The IGF-Independent Role of IRS-2 in the Secretion of MMP-9 Enhances the Growth of Prostate Carcinoma Cell Line PC3
Abstract
1. Introduction
2. Results
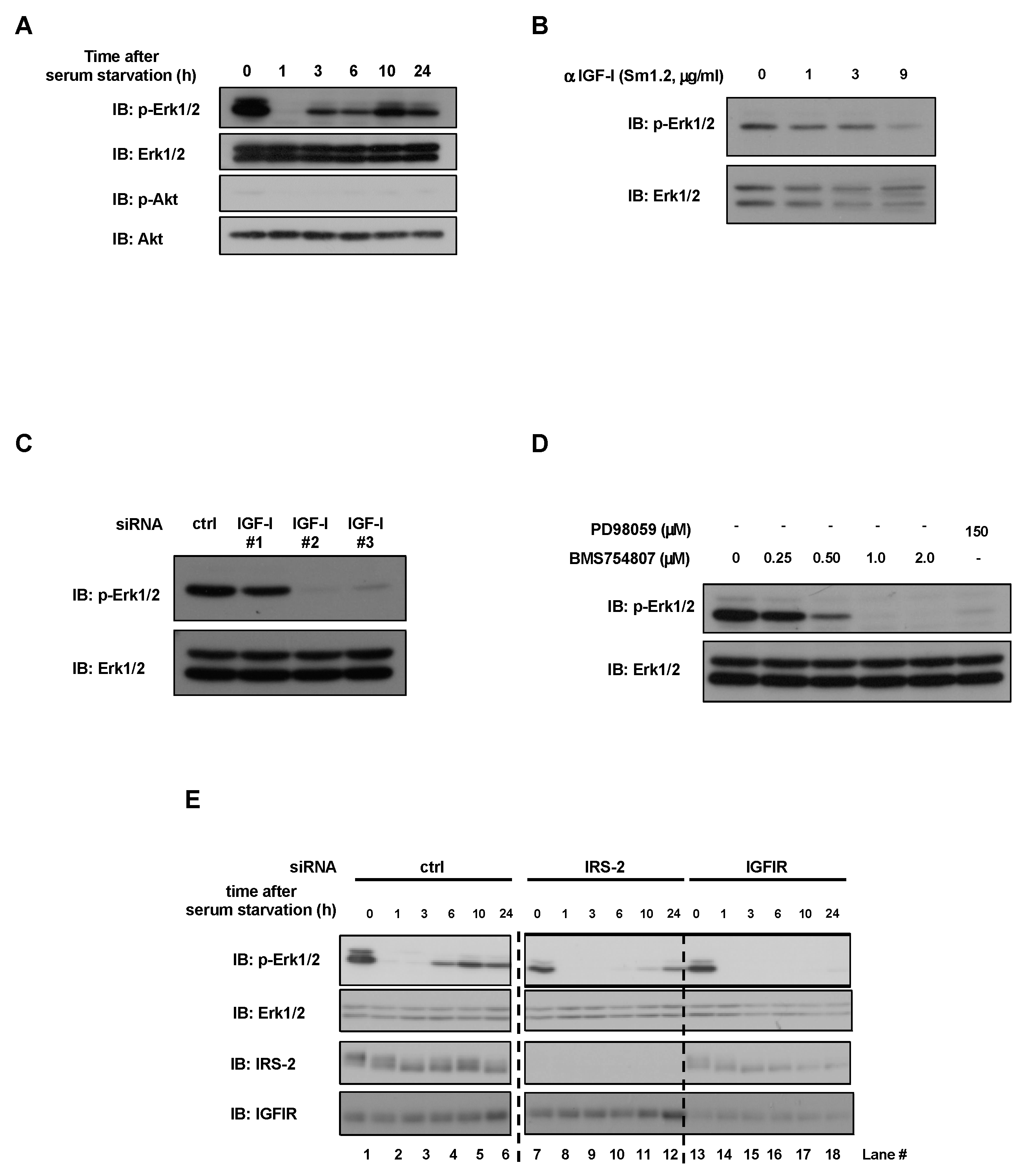
2.1. Inhibition of IGF-I or IGFIR Decreased Erk1/2 Phosphorylation under Serum-Free Conditions in PC3 Cells
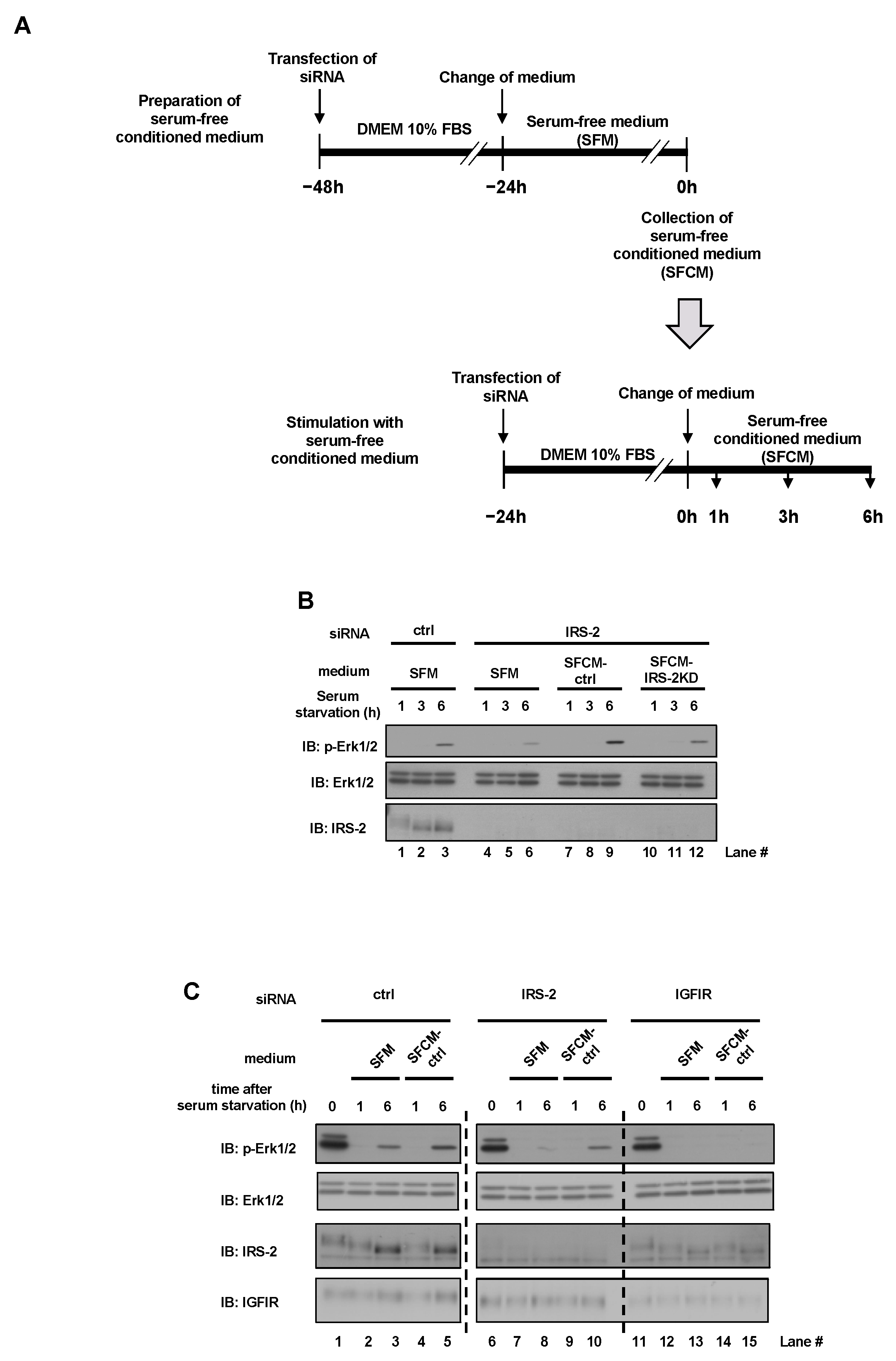
2.2. Decrease of Erk1/2 Phosphorylation in IRS-2 Knockdown Cells Was Rescued by Culturing with Conditioned Medium from PC3 Cells
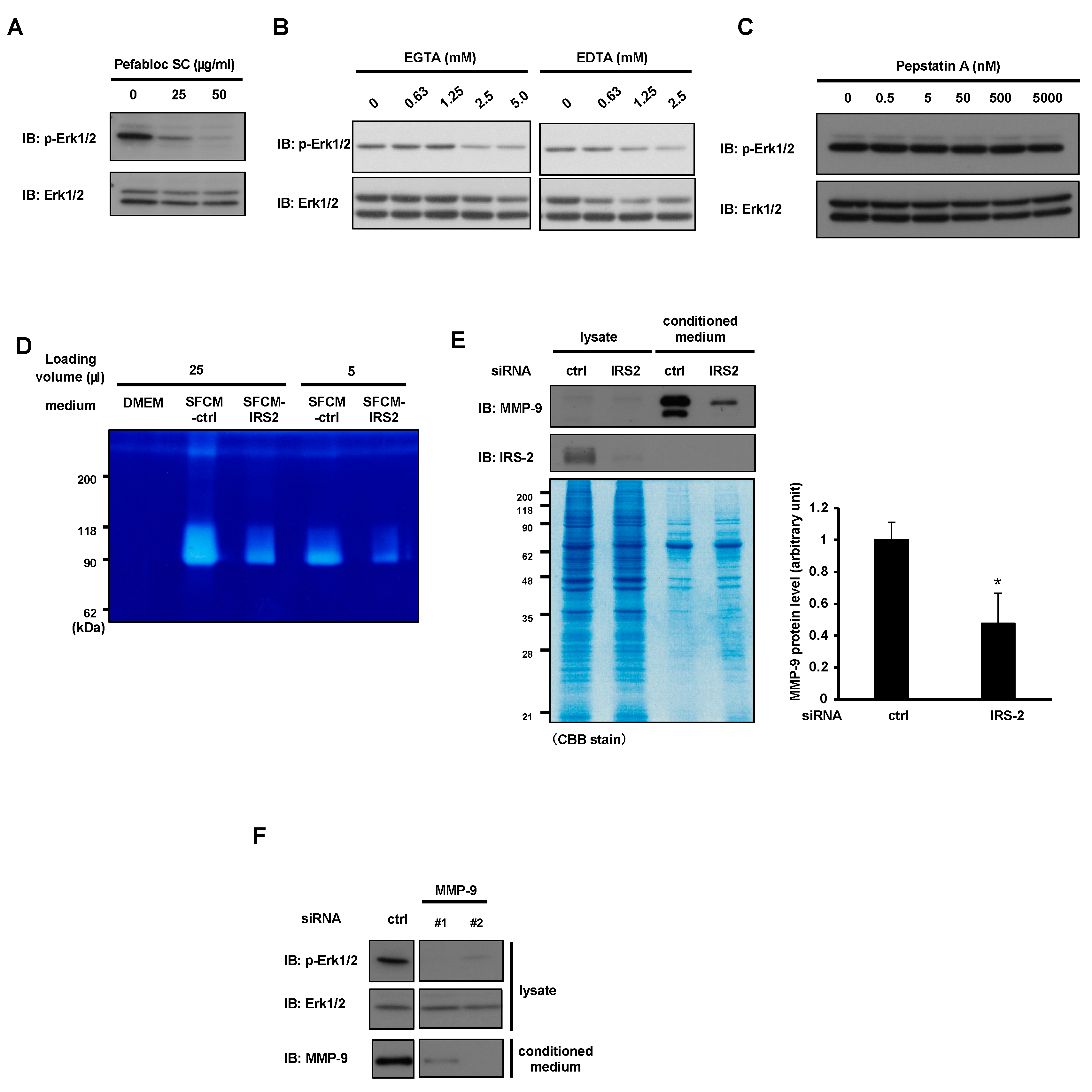
2.3. MMP-9 Secretion from PC3 Cells Is Required for Erk1/2 Activation under Serum-Free Conditions
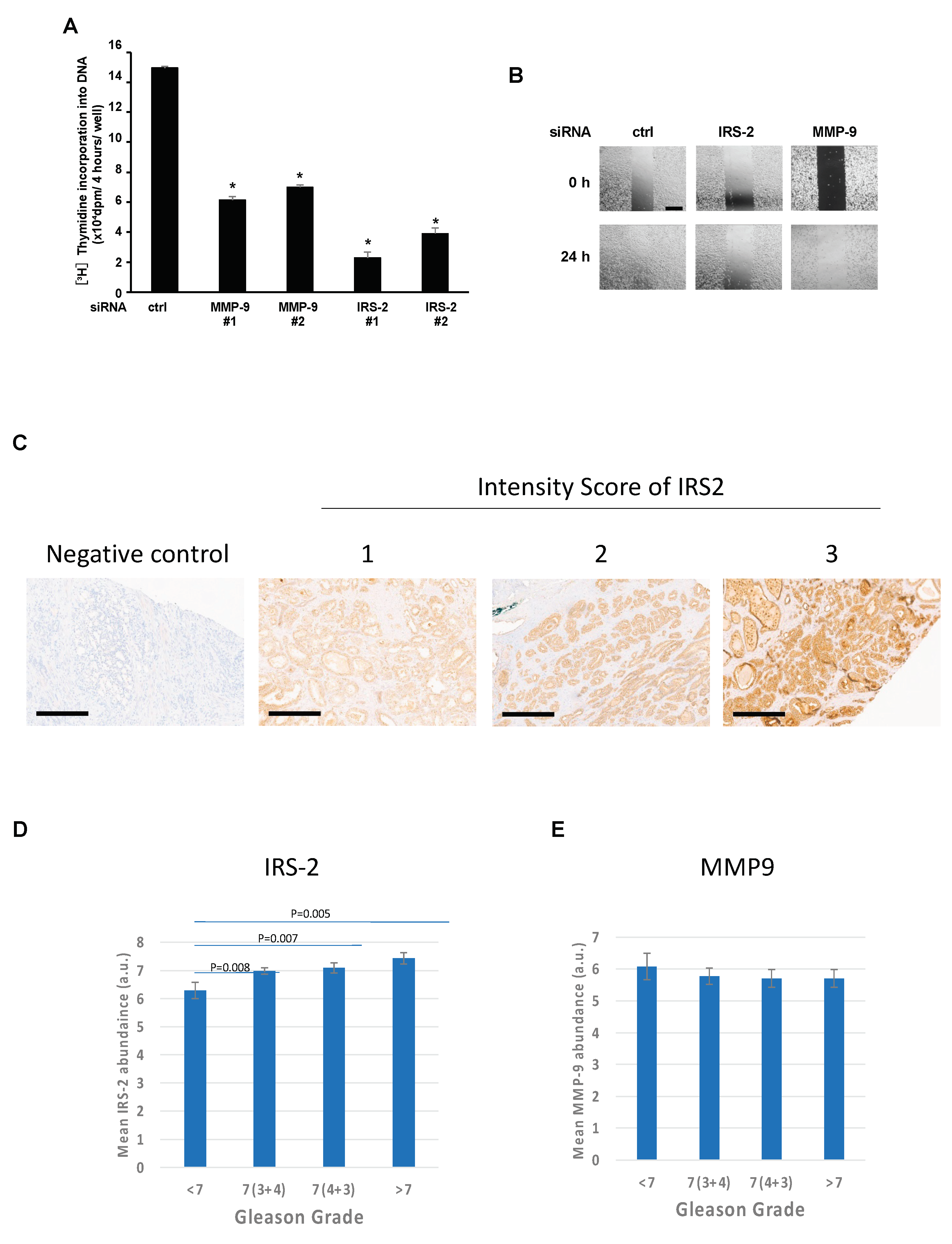
2.4. IRS-2 Is Required for MMP-9 Secretion from the PC3 Cells
2.5. IRS-2 Expression Levels Are Positively Correlated with the Gleason Grade in Prostate Cancer Tissues
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture
4.3. Immunoblotting Analysis
4.4. siRNAs
4.5. Transfection with Plasmids or siRNA
4.6. Preparation of Conditioned Medium and Stimulation with Conditioned Medium
4.7. Gelatin Zymography Assay
4.8. DNA Synthesis Assay
4.9. Wound Assay
4.10. Real-Time PCR
4.11. Immunostaining Analysis
4.12. Prostate Tissue Study
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jones, J.I.; Clemmons, D.R. Insulin-like growth factors and their binding proteins: Biological actions. Endocr. Rev. 1995, 16, 3–34. [Google Scholar]
- Heidegger, I.; Ofer, P.; Doppler, W.; Rotter, V.; Klocker, H.; Massoner, P. Diverse functions of IGF/insulin signaling in malignant and noncancerous prostate cells: Proliferation in cancer cells and differentiation in noncancerous cells. Endocrinology 2012, 153, 4633–4643. [Google Scholar] [CrossRef] [PubMed]
- Heidegger, I.; Kern, J.; Ofer, P.; Klocker, H.; Massoner, P. Oncogenic functions of IGF1R and INSR in prostate cancer include enhanced tumor growth, cell migration and angiogenesis. Oncotarget 2014, 5, 2723–2735. [Google Scholar] [CrossRef]
