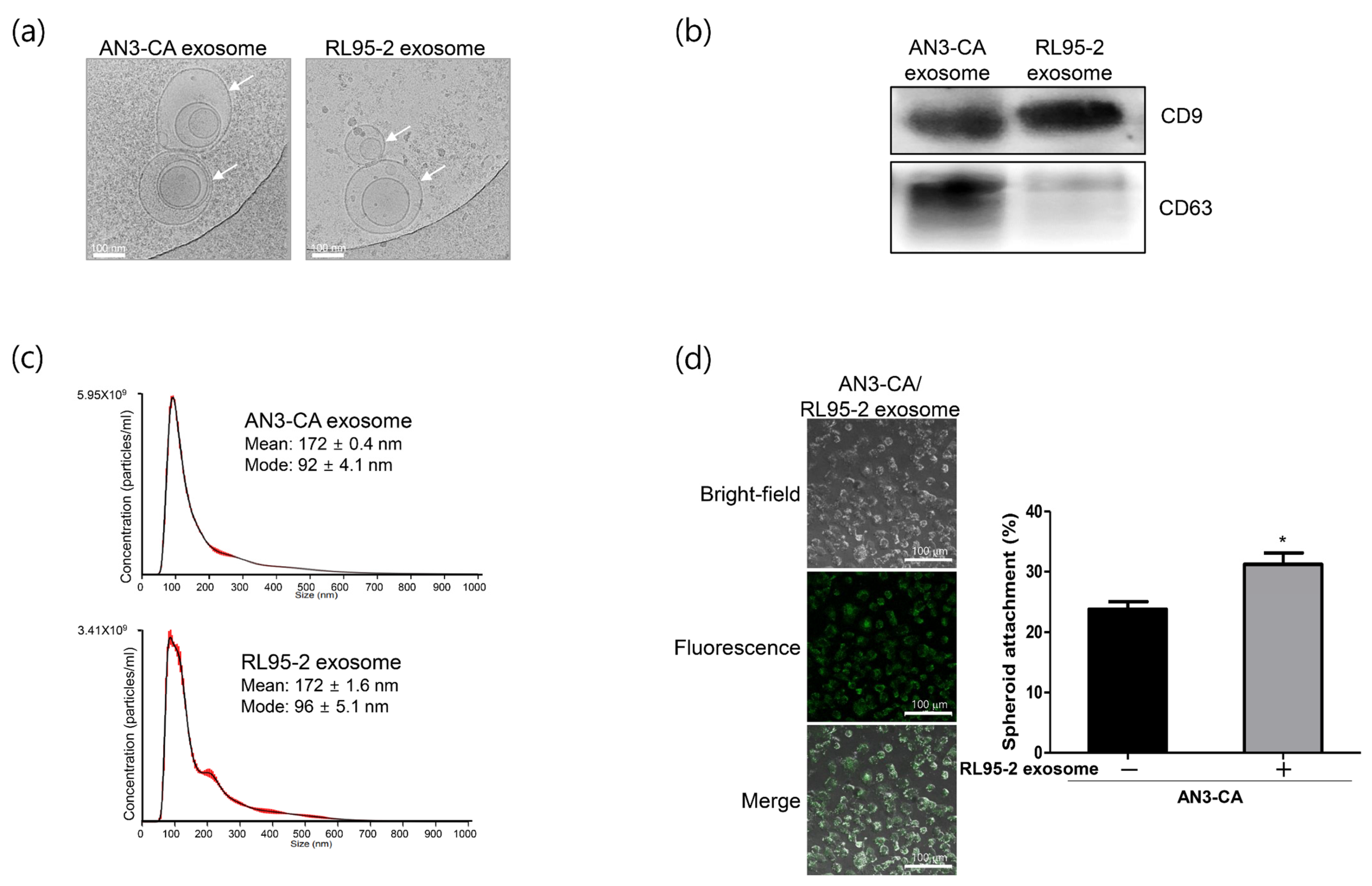
2.1. Characterization of Exosomes Derived from EECs Reveals Differences in Endometrial Receptivity
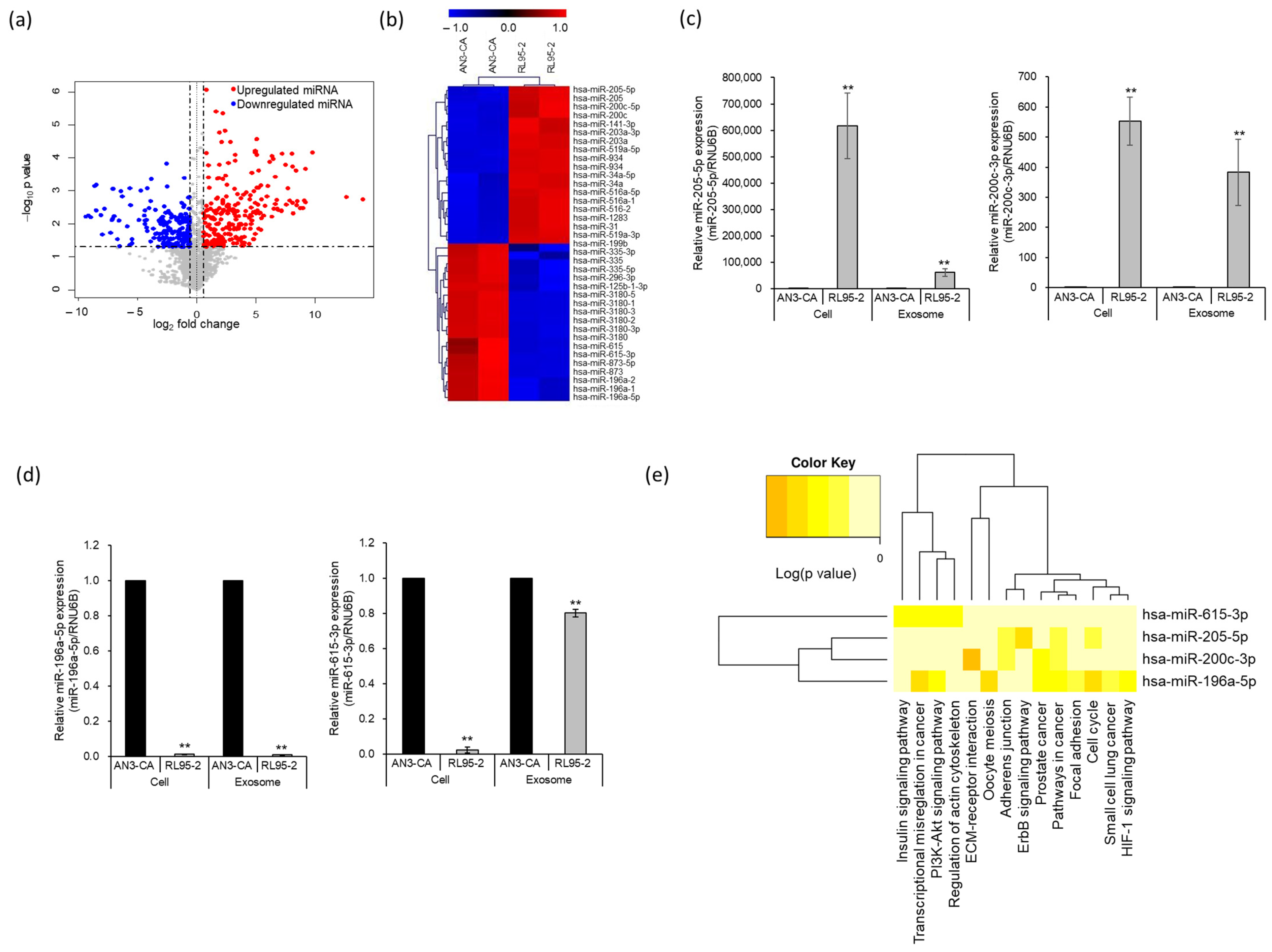
2.2. Identification of Differentially Expressed miRNAs in EEC-Derived Exosomes
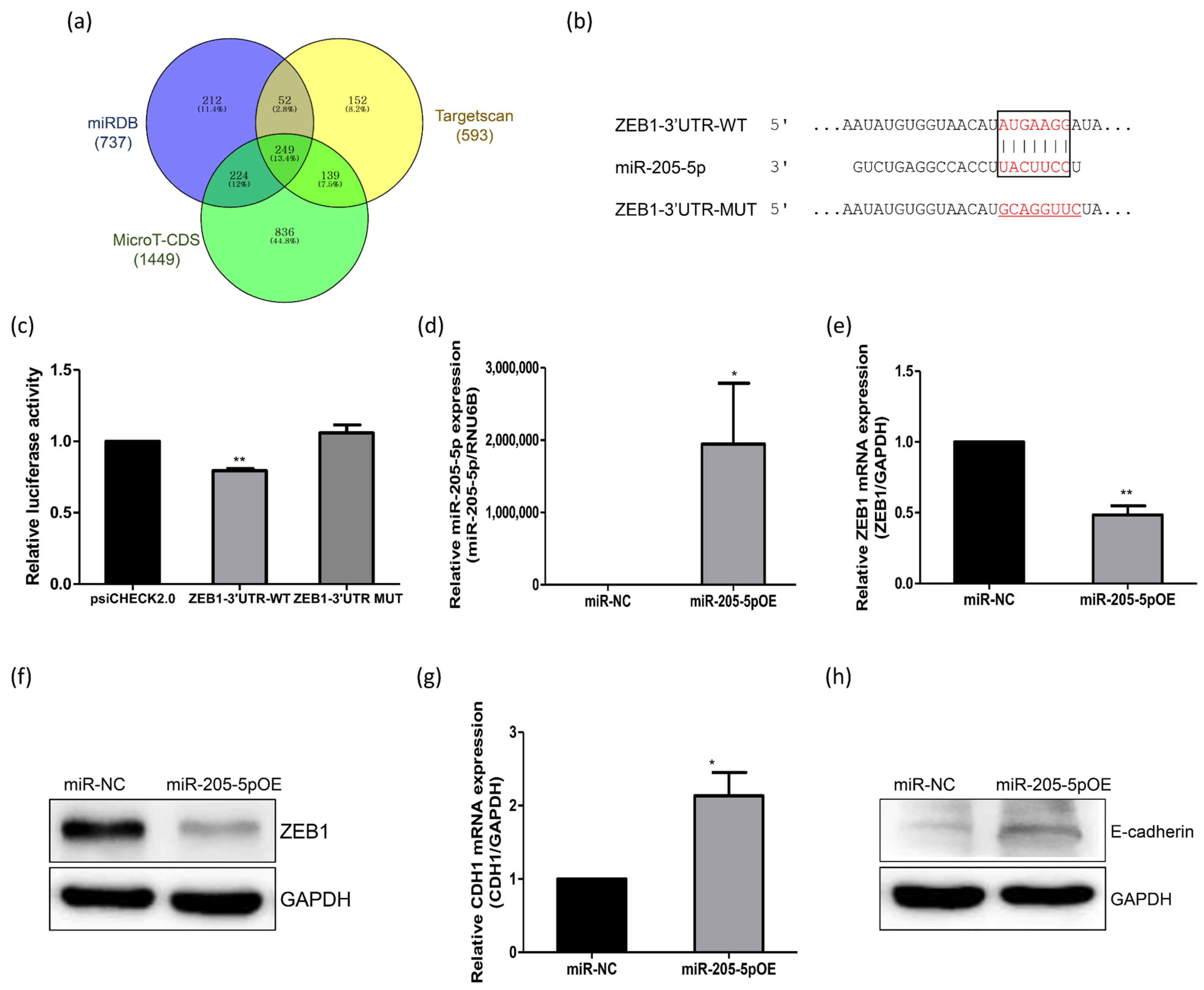
2.3. Identification of miR-205-5p Target Genes Related to the Adhesion of Endometrial Cells
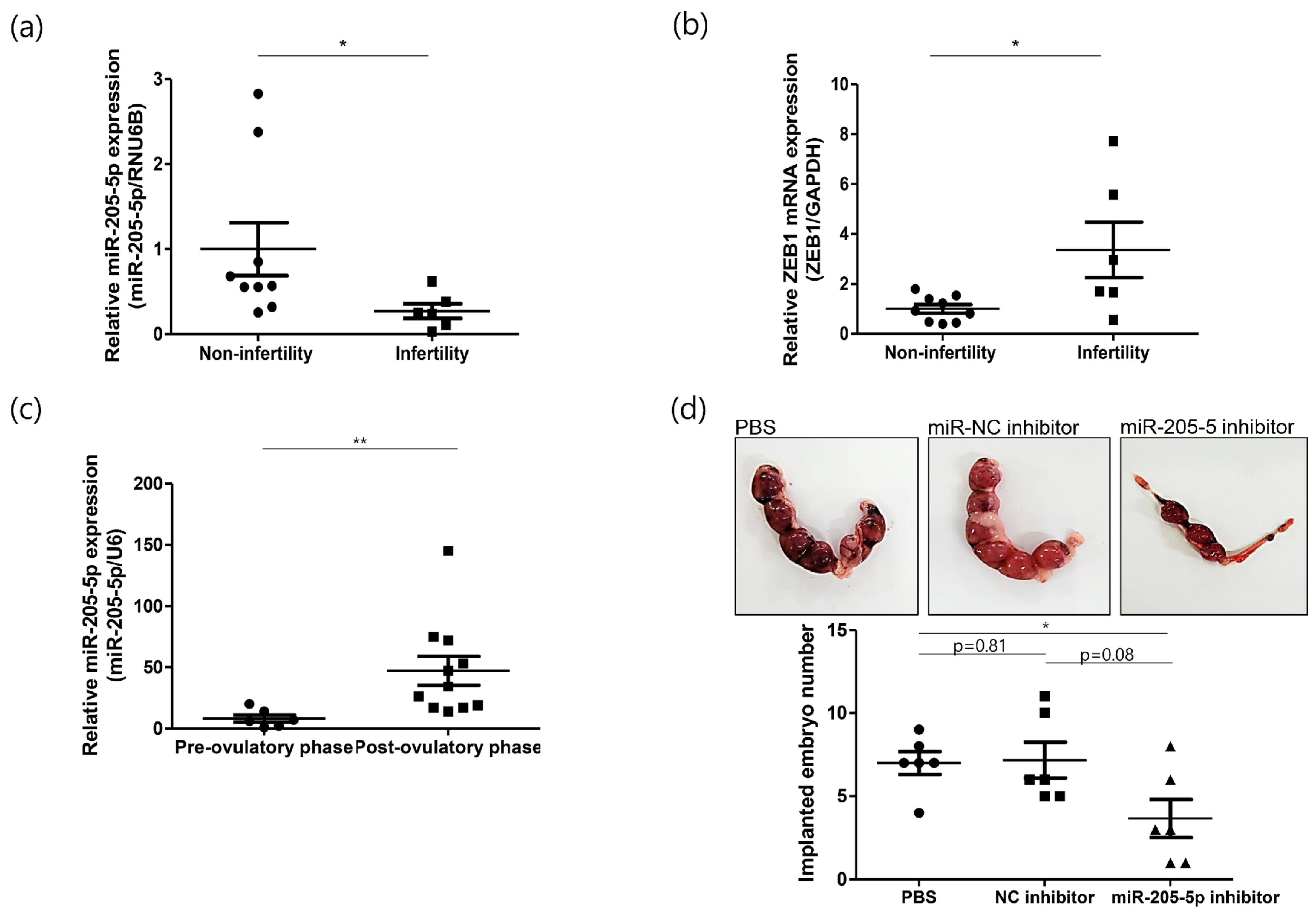
2.4. Role of miR-205-5p in Embryo Implantation
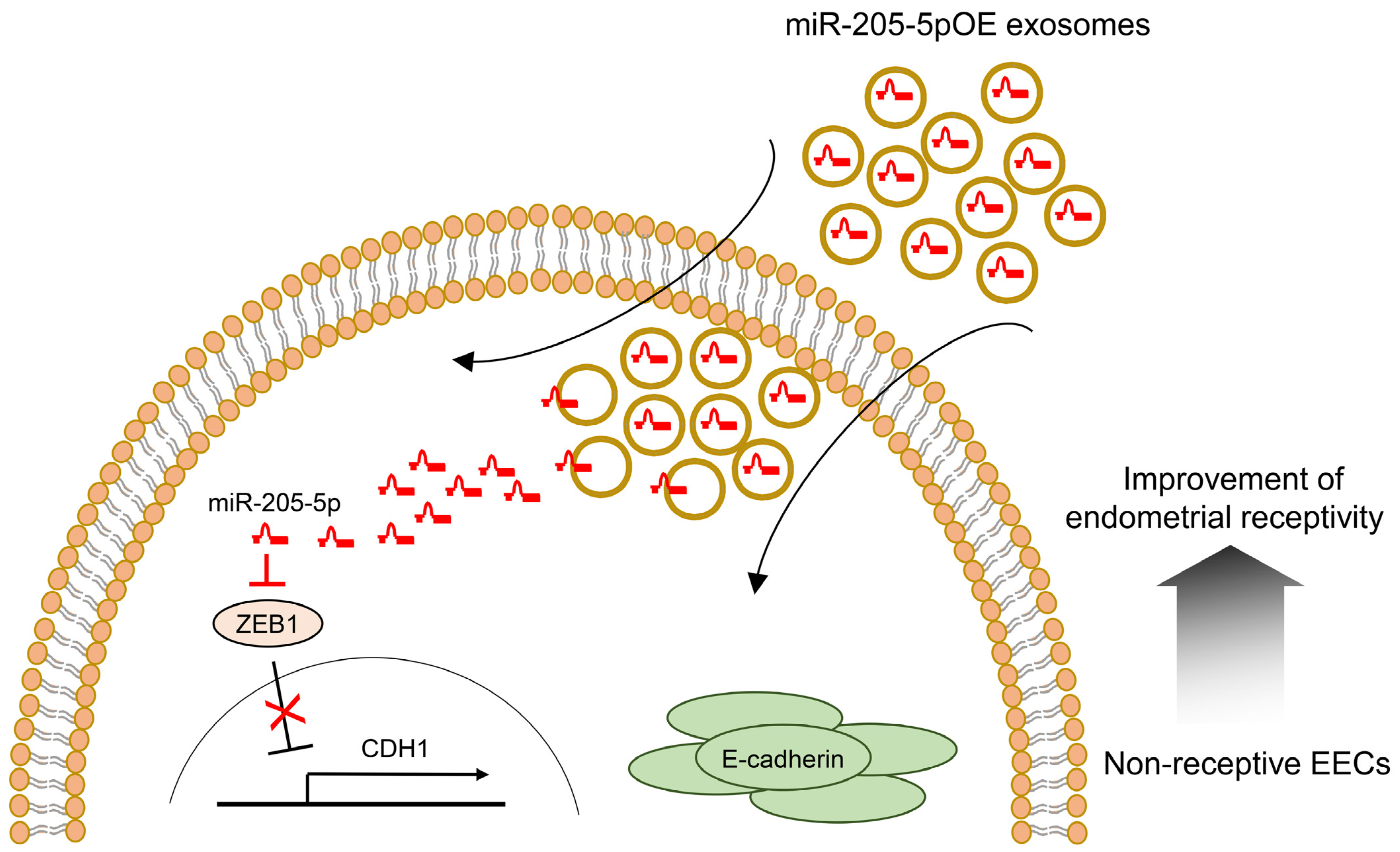
2.5. Role of Modified Exosomes Overexpressing miR-205-5p Mimics in Endometrial Receptivity
3. Discussion
4. Materials and Methods
4.1. Collection of Human Endometrial Tissue
4.2. Cell Culture
4.3. Exosome Isolation, Uptake, and Transfection
4.4. Cryo-EM and NTA
4.5. NGS Library Generation and Sequencing
4.6. Immunoblot Analysis
4.7. Total and Exosomal RNA Isolation and qRT-PCR Analysis
4.8. In Vitro Assay for JAr Spheroid Implantation
4.9. Plasmid Construction and Luciferase Reporter Assay
4.10. Animal Experiments
4.11. Statistical Analyses
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Evans, J.; Hannan, N.J.; Hincks, C.; Rombauts, L.J.; Salamonsen, L.A. Defective soil for a fertile seed? Altered endometrial development is detrimental to pregnancy success. PLoS ONE 2012, 7, e53098. [Google Scholar] [CrossRef]
- Ochoa-Bernal, M.A.; Fazleabas, A.T. Physiologic Events of Embryo Implantation and Decidualization in Human and Non-Human Primates. Int. J. Mol. Sci. 2020, 21, 1973. [Google Scholar] [CrossRef]
- Psychoyos, A. Uterine receptivity for nidation. Ann. N. Y. Acad. Sci. 1986, 476, 36–42. [Google Scholar] [CrossRef]
- Shekibi, M.; Heng, S.; Nie, G. MicroRNAs in the Regulation of Endometrial Receptivity for Embryo Implantation. Int. J. Mol. Sci. 2022, 23, 6210. [Google Scholar] [CrossRef]
- Benkhalifa, M.; Joao, F.; Duval, C.; Montjean, D.; Bouricha, M.; Cabry, R.; Belanger, M.C.; Bahri, H.; Miron, P.; Benkhalifa, M. Endometrium Immunomodulation to Prevent Recurrent Implantation Failure in Assisted Reproductive Technology. Int. J. Mol. Sci. 2022, 23, 12787. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Liang, J.; Wang, S.; Wang, Z. Role of microRNAs in embryo implantation. Reprod. Biol. Endocrinol. 2017, 15, 90. [Google Scholar] [CrossRef]
