Cajanus platycarpus Flavonoid 3′5′ Hydroxylase_2 (CpF3′5′H_2) Confers Resistance to Helicoverpa armigera by Modulating Total Polyphenols and Flavonoids in Transgenic Tobacco
Abstract
1. Introduction
2. Results and Discussion
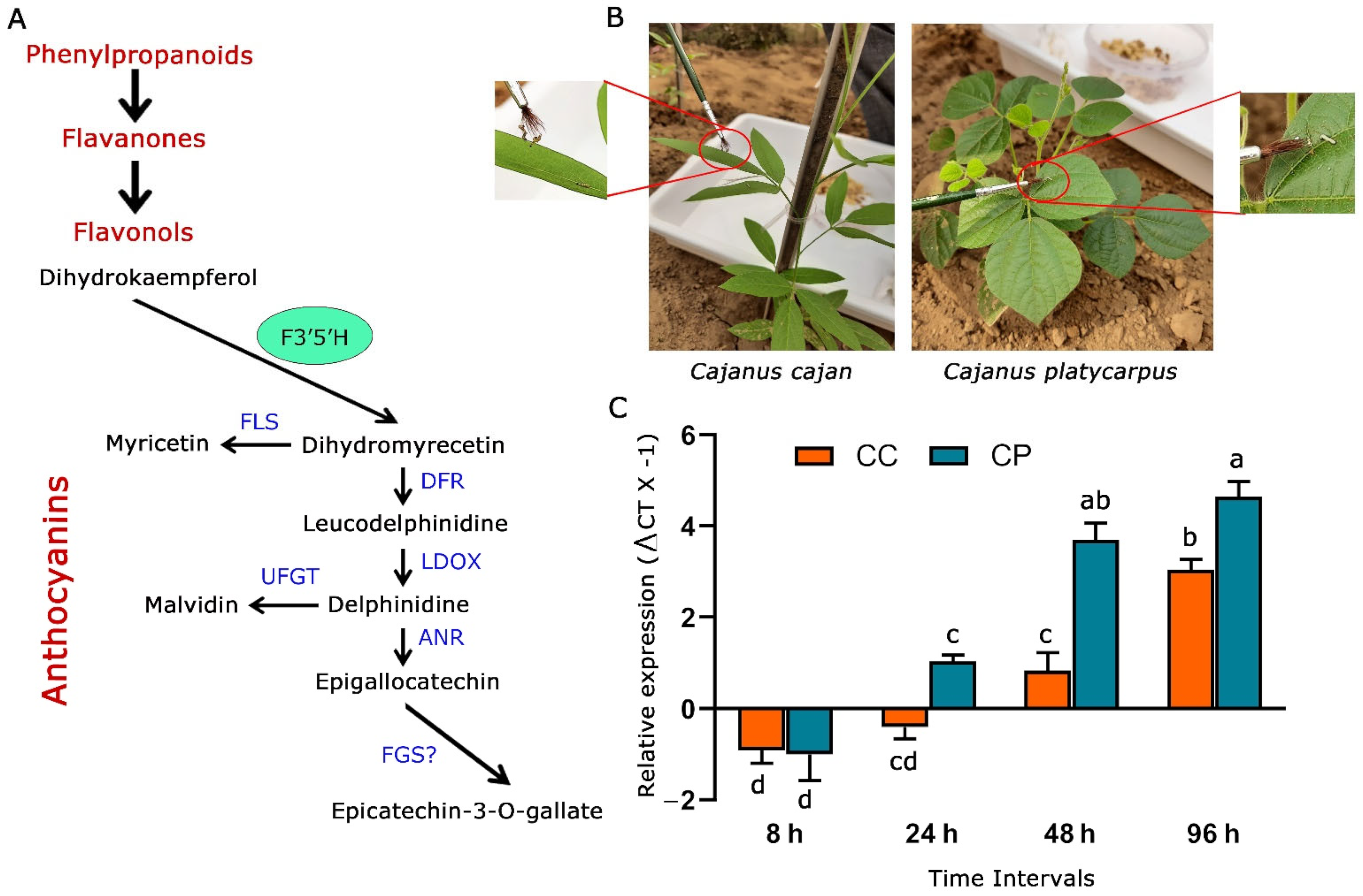
2.1. F3′5′H_2 Gene was Upregulated at Varying Levels in C. cajan and C. platycarpus in Response to H. armigera
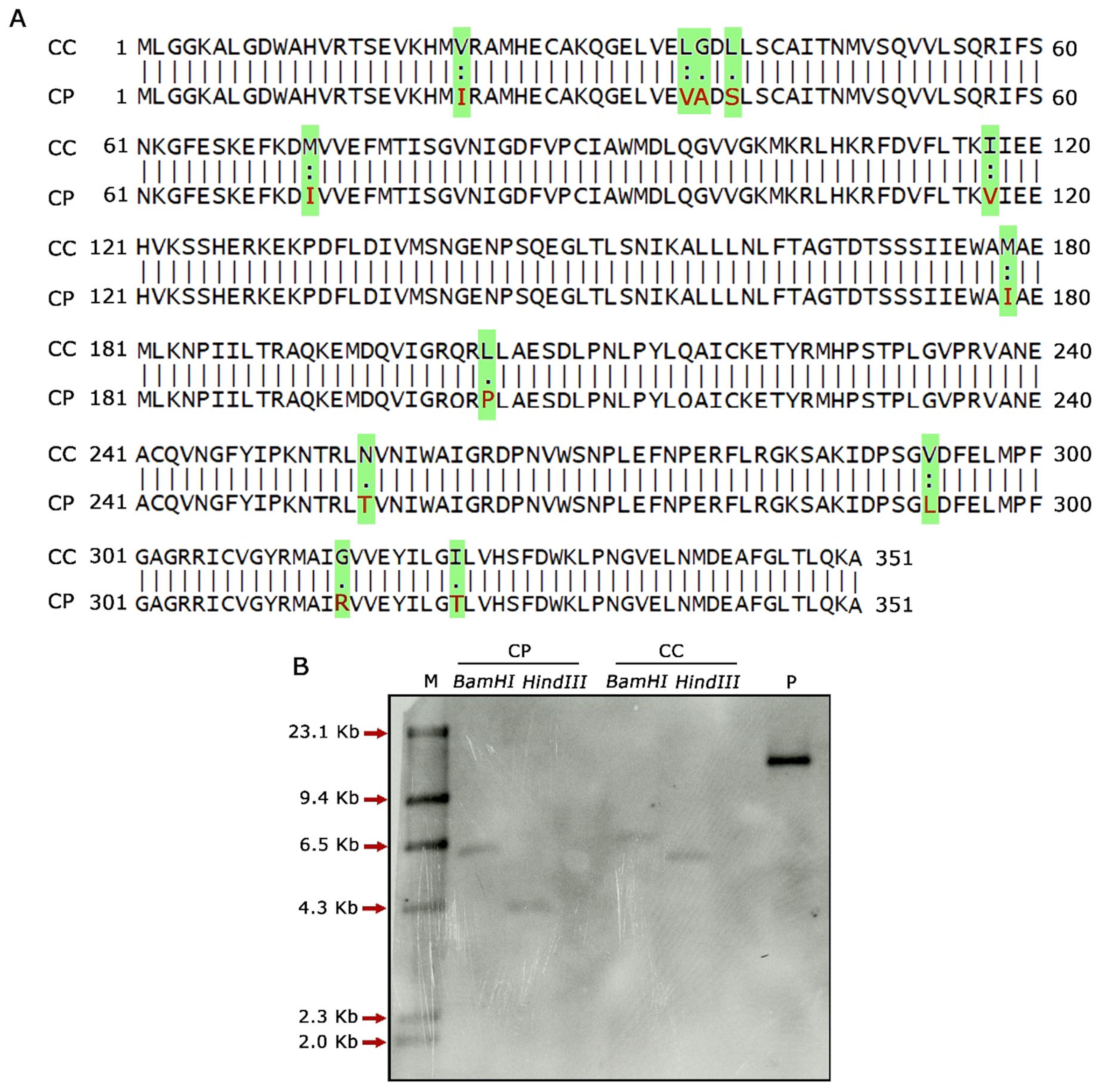
2.2. Assessment of CcF3′5′H_2 and CpF3′5′H_2 for Variation in Gene Sequence and Copy Number
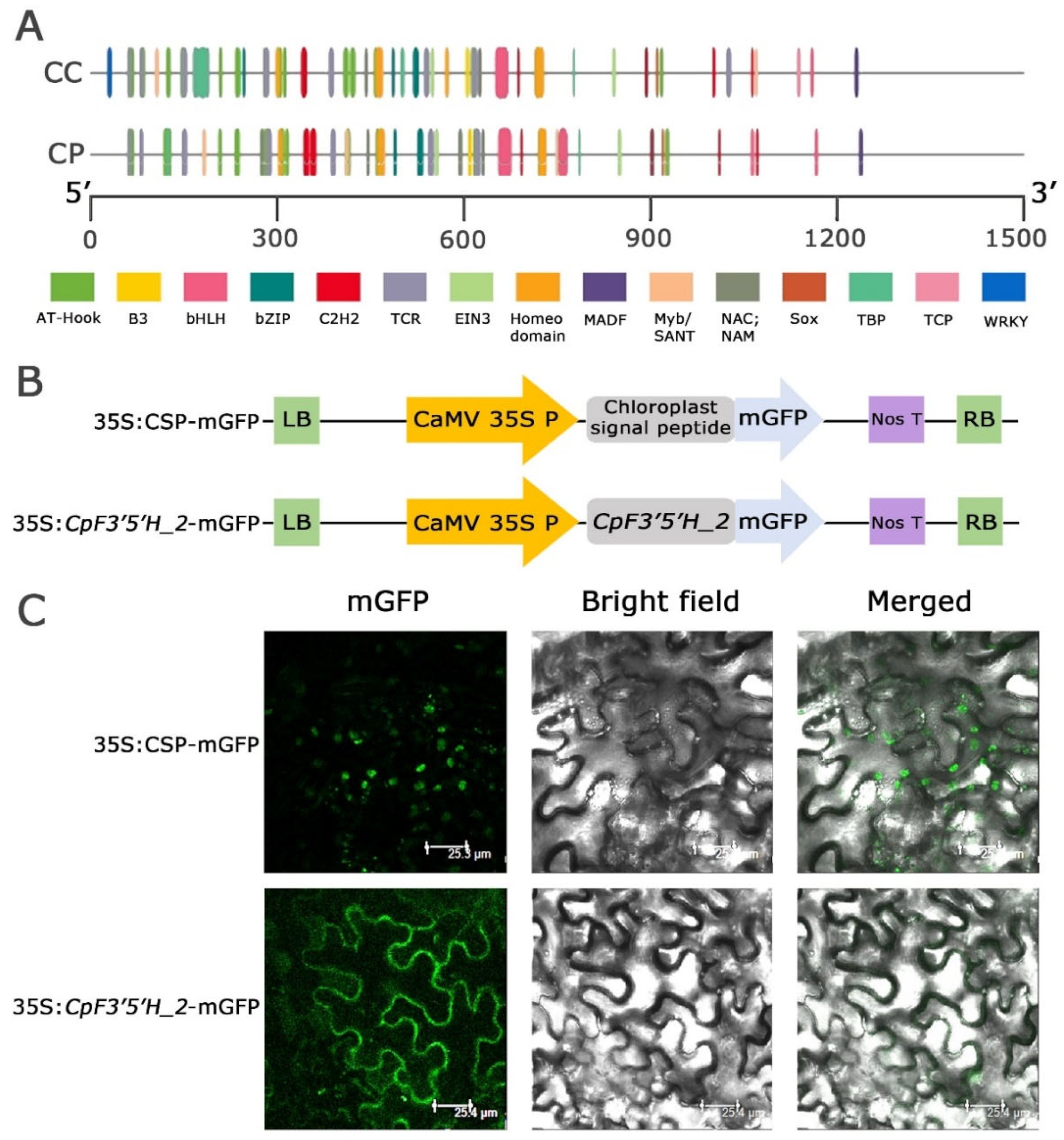
2.3. Assessment of Upstream Regulatory Elements of CcF3′5′H_2 and CpF3′5′H_2
2.4. Subcellular Localisation of CpF3′5′H_2 through Transient Expression in N. benthamiana
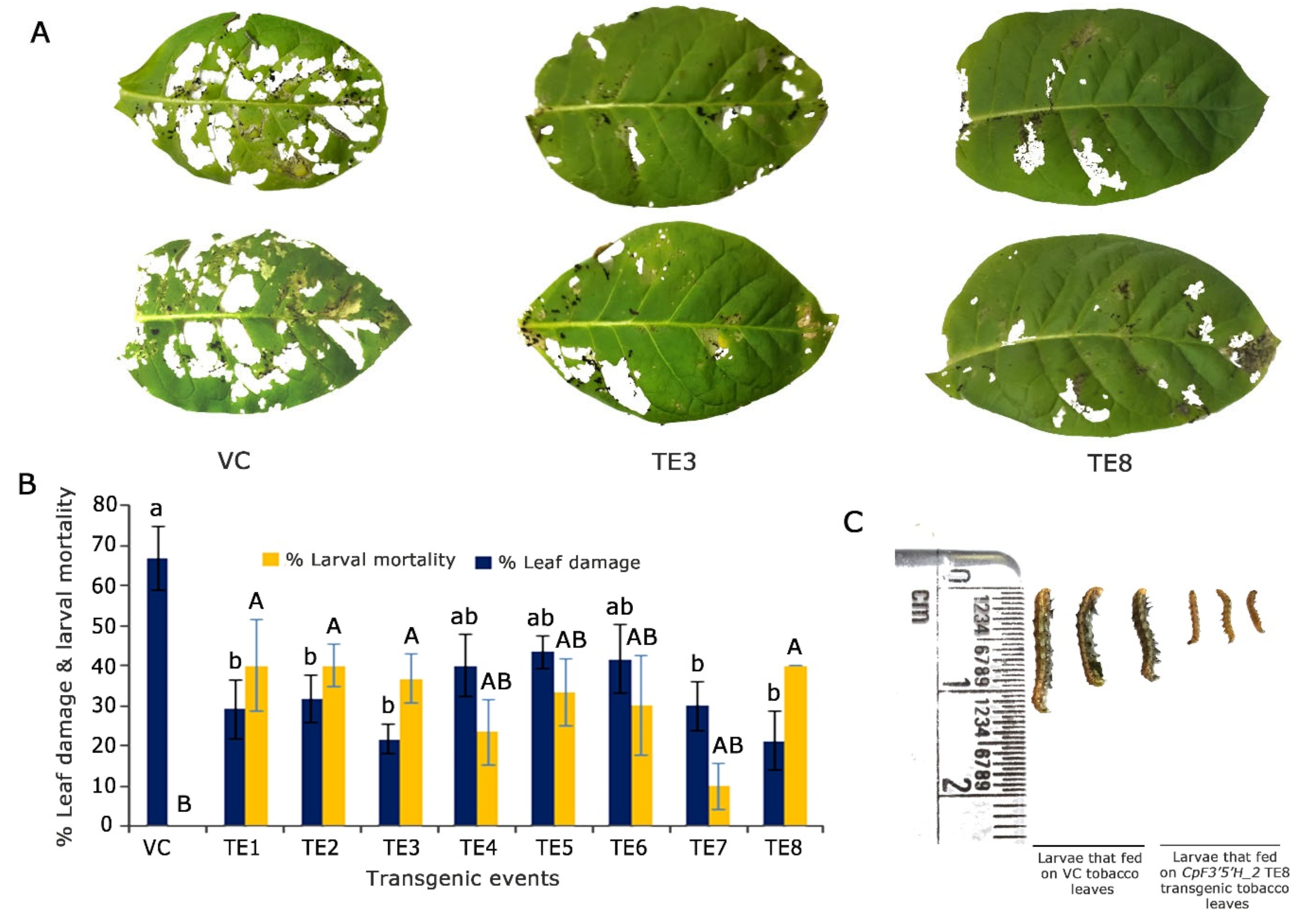
2.5. Overexpression and Functional Validation of CpF3′5′H_2 in Tobacco for Resistance against H. armigera
2.6. Characterisation of Transgenic Tobacco for T-DNA Integration
2.7. Expression Analysis of CpF3′5′H_2 in Transgenic Tobacco
2.8. CpF3′5′H_2 Aids in the Management of H. armigera by Improving the ROS Scavenging Ability and Increasing Total Polyphenol and Flavonoid Content in Transgenic Tobacco
3. Methods and Materials
3.1. Gene Source
3.2. Plant Material and Herbivore Challenge by H. armigera
3.3. Expression Analysis of CcF3′5′H_2 and CpF3′5′H_2 after Herbivore Challenge
Total RNA Isolation and cDNA Synthesis
3.4. Quantitative Real-Time PCR(qRT-PCR)