- Pollak, M. Insulin and insulin-like growth factor signalling in neoplasia. Nat. Rev. Cancer 2008, 8, 915–928. [Google Scholar] [CrossRef] [PubMed]
- Grønborg, M.; Wulff, B.S.; Rasmussen, J.S.; Kjeldsen, T.; Gammeltoft, S. Structure-function relationship of the insulin-like growth factor-I receptor tyrosine kinase. J. Biol. Chem. 1993, 268, 23435–23440. [Google Scholar] [CrossRef]
- Ullrich, A.; Schlessinger, J. Signal transduction by receptors with tyrosine kinase activity. Cell 1990, 61, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Yenush, L.; White, M.F. The IRS-signalling system during insulin and cytokine action. Bioessays 1997, 19, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Backer, J.; Myers, M.; Shoelson, S.; Chin, D.; Sun, X.; Miralpeix, M.; Hu, P.; Margolis, B.; Skolnik, E.; Schlessinger, J. Phosphatidylinositol 3′-kinase is activated by association with IRS-1 during insulin stimulation. EMBO J. 1992, 11, 3469–3479. [Google Scholar] [CrossRef]
- Myers, M.G., Jr.; Backer, J.M.; Sun, X.J.; Shoelson, S.P.; Hu, P.; Schlessinger, J.; Yoakim, M.; Schaffhausen, B.; White, M.F. IRS-1 activates phosphatidylinositol 3′-kinase by associating with src homology 2 domains of p85. Proc. Natl. Acad. Sci. USA 1992, 89, 10350–10354. [Google Scholar] [CrossRef]
- Dearth, R.K.; Cui, X.; Kim, H.-J.; Kuiatse, I.; Lawrence, N.A.; Zhang, X.; Divisova, J.; Britton, O.L.; Mohsin, S.; Allred, D.C.; et al. Mammary tumorigenesis and metastasis caused by overexpression of insulin receptor substrate 1 (IRS-1) or IRS-2. Mol. Cell Biol. 2006, 26, 9302–9314. [Google Scholar] [CrossRef]
- Nagle, J.A.; Ma, Z.; Byrne, M.A.; White, M.F.; Shaw, L.M. Involvement of insulin receptor substrate 2 in mammary tumor metastasis. Mol. Cell Biol. 2004, 24, 9726–9735. [Google Scholar] [CrossRef]