- Tesfaye, D.; Salilew-Wondim, D.; Gebremedhn, S.; Sohel, M.M.; Pandey, H.O.; Hoelker, M.; Schellander, K. Potential role of microRNAs in mammalian female fertility. Reprod. Fertil. Dev. 2016, 29, 8–23. [Google Scholar] [CrossRef]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell. Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Izumi, H.; Sekine, K.; Ochiya, T. microRNA as a new immune-regulatory agent in breast milk. Silence 2010, 1, 7. [Google Scholar] [CrossRef]
- Thery, C. Exosomes: Secreted vesicles and intercellular communications. F1000 Biol. Rep. 2011, 3, 15. [Google Scholar] [CrossRef] [PubMed]
- Ng, Y.H.; Rome, S.; Jalabert, A.; Forterre, A.; Singh, H.; Hincks, C.L.; Salamonsen, L.A. Endometrial exosomes/microvesicles in the uterine microenvironment: A new paradigm for embryo-endometrial cross talk at implantation. PLoS ONE 2013, 8, e58502. [Google Scholar] [CrossRef]
- Greening, D.W.; Nguyen, H.P.; Elgass, K.; Simpson, R.J.; Salamonsen, L.A. Human Endometrial Exosomes Contain Hormone-Specific Cargo Modulating Trophoblast Adhesive Capacity: Insights into Endometrial-Embryo Interactions. Biol. Reprod. 2016, 94, 38. [Google Scholar] [CrossRef] [PubMed]
- Kusama, K.; Nakamura, K.; Bai, R.; Nagaoka, K.; Sakurai, T.; Imakawa, K. Intrauterine exosomes are required for bovine conceptus implantation. Biochem. Biophys. Res. Commun. 2018, 495, 1370–1375. [Google Scholar] [CrossRef]
- Tan, Q.; Shi, S.; Liang, J.; Cao, D.; Wang, S.; Wang, Z. Endometrial cell-derived small extracellular vesicle miR-100-5p promotes functions of trophoblast during embryo implantation. Mol. Ther. Nucleic Acids 2021, 23, 217–231. [Google Scholar] [CrossRef] [PubMed]
- Tan, Q.; Shi, S.; Liang, J.; Zhang, X.; Cao, D.; Wang, Z. MicroRNAs in Small Extracellular Vesicles Indicate Successful Embryo Implantation during Early Pregnancy. Cells 2020, 9, 645. [Google Scholar] [CrossRef]
- Ho, H.; Singh, H.; Aljofan, M.; Nie, G. A high-throughput in vitro model of human embryo attachment. Fertil. Steril. 2012, 97, 974–978. [Google Scholar] [CrossRef]