3.5. Copy Number Assessment of F3′5′H_2 Gene in C. cajan and C. platycarpus
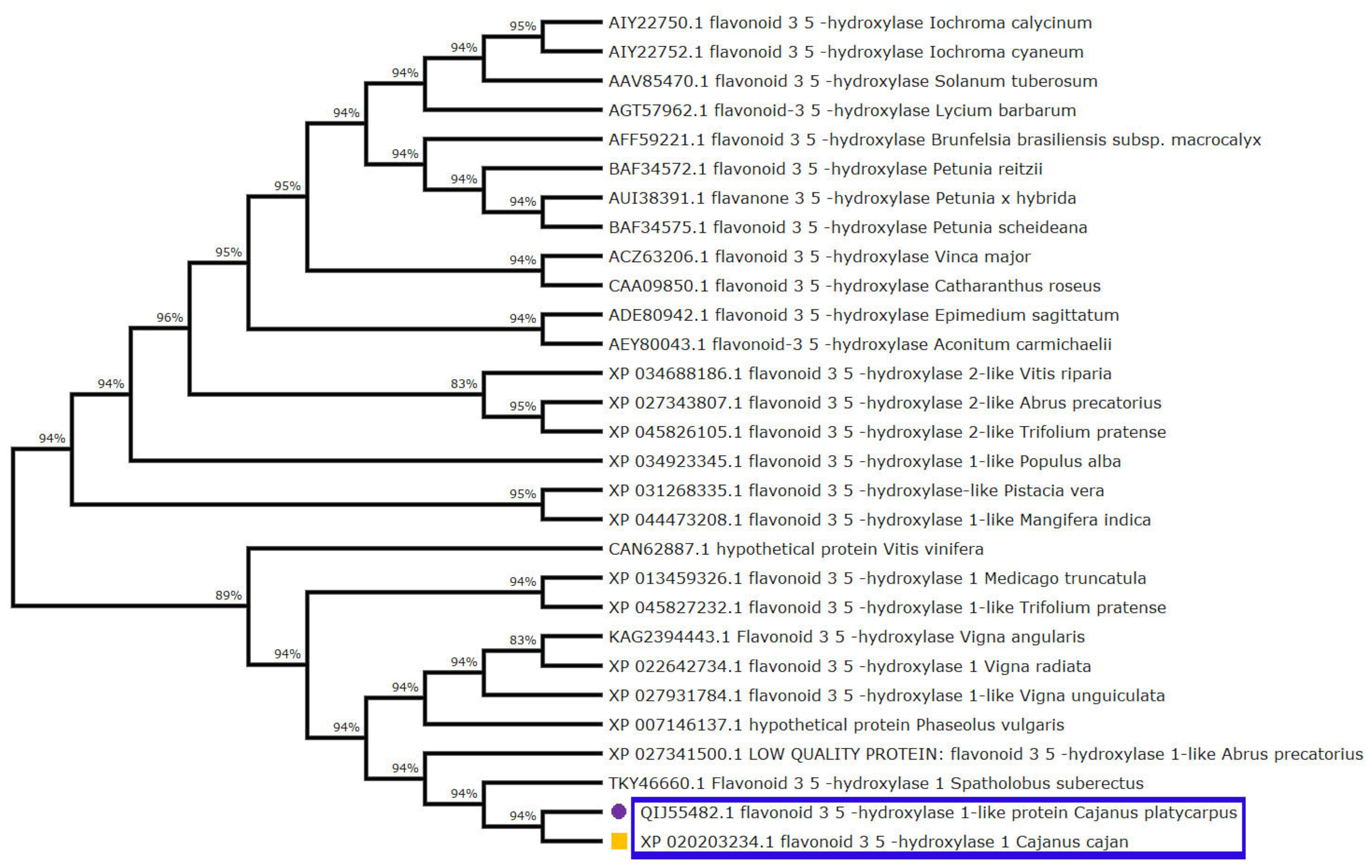
3.6. Multiple Sequence Alignment and Phylogenetic Analysis of F3′5′H_2 in C. cajan, C. Platycarpus and Other Closely Related Species
3.7. Promoter Analyses of F3′5′H_2 in C. cajan and C. platycarpus
3.8. Transient Expression and Subcellular Localisation of CpF3′5′H_2 in Nicotiana benthamiana
3.9. Validation of CpF3′5′H_2 in the Model Plant Tobacco (Nicotiana tabacum L. cv petit Havana) through a Transgenic Approach
Cloning of CpF3′5′H_2 from C. platycarpus
3.10. A. tumefaciens-Mediated Transformation of Tobacco
3.11. In-Vitro Bio-Efficacy Analysis of Tobacco Transformants to H. armigera Challenge
3.12. Molecular Characterisation of Transgenic Tobacco Plants
3.13. Expression Analyses of CpF3′5′H_2 in Tobacco Transgenics
3.14. Biochemical Analysis of Tobacco Transgenics Overexpressing CpF3′5′H_2
2,2-diphenyl-1-picrylhydrazyl (DPPH) Free Radical Scavenging Activity
3.15. Estimation of Total Flavonoid Content
3.16. Estimation of Total Polyphenolic Content
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sharma, D.; Reddy, L.J.; Srivastava, R.K.; Saxena, K.B. The unique pigeonpea landrace with multiple properties. J. Food Legume 2021, 34, 132–135. [Google Scholar]
- Sultana, R.; Saxena, K.B.; Kumar, R.R.; Kumar, D.; Kirti, M. Pigeonpea. In The Beans and the Peas; Elsevier: Amsterdam, The Netherlands, 2021; pp. 217–240. [Google Scholar]