- Szabolcs, M.; Keniry, M.; Simpson, L.; Reid, L.J.; Koujak, S.; Schiff, S.C.; Davidian, G.; Licata, S.; Gruvberger-Saal, S.; Murty, V.V.; et al. Irs2 inactivation suppresses tumor progression in Pten+/− mice. Am. J. Pathol. 2009, 174, 276–286. [Google Scholar] [CrossRef] [PubMed]
- Jabłońska-Trypuć, A.; Matejczyk, M.; Rosochacki, S. Matrix metalloproteinases (MMPs), the main extracellular matrix (ECM) enzymes in collagen degradation, as a target for anticancer drugs. J. Enzyme Inhib. Med. Chem. 2016, 31, 177–183. [Google Scholar] [CrossRef] [PubMed]
- George, S.J.; Dwivedi, A. MMPs, cadherins, and cell proliferation. Trends Cardiovasc. Med. 2004, 14, 100–105. [Google Scholar] [CrossRef]
- Rao, J.S.; Bhoopathi, P.; Chetty, C.; Gujrati, M.; Lakka, S.S. MMP-9 short interfering RNA induced senescence resulting in inhibition of medulloblastoma growth via p16(INK4a) and mitogen-activated protein kinase pathway. Cancer Res. 2007, 67, 4956–4964. [Google Scholar] [CrossRef] [PubMed]
- Shay, G.; Lynch, C.C.; Fingleton, B. Moving targets: Emerging roles for MMPs in cancer progression and metastasis. Matrix Biol. 2015, 44–46, 200–206. [Google Scholar] [CrossRef]
- Augoff, K.; Hryniewicz-Jankowska, A.; Tabola, R.; Stach, K. MMP9: A Tough Target for Targeted Therapy for Cancer. Cancers 2022, 14, 1847. [Google Scholar] [CrossRef]
- Gennigens, C.; Menetrier-Caux, C.; Droz, J.P. Insulin-Like Growth Factor (IGF) family and prostate cancer. Crit. Rev. Oncol. Hematol. 2006, 58, 124–145. [Google Scholar] [CrossRef]
- Chan, J.M.; Stampfer, M.J.; Giovannucci, E.; Gann, P.H.; Ma, J.; Wilkinson, P.; Hennekens, C.H.; Pollak, M. Plasma insulin-like growth factor-I and prostate cancer risk: A prospective study. Science 1998, 279, 563–566. [Google Scholar] [CrossRef]
- Pollak, M.; Beamer, W.; Zhang, J.C. Insulin-like growth factors and prostate cancer. Cancer Metastasis Rev. 1998, 17, 383–390. [Google Scholar] [CrossRef]