- John, N.J.; Linke, M.; Denker, H.W. Quantitation of human choriocarcinoma spheroid attachment to uterine epithelial cell monolayers. In Vitro Cell. Dev. Biol. Anim. 1993, 29, 461–468. [Google Scholar] [CrossRef]
- Rahnama, F.; Thompson, B.; Steiner, M.; Shafiei, F.; Lobie, P.E.; Mitchell, M.D. Epigenetic regulation of E-cadherin controls endometrial receptivity. Endocrinology 2009, 150, 1466–1472. [Google Scholar] [CrossRef]
- Yu, S.L.; Kang, Y.; Jeong, D.U.; Lee, D.C.; Jeon, H.J.; Kim, T.H.; Lee, S.K.; Han, A.R.; Kang, J.; Park, S.R. The miR-182-5p/NDRG1 Axis Controls Endometrial Receptivity through the NF-kappaB/ZEB1/E-Cadherin Pathway. Int. J. Mol. Sci. 2022, 23, 12303. [Google Scholar] [CrossRef]
- Eger, A.; Aigner, K.; Sonderegger, S.; Dampier, B.; Oehler, S.; Schreiber, M.; Berx, G.; Cano, A.; Beug, H.; Foisner, R. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene 2005, 24, 2375–2385. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Aplin, J.D. Adhesion molecules in endometrial epithelium: Tissue integrity and embryo implantation. J. Anat. 2009, 215, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Cha, J.; Sun, X.; Dey, S.K. Mechanisms of implantation: Strategies for successful pregnancy. Nat. Med. 2012, 18, 1754–1767. [Google Scholar] [CrossRef] [PubMed]
- Governini, L.; Luongo, F.P.; Haxhiu, A.; Piomboni, P.; Luddi, A. Main actors behind the endometrial receptivity and successful implantation. Tissue Cell. 2021, 73, 101656. [Google Scholar] [CrossRef]
- Borthwick, J.M.; Charnock-Jones, D.S.; Tom, B.D.; Hull, M.L.; Teirney, R.; Phillips, S.C.; Smith, S.K. Determination of the transcript profile of human endometrium. Mol. Hum. Reprod. 2003, 9, 19–33. [Google Scholar] [CrossRef]
- Carson, D.D.; Lagow, E.; Thathiah, A.; Al-Shami, R.; Farach-Carson, M.C.; Vernon, M.; Yuan, L.; Fritz, M.A.; Lessey, B. Changes in gene expression during the early to mid-luteal (receptive phase) transition in human endometrium detected by high-density microarray screening. Mol. Hum. Reprod. 2002, 8, 871–879. [Google Scholar] [CrossRef]