- Shah, A.; Tyagi, S.; Saratale, G.D.; Guzik, U.; Hu, A.; Sreevathsa, R.; Reddy, V.D.; Rai, V.; Mulla, S.I. A comprehensive review on the influence of light on signaling cross-talk and molecular communication against phyto-microbiome interactions. Crit. Rev. Biotechnol. 2021, 41, 370–393. [Google Scholar] [CrossRef]
- Singh, S.P.; Singh, Y. Control of pod borers on pigeonpea. Indian J. Entomol. 2001, 63, 356–359. [Google Scholar]
- Tyagi, S.; Shah, A.; Karthik, K.; Rathinam, M.; Rai, V.; Chaudhary, N.; Sreevathsa, R. Reactive oxygen species in plants: An invincible fulcrum for biotic stress mitigation. Appl. Microbiol. Biotechnol. 2022, 106, 5945–5955. [Google Scholar] [CrossRef] [PubMed]
- Gandhi Gracy, R.; Mani, M.; Swathi, R.S.; Venkatesan, T.; Mohan, M. Biotechnological Applications in Horticultural Entomology. In Trends in Horticultural Entomology; Springer: Berlin/Heidelberg, Germany, 2022; pp. 185–209. [Google Scholar]
- Rathinam, M.; Rao, U.; Sreevathsa, R. Novel biotechnological strategies to combat biotic stresses: Polygalacturonase inhibitor (PGIP) proteins as a promising comprehensive option. Appl. Microbiol. Biotechnol. 2020, 104, 2333–2342. [Google Scholar] [CrossRef] [PubMed]