- Cox, M.E.; Gleave, M.E.; Zakikhani, M.; Bell, R.H.; Piura, E.; Vickers, E.; Cunningham, M.; Larsson, O.; Fazli, L.; Pollak, M. Insulin receptor expression by human prostate cancers. Prostate 2009, 69, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Furuta, H.; Yoshihara, H.; Fukushima, T.; Yoneyama, Y.; Ito, A.; Worrall, C.; Girnita, A.; Girnita, L.; Yoshida, M.; Asano, T.; et al. IRS-2 deubiquitination by USP9X maintains anchorage-independent cell growth via Erk1/2 activation in prostate carcinoma cell line. Oncotarget 2018, 9, 33871–33883. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Collett-Solberg, P.F.; Cohen, P. Genetics, chemistry, and function of the IGF/IGFBP system. Endocrine 2000, 12, 121–136. [Google Scholar] [CrossRef] [PubMed]
- Cudic, M.; Fields, G.B. Extracellular proteases as targets for drug development. Curr. Protein Pept. Sci. 2009, 10, 297–307. [Google Scholar] [CrossRef]
- Nalla, A.K.; Gorantla, B.; Gondi, C.S.; Lakka, S.S.; Rao, J.S. Targeting MMP-9, uPAR, and cathepsin B inhibits invasion, migration and activates apoptosis in prostate cancer cells. Cancer Gene Ther. 2010, 17, 599–613. [Google Scholar] [CrossRef]
- Robinson, D.; Van Allen, E.M.; Wu, Y.-M.; Schultz, N.; Lonigro, R.J.; Mosquera, J.-M.; Montgomery, B.; Taplin, M.-E.; Pritchard, C.C.; Attard, G.; et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015, 161, 1215–1228. [Google Scholar] [CrossRef]
- Stopsack, K.H.; Nandakumar, S.; Arora, K.; Nguyen, B.; Vasselman, S.E.; Nweji, B.; McBride, S.M.; Morris, M.J.; Rathkopf, D.E.; Slovin, S.F.; et al. Differences in Prostate Cancer Genomes by Self-reported Race: Contributions of Genetic Ancestry, Modifiable Cancer Risk Factors, and Clinical Factors. Clin. Cancer Res. 2022, 28, 318–326. [Google Scholar] [CrossRef]