- Hu, S.; Yao, G.; Wang, Y.; Xu, H.; Ji, X.; He, Y.; Zhu, Q.; Chen, Z.; Sun, Y. Transcriptomic changes during the pre-receptive to receptive transition in human endometrium detected by RNA-Seq. J. Clin. Endocrinol. Metab. 2014, 99, E2744–E2753. [Google Scholar] [CrossRef]
- Kao, L.C.; Tulac, S.; Lobo, S.; Imani, B.; Yang, J.P.; Germeyer, A.; Osteen, K.; Taylor, R.N.; Lessey, B.A.; Giudice, L.C. Global gene profiling in human endometrium during the window of implantation. Endocrinology 2002, 143, 2119–2138. [Google Scholar] [CrossRef]
- Mirkin, S.; Arslan, M.; Churikov, D.; Corica, A.; Diaz, J.I.; Williams, S.; Bocca, S.; Oehninger, S. In search of candidate genes critically expressed in the human endometrium during the window of implantation. Hum. Reprod. 2005, 20, 2104–2117. [Google Scholar] [CrossRef]
- Riesewijk, A.; Martin, J.; van Os, R.; Horcajadas, J.A.; Polman, J.; Pellicer, A.; Mosselman, S.; Simon, C. Gene expression profiling of human endometrial receptivity on days LH+2 versus LH+7 by microarray technology. Mol. Hum. Reprod. 2003, 9, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Sigurgeirsson, B.; Amark, H.; Jemt, A.; Ujvari, D.; Westgren, M.; Lundeberg, J.; Gidlof, S. Comprehensive RNA sequencing of healthy human endometrium at two time points of the menstrual cycle. Biol. Reprod. 2017, 96, 24–33. [Google Scholar] [PubMed]
- Suhorutshenko, M.; Kukushkina, V.; Velthut-Meikas, A.; Altmae, S.; Peters, M.; Magi, R.; Krjutskov, K.; Koel, M.; Codoner, F.M.; Martinez-Blanch, J.F.; et al. Endometrial receptivity revisited: Endometrial transcriptome adjusted for tissue cellular heterogeneity. Hum. Reprod. 2018, 33, 2074–2086. [Google Scholar] [CrossRef]
- Yu, S.L.; Kim, T.H.; Han, Y.H.; Kang, Y.; Jeong, D.U.; Lee, D.C.; Kang, J.; Park, S.R. Transcriptomic analysis and competing endogenous RNA network in the human endometrium between proliferative and mid-secretory phases. Exp. Ther. Med. 2021, 21, 660. [Google Scholar] [CrossRef]
- Yang, J.; Barkley, J.E.; Bhattarai, B.; Firouzi, K.; Monk, B.J.; Coonrod, D.V.; Zenhausern, F. Identification of endometrial cancer-specific microRNA biomarkers in endometrial Fluid. Int. J. Mol. Sci. 2023, 24, 8683. [Google Scholar] [CrossRef] [PubMed]