- Hossain, A.; Maitra, S.; Pramanick, B.; Bhutia, K.L.; Ahmad, Z.; Moulik, D.; Syed, M.A.; Shankar, T.; Adeel, M.; Hassan, M.M. Wild relatives of plants as sources for the development of abiotic stress tolerance in plants. In Plant Perspectives to Global Climate Changes; Elsevier: Amsterdam, The Netherlands, 2022; pp. 471–518. [Google Scholar]
- Wani, S.H.; Samantara, K.; Razzaq, A.; Kakani, G.; Kumar, P. Back to the wild: Mining maize (Zea mays l.) disease resistance using advanced breeding tools. Mol. Biol. Rep. 2022, 49, 5787–5803. [Google Scholar] [CrossRef]
- Sharma, S.; Jaba, J.; Rao, P.J.; Prasad, S.; Gopal, N.T.V.V.; Sharma, H.C.; Kilian, B. Reaping the potential of wild cajanus species through pre-breeding for improving resistance to pod borer, Helicoverpa armigera, in cultivated pigeonpea (Cajanus cajan (l.) millsp.). Biology 2022, 11, 485. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, S.; Rathinam, M.; Shashank, P.R.; Chaudhary, N.; Shasany, A.K.; Sreevathsa, R. Deciphering of pod borer [Helicoverpa armigera (hübner)] resistance in Cajanus platycarpus (benth.) offers novel insights on the reprogramming and role of flavonoid biosynthesis pathway. Toxins 2022, 14, 455. [Google Scholar] [CrossRef] [PubMed]
- Rathinam, M.; Tyagi, S.; Konda, A.K.; Rengarajan, D.; Prashat, G.R.; Sreevathsa, R. Relevance of methionine sulfoxide reductase (s)(MSR) as candidate proteins in redox homeostasis-mediated resistance response to Helicoverpa armigera (hübner) in the pigeonpea wild relative Cajanus platycarpus (benth.) Maesen. Int. J. Biol. Macromol. 2022, 215, 290–302. [Google Scholar] [CrossRef]