- Abida, W.; Armenia, J.; Gopalan, A.; Brennan, R.; Walsh, M.; Barron, D.; Danila, D.; Rathkopf, D.; Morris, M.; Slovin, S.; et al. Prospective Genomic Profiling of Prostate Cancer Across Disease States Reveals Germline and Somatic Alterations That May Affect Clinical Decision Making. JCO Precis. Oncol. 2017, 1, 1–16. [Google Scholar] [CrossRef]
- Twigg, S.M.; Kiefer, M.C.; Zapf, J.; Baxter, R.C. Insulin-like growth factor-binding protein 5 complexes with the acid-labile subunit. Role of the carboxyl-terminal domain. J. Biol. Chem. 1998, 273, 28791–28798. [Google Scholar] [CrossRef]
- Coppock, H.A.; White, A.; Aplin, J.D.; Westwood, M. Matrix metalloprotease-3 and -9 proteolyze insulin-like growth factor-binding protein-1. Biol. Reprod. 2004, 71, 438–443. [Google Scholar] [CrossRef][Green Version]
- Mañes, S.; Llorente, M.; Lacalle, R.A.; Gómez-Moutón, C.; Kremer, L.; Mira, E.; Martínez-A, C. The matrix metalloproteinase-9 regulates the insulin-like growth factor-triggered autocrine response in DU-145 carcinoma cells. J. Biol. Chem. 1999, 274, 6935–6945. [Google Scholar] [CrossRef]
- Miyamoto, S.; Yano, K.; Sugimoto, S.; Ishii, G.; Hasebe, T.; Endoh, Y.; Kodama, K.; Goya, M.; Chiba, T.; Ochiai, A. Matrix metalloproteinase-7 facilitates insulin-like growth factor bioavailability through its proteinase activity on insulin-like growth factor binding protein 3. Cancer Res. 2004, 64, 665–671. [Google Scholar] [CrossRef] [PubMed]
- Prudova, A.; auf dem Keller, U.; Butler, G.S.; Overall, C.M. Multiplex N-terminome analysis of MMP-2 and MMP-9 substrate degradomes by iTRAQ-TAILS quantitative proteomics. Mol. Cell Proteom. 2010, 9, 894–911. [Google Scholar] [CrossRef]
- Rajah, R.; Katz, L.; Nunn, S.; Solberg, P.; Beers, T.; Cohen, P. Insulin-like growth factor binding protein (IGFBP) proteases: Functional regulators of cell growth. Prog. Growth Factor Res. 1995, 6, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Sawaguchi, S.; Twining, S.S.; Sugar, J.; Feder, R.S.; Yue, B.Y. Expression of degradative enzymes and protease inhibitors in corneas with keratoconus. Investig. Ophthalmol. Vis. Sci. 1998, 39, 1117–1124. [Google Scholar]