- Aberle, H.; Butz, S.; Stappert, J.; Weissig, H.; Kemler, R.; Hoschuetzky, H. Assembly of the cadherin-catenin complex in vitro with recombinant proteins. J. Cell. Sci. 1994, 107, 3655–3663. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, J.; Ichigo, S.; Hori, M.; Tamaya, T. Alteration of E-cadherin, alpha- and beta-catenin mRNA expression in human uterine endometrium during the menstrual cycle. Gynecol. Endocrinol. 1996, 10, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Rowlands, T.M.; Symonds, J.M.; Farookhi, R.; Blaschuk, O.W. Cadherins: Crucial regulators of structure and function in reproductive tissues. Rev. Reprod. 2000, 5, 53–61. [Google Scholar] [CrossRef]
- Achache, H.; Revel, A. Endometrial receptivity markers, the journey to successful embryo implantation. Hum. Reprod. Update 2006, 12, 731–746. [Google Scholar] [CrossRef]
- Dai, B.; Zhou, G.; Hu, Z.; Zhu, G.; Mao, B.; Su, H.; Jia, Q. MiR-205 suppresses epithelial-mesenchymal transition and inhibits tumor growth of human glioma through down-regulation of HOXD9. Biosci. Rep. 2019, 39, BSR20181989. [Google Scholar] [CrossRef]
- Fan, Y.; Wang, K. miR-205 suppresses cell migration, invasion and EMT of colon cancer by targeting mouse double minute 4. Mol. Med. Rep. 2020, 22, 633–642. [Google Scholar] [CrossRef]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell. Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef]
- Gulei, D.; Magdo, L.; Jurj, A.; Raduly, L.; Cojocneanu-Petric, R.; Moldovan, A.; Moldovan, C.; Florea, A.; Pasca, S.; Pop, L.A.; et al. The silent healer: miR-205-5p up-regulation inhibits epithelial to mesenchymal transition in colon cancer cells by indirectly up-regulating E-cadherin expression. Cell. Death Dis. 2018, 9, 66. [Google Scholar] [CrossRef]
- Vosgha, H.; Ariana, A.; Smith, R.A.; Lam, A.K. miR-205 targets angiogenesis and EMT concurrently in anaplastic thyroid carcinoma. Endocr. Relat. Cancer 2018, 25, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Kang, F.B.; Wang, J.; Yang, C.; He, D.W. Downregulation of miR-205 contributes to epithelial-mesenchymal transition and invasion in triple-negative breast cancer by targeting HMGB1-RAGE signaling pathway. Anticancer. Drugs 2019, 30, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.; Wang, P.; Xu, J.; Zhu, Y.; Sun, G.; Pang, Q.; Tao, R. MicroRNA-205 functions as a tumor suppressor in human glioblastoma cells by targeting VEGF-A. Oncol. Rep. 2012, 27, 1200–1206. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 2009, 119, 1420–1428. [Google Scholar] [CrossRef]