- Rathinam, M.; Mishra, P.; Mahato, A.K.; Singh, N.K.; Rao, U.; Sreevathsa, R. Comparative transcriptome analyses provide novel insights into the differential response of Pigeonpea (Cajanus cajan L.) and its wild relative (Cajanus platycarpus (Benth.) Maesen) to herbivory by Helicoverpa armigera (Hübner). Plant Mol. Biol. 2019, 101, 163–182. [Google Scholar] [CrossRef]
- Rathinam, M.; Roschitzki, B.; Grossmann, J.; Mishra, P.; Kunz, L.; Wolski, W.; Panse, C.; Tyagi, S.; Rao, U.; Schlapbach, R.; et al. Unraveling the proteomic changes involved in the resistance response of Cajanus platycarpus to herbivory by Helicoverpa armigera. Appl. Microbiol. Biotechnol. 2020, 104, 7603–7618. [Google Scholar] [CrossRef] [PubMed]
- Divekar, P.A.; Narayana, S.; Divekar, B.A.; Kumar, R.; Gadratagi, B.G.; Ray, A.; Singh, A.K.; Rani, V.; Singh, V.; Singh, A.K. Plant secondary metabolites as defense tools against herbivores for sustainable crop protection. Int. J. Mol. Sci. 2022, 23, 2690. [Google Scholar] [CrossRef] [PubMed]
- Vanambathina, P.; Rachaputi, R.C.; Sultanbawa, Y.; Phan, A.D.T.; Henry, R.J.; Brier, H. Biochemical Basis of Resistance to Pod Borer (Helicoverpa armigera) in Australian wild relatives of pigeonpea. Legume Sci. 2021, 3, e101. [Google Scholar] [CrossRef]