- Rajah, R.; Nunn, S.E.; Herrick, D.J.; Grunstein, M.M.; Cohen, P. Leukotriene D4 induces MMP-1, which functions as an IGFBP protease in human airway smooth muscle cells. Am. J. Physiol. 1996, 271, L1014–L1022. [Google Scholar] [CrossRef]
- Yamada, P.M.; Lee, K.W. Perspectives in mammalian IGFBP-3 biology: Local vs. systemic action. Am. J. Physiol. Cell Physiol. 2009, 296, C954–C976. [Google Scholar] [CrossRef]
- Mondal, S.; Adhikari, N.; Banerjee, S.; Amin, S.A.; Jha, T. Matrix metalloproteinase-9 (MMP-9) and its inhibitors in cancer: A minireview. Eur. J. Med. Chem. 2020, 194, 112260. [Google Scholar] [CrossRef]
- Hakuno, F.; Takahashi, S.I. IGF1 receptor signaling pathways. J. Mol. Endocrinol. 2018, 61, T69–T86. [Google Scholar] [CrossRef]
- Fukushima, T.; Arai, T.; Ariga-Nedachi, M.; Okajima, H.; Ooi, Y.; Iijima, Y.; Sone, M.; Cho, Y.; Ando, Y.; Kasahara, K.; et al. Insulin receptor substrates form high-molecular-mass complexes that modulate their availability to insulin/insulin-like growth factor-I receptor tyrosine kinases. Biochem. Biophys. Res. Commun. 2011, 404, 767–773. [Google Scholar] [CrossRef]
- Hakuno, F.; Fukushima, T.; Yoneyama, Y.; Kamei, H.; Ozoe, A.; Yoshihara, H.; Yamanaka, D.; Shibano, T.; Sone-Yonezawa, M.; Yu, B.-C.; et al. The Novel Functions of High-Molecular-Mass Complexes Containing Insulin Receptor Substrates in Mediation and Modulation of Insulin-Like Activities: Emerging Concept of Diverse Functions by IRS-Associated Proteins. Front. Endocrinol. 2015, 6, 73. [Google Scholar] [CrossRef]
- Fukushima, T.; Yoshihara, H.; Furuta, H.; Hakuno, F.; Iemura, S.-I.; Natsume, T.; Nakatsu, Y.; Kamata, H.; Asano, T.; Komada, M.; et al. USP15 attenuates IGF-I signaling by antagonizing Nedd4-induced IRS-2 ubiquitination. Biochem. Biophys. Res. Commun. 2017, 484, 522–528. [Google Scholar] [CrossRef]