- Xiao, Y.Y.; Lin, L.; Li, Y.H.; Jiang, H.P.; Zhu, L.T.; Deng, Y.R.; Lin, D.; Chen, W.; Zeng, C.Y.; Wang, L.J.; et al. ZEB1 promotes invasion and metastasis of endometrial cancer by interacting with HDGF and inducing its transcription. Am. J. Cancer Res. 2019, 9, 2314–2330. [Google Scholar]
- Jimenez, P.T.; Mainigi, M.A.; Word, R.A.; Kraus, W.L.; Mendelson, C.R. miR-200 Regulates Endometrial Development During Early Pregnancy. Mol. Endocrinol. 2016, 30, 977–987. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Chen, Y.; Zhao, Y.; Xu, C.; Zhang, A.; Zhang, Q.; Wang, D.; He, J.; Hua, W.; Duan, P. miR-200c suppresses endometriosis by targeting MALAT1 in vitro and in vivo. Stem Cell. Res. Ther. 2017, 8, 251. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Zhang, D.; Yang, Y.U.; Cui, X.; Sun, J.; Liang, C.; Qin, H.; Yang, X.; Liu, S.; Yan, Q. MicroRNA-200c impairs uterine receptivity formation by targeting FUT4 and alpha1,3-fucosylation. Cell Death Differ. 2017, 24, 2161–2172. [Google Scholar] [CrossRef]
- Zhou, M.; Fu, J.; Xiao, L.; Yang, S.; Song, Y.; Zhang, X.; Feng, X.; Sun, H.; Xu, W.; Huang, W. miR-196a overexpression activates the MEK/ERK signal and represses the progesterone receptor and decidualization in eutopic endometrium from women with endometriosis. Hum. Reprod. 2016, 31, 2598–2608. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Tang, Y.; Fan, Y.; Wu, D. MiR-196a-5p facilitates progression of estrogen-dependent endometrial cancer by regulating FOXO1. Histol. Histopathol. 2022, 18572. [Google Scholar]
- Zhang, S.; Wang, D.; Yang, F.; Shen, Y.; Li, D.; Deng, X. Intrauterine Injection of Umbilical Cord Mesenchymal Stem Cell Exosome Gel Significantly Improves the Pregnancy Rate in Thin Endometrium Rats. Cell. Transplant. 2022, 31, 9636897221133345. [Google Scholar] [CrossRef]