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z.; Li, X.; Li, J.; Yin, H. The flavonoid biosynthesis network in plants. Int. J. Mol. Sci. 2021, 22, 12824. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lui, A.C.; Lam, P.Y.; Liu, G.; Godwin, I.D.; Lo, C. Transgenic expression of flavanone 3-hydroxylase redirects flavonoid biosynthesis and alleviates anthracnose susceptibility in sorghum. Plant Biotechnol. J. 2020, 18, 2170. [Google Scholar] [CrossRef]
- He, H.; Ke, H.; Keting, H.; Qiaoyan, X.; Silan, D. Flower colour modification of chrysanthemum by suppression of F3’H and overexpression of the exogenous Senecio cruentus F3’5’H Gene. PLoS ONE 2013, 8, e74395. [Google Scholar] [CrossRef]
- Qi, Y.; Lou, Q.; Quan, Y.; Liu, Y.; Wang, Y. Flower-specific expression of the Phalaenopsis flavonoid 3′,5′-hydoxylase modifies flower color pigmentation in Petunia and Lilium. Plant Cell Tissue Organ Cult. 2013, 115, 263–273. [Google Scholar] [CrossRef]
- Nguyen, Y.T.H.; Hoang, H.T.T.; Mai, A.T.H.; Nguyen, L.T.N.; Nguyen, Q.H.; Pham, N.T.T.; Sy, T.D.; Chu, M.H. The Aconitum carmichaelii f3′ 5′ h gene overexpression increases flavonoid accumulation in transgenic tobacco plants. Horticulturae 2021, 7, 384. [Google Scholar] [CrossRef]
- Ahmad, N.; Jianyu, L.; Xu, T.; Noman, M.; Jameel, A.; Na, Y.; Yuanyuan, D.; Nan, W.; Xiaowei, L.; Fawei, W. Overexpression of a novel cytochrome p450 promotes flavonoid biosynthesis and osmotic stress tolerance in transgenic Arabidopsis. Genes 2019, 10, 756. [Google Scholar] [CrossRef]