- Fukushima, T.; Yoshihara, H.; Furuta, H.; Kamei, H.; Hakuno, F.; Luan, J.; Duan, C.; Saeki, Y.; Tanaka, K.; Iemura, S.-I.; et al. Nedd4-induced monoubiquitination of IRS-2 enhances IGF signalling and mitogenic activity. Nat. Commun. 2015, 6, 6780. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, T.; Okajima, H.; Yamanaka, D.; Ariga, M.; Nagata, S.; Ito, A.; Yoshida, M.; Asano, T.; Chida, K.; Hakuno, F.; et al. HSP90 interacting with IRS-2 is involved in cAMP-dependent potentiation of IGF-I signals in FRTL-5 cells. Mol. Cell Endocrinol. 2011, 344, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Trifaró, J.M.; Lejen, T.; Rosé, S.D.; Pene, T.D.; Barkar, N.D.; Seward, E.P. Pathways that control cortical F-actin dynamics during secretion. Neurochem. Res. 2002, 27, 1371–1385. [Google Scholar] [CrossRef]
- Inoue, T.; Yoshida, T.; Shimizu, Y.; Kobayashi, T.; Yamasaki, T.; Toda, Y.; Segawa, T.; Kamoto, T.; Nakamura, E.; Ogawa, O. Requirement of androgen-dependent activation of protein kinase Czeta for androgen-dependent cell proliferation in LNCaP Cells and its roles in transition to androgen-independent cells. Mol. Endocrinol. 2006, 20, 3053–3069. [Google Scholar] [CrossRef] [PubMed]
- Hakuno, F.; Yamauchi, Y.; Kaneko, G.; Yoneyama, Y.; Nakae, J.; Chida, K.; Kadowaki, T.; Yamanouchi, K.; Nishihara, M.; Takahashi, S.-I. Constitutive expression of insulin receptor substrate (IRS)-1 inhibits myogenic differentiation through nuclear exclusion of Foxo1 in L6 myoblasts. PLoS ONE 2011, 6, e25655. [Google Scholar] [CrossRef]
- Hackshaw-McGeagh, L.E.; Penfold, C.; Shingler, E.; Robles, L.A.; Perks, C.M.; Holly, J.M.P.; Rowe, E.; Koupparis, A.; Bahl, A.; Persad, R.; et al. Phase II randomised control feasibility trial of a nutrition and physical activity intervention after radical prostatectomy for prostate cancer. BMJ Open 2019, 9, e029480. [Google Scholar] [CrossRef]