- Zhu, Q.; Tang, S.; Zhu, Y.; Chen, D.; Huang, J.; Lin, J. Exosomes Derived From CTF1-Modified Bone Marrow Stem Cells Promote Endometrial Regeneration and Restore Fertility. Front. Bioeng. Biotechnol. 2022, 10, 868734. [Google Scholar] [CrossRef]
- Kamel, R.M. Management of the infertile couple: An evidence-based protocol. Reprod. Biol. Endocrinol. 2010, 8, 21. [Google Scholar] [CrossRef] [PubMed]
- Noyes, R.W.; Hertig, A.T.; Rock, J. Reprint of: Dating the Endometrial Biopsy. Fertil. Steril. 2019, 112, e93–e115. [Google Scholar] [CrossRef]

| miRNA ID | Fold Change | p | |
|---|---|---|---|
| Upregulated | hsa-miR-205-5p | 14.02 | 1.77 × 10−3 |
| hsa-miR-205 | 12.61 | 1.51 × 10−3 | |
| hsa-miR-934 | 9.78 | 6.91 × 10−5 | |
| hsa-miR-203a-3p | 9.21 | 2.14 × 10−4 | |
| hsa-miR-141-3p | 9.21 | 2.18 × 10−3 | |
| hsa-miR-31-5p | 9.04 | 1.94 × 10−3 | |
| hsa-miR-200c-3p | 8.97 | 3.33 × 10−3 | |
| hsa-miR-200c | 8.73 | 3.35 × 10−3 | |
| hsa-miR-141 | 8.47 | 2.02 × 10−3 | |
| hsa-miR-516-5p | 8.44 | 2.73 × 10−3 | |
| hsa-miR-203a | 8.37 | 2.38 × 10−4 | |
| hsa-miR-34a-5p | 8.14 | 4.69 × 10−3 | |
| hsa-miR-519a-5p | 8.07 | 1.81 × 10−4 | |
| hsa-miR-516a-1 | 8.06 | 2.32 × 10−3 | |
| hsa-miR-516a-2 | 8.06 | 2.32 × 10−3 | |
| hsa-miR-934 | 8.01 | 7.33 × 10−5 | |
| hsa-miR-1283 | 7.83 | 1.98 × 10−3 | |
| hsa-miR-31 | 7.65 | 8.52 × 10−4 | |
| hsa-miR-34a | 7.56 | 2.58 × 10−3 | |
| hsa-miR-519a-3p | 7.52 | 1.60 × 10−4 | |
| Downregulated | hsa-miR3180-5 | −6.38 | 1.02 × 10−3 |
| hsa-miR-199b | −6.50 | 4.71 × 10−2 | |
| hsa-miR-873-3p | −6.56 | 7.86 × 10−3 | |
| hsa-miR-615 | −6.70 | 2.89 × 10−2 | |
| hsa-miR-873 | −6.97 | 5.75 × 10−3 | |
| hsa-miR-3180-3 | −7.01 | 8.69 × 10−4 | |
| hsa-miR-3180-1 | −7.01 | 8.69 × 10−4 | |
| hsa-miR-3180-2 | −7.09 | 8.56 × 10−4 | |
| hsa-miR-335 | −7.10 | 7.12 × 10−3 | |
| hsa-miR-335-5p | −7.12 | 7.80 × 10−3 | |
| hsa-miR-335-3p | −7.26 | 1.90 × 10−2 | |
| hsa-miR-296-3p | −7.80 | 3.42 × 10−3 | |
| hsa-miR-615-3p | −7.98 | 2.11 × 10−2 | |
| hsa-miR-125b-1-3p | −8.06 | 5.32 × 10−3 | |
| hsa-miR-3180-3p | −8.45 | 6.71 × 10−4 | |
| hsa-miR-873-5p | −8.49 | 3.82 × 10−3 | |
| hsa-miR-3180 | −8.57 | 7.17 × 10−4 | |
| hsa-miR-196a-2 | −8.96 | 6.22 × 10−3 | |
| hsa-miR-196a-1 | −9.14 | 5.60 × 10−3 | |
| hsa-miR-196a-5p | −9.33 | 5.93 × 10−3 |
| Variable/Group | Non-Infertility (20–24 mcd; n = 9) | Infertility (20–22 mcd; n = 6) | p |
|---|---|---|---|
| Age (y) | 37.8 ± 2.6 | 38.5 ± 3.7 | 0.76 |
| BMI (kg/m2) | 22.7 ± 2.8 | 23.0 ± 4.4 | 0.95 |
| No. of live births (n) | 1.9 ± 0.4 | 0.2 ± 0.4 | 0.0015 |
| No. of abortions (n) | 0.4 ± 0.5 | 0.8 ± 1.3 | 0.82 |
| Gene | Primer Sequence | Annealing Temperature | Size of Amplicon (bp) |
|---|---|---|---|
| ZEB1 | Forward: 5′-AAGAATTCACAGTGGAGAGAAGCC-3′ Reverse: 5′-CGTTTCTTGCAGTTTGGGCAT-3′ | 56 °C | 51 |
| CDH1 | Forward: 5′-TCAGCGTGTGTGACTGTGAA-3′ Reverse: 5′-CCTCCAAGAATCCCCAGAAT-3′ | 56 °C | 100 |
| GAPDH | Forward: 5′-ACAGTCAGCCGCATCTTCTT-3′ Reverse: 5′-ACGACCAAATCCGTTGACTC-3′ | 56 °C | 94 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, S.-L.; Jeong, D.-U.; Noh, E.-J.; Jeon, H.J.; Lee, D.C.; Kang, M.; Kim, T.-H.; Lee, S.K.; Han, A.R.; Kang, J.; et al. Exosomal miR-205-5p Improves Endometrial Receptivity by Upregulating E-Cadherin Expression through ZEB1 Inhibition. Int. J. Mol. Sci. 2023, 24, 15149. https://doi.org/10.3390/ijms242015149
Yu S-L, Jeong D-U, Noh E-J, Jeon HJ, Lee DC, Kang M, Kim T-H, Lee SK, Han AR, Kang J, et al. Exosomal miR-205-5p Improves Endometrial Receptivity by Upregulating E-Cadherin Expression through ZEB1 Inhibition. International Journal of Molecular Sciences. 2023; 24(20):15149. https://doi.org/10.3390/ijms242015149
Chicago/Turabian StyleYu, Seong-Lan, Da-Un Jeong, Eui-Jeong Noh, Hye Jin Jeon, Dong Chul Lee, Minho Kang, Tae-Hyun Kim, Sung Ki Lee, Ae Ra Han, Jaeku Kang, and et al. 2023. "Exosomal miR-205-5p Improves Endometrial Receptivity by Upregulating E-Cadherin Expression through ZEB1 Inhibition" International Journal of Molecular Sciences 24, no. 20: 15149. https://doi.org/10.3390/ijms242015149
APA StyleYu, S.-L., Jeong, D.-U., Noh, E.-J., Jeon, H. J., Lee, D. C., Kang, M., Kim, T.-H., Lee, S. K., Han, A. R., Kang, J., & Park, S.-R. (2023). Exosomal miR-205-5p Improves Endometrial Receptivity by Upregulating E-Cadherin Expression through ZEB1 Inhibition. International Journal of Molecular Sciences, 24(20), 15149. https://doi.org/10.3390/ijms242015149