- Dean, S.J.R.; Perks, C.M.; Holly, J.M.P.; Bhoo-Pathy, N.; Looi, L.-M.; Mohammed, N.A.T.; Mun, K.-S.; Teo, S.-H.; Koobotse, M.O.; Yip, C.-H.; et al. Loss of PTEN expression is associated with IGFBP2 expression, younger age, and late stage in triple-negative breast cancer. Am. J. Clin. Pathol. 2014, 141, 323–333. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Furuta, H.; Sheng, Y.; Takahashi, A.; Nagano, R.; Kataoka, N.; Perks, C.M.; Barker, R.; Hakuno, F.; Takahashi, S.-I. The IGF-Independent Role of IRS-2 in the Secretion of MMP-9 Enhances the Growth of Prostate Carcinoma Cell Line PC3. Int. J. Mol. Sci. 2023, 24, 15065. https://doi.org/10.3390/ijms242015065
Furuta H, Sheng Y, Takahashi A, Nagano R, Kataoka N, Perks CM, Barker R, Hakuno F, Takahashi S-I. The IGF-Independent Role of IRS-2 in the Secretion of MMP-9 Enhances the Growth of Prostate Carcinoma Cell Line PC3. International Journal of Molecular Sciences. 2023; 24(20):15065. https://doi.org/10.3390/ijms242015065
Chicago/Turabian StyleFuruta, Haruka, Yina Sheng, Ayaka Takahashi, Raku Nagano, Naoyuki Kataoka, Claire Marie Perks, Rachel Barker, Fumihiko Hakuno, and Shin-Ichiro Takahashi. 2023. "The IGF-Independent Role of IRS-2 in the Secretion of MMP-9 Enhances the Growth of Prostate Carcinoma Cell Line PC3" International Journal of Molecular Sciences 24, no. 20: 15065. https://doi.org/10.3390/ijms242015065
APA StyleFuruta, H., Sheng, Y., Takahashi, A., Nagano, R., Kataoka, N., Perks, C. M., Barker, R., Hakuno, F., & Takahashi, S.-I. (2023). The IGF-Independent Role of IRS-2 in the Secretion of MMP-9 Enhances the Growth of Prostate Carcinoma Cell Line PC3. International Journal of Molecular Sciences, 24(20), 15065. https://doi.org/10.3390/ijms242015065