- Jan, R.; Asaf, S.; Numan, M.; Kim, K.-M. Plant secondary metabolite biosynthesis and transcriptional regulation in response to biotic and abiotic stress conditions. Agronomy 2021, 11, 968. [Google Scholar] [CrossRef]
- Nguyen, T.N.L.; Hoang, T.T.H.; Nguyen, H.Q.; Tu, Q.T.; Tran, T.H.; Lo, T.M.T.; Vu, T.T.T.; Chu, H.M. Agrobacterium tumefaciens–mediated genetic transformation and overexpression of the flavonoid 3′ 5′-hydroxylase gene increases the flavonoid content of the transgenic Aconitum carmichaelii Debx. Vitr. Cell. Dev. Biol. Plant 2022, 58, 93–102. [Google Scholar] [CrossRef]
- Wang, S.; Moustaid-Moussa, N.; Chen, L.; Mo, H.; Shastri, A.; Su, R.; Bapat, P.; Kwun, I.; Shen, C.L. Novel insights of dietary polyphenols and obesity. J. Nutr. Biochem. 2014, 25, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Tang, H.; Reichelt, M.; Piirtola, E.-M.; Salminen, J.-P.; Gershenzon, J.; Constabel, C.P. Poplar MYB117 promotes anthocyanin synthesis and enhances flavonoid B-ring hydroxylation by up-regulating the flavonoid 3′,5′-hydroxylase gene. J. Exp. Bot. 2021, 72, 3864–3880. [Google Scholar] [CrossRef]
- Naing, A.H.; Kim, C.K. Roles of R2R3-MYB transcription factors in transcriptional regulation of anthocyanin biosynthesis in horticultural plants. Plant Mol. Biol. 2018, 98, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Chen, L.; Ma, A.; Wang, D.; Lu, H.; Chen, L.; Wang, H.; Qin, Y.; Hu, G. R3-MYB transcription factor LcMYBx from Litchi chinensis negatively regulates anthocyanin biosynthesis by ectopic expression in tobacco. Gene 2022, 812, 146105. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, T.; Xin, Y.; Wang, G.; Xu, L.-A. Overexpression of the GbF3′ H1 gene enhanced the epigallocatechin, gallocatechin, and catechin contents in transgenic Populus. J. Agric. Food Chem. 2020, 68, 998–1006. [Google Scholar] [CrossRef]
- Dasilva, L.L.; Foresti, O.; Denecke, J. Targeting of the plant vacuolar sorting receptor BP80 is dependent on multiple sorting signals in the cytosolic tail. Plant Cell 2006, 18, 1477–1497. [Google Scholar] [CrossRef]
- Rathinam, M.; Marimuthu, S.K.; Tyagi, S.; Kesiraju, K.; Alagiamanavalan, L.P.; Rao, U.; Sreevathsa, R. Characterization and in planta validation of a CHI4 chitinase from Cajanus platycarpus (Benth.) Maesen for its efficacy against pod borer, Helicoverpa armigera (Hübner). Pest Manag. Sci. 2021, 77, 2337–2349. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-García, C.; Ahmed, S.S.; Ramalingam, S.; Selvaraj, D.; Srivastava, A.; Paul, S.; Sharma, A. Identification of microRNAs from medicinal plant Murraya koenigii by high-throughput sequencing and their functional implications in secondary metabolite biosynthesis. Plants 2021, 11, 46. [Google Scholar] [CrossRef]
- Marone, D.; Mastrangelo, A.M.; Borrelli, G.M.; Mores, A.; Laidò, G.; Russo, M.A.; Ficco, D.B.M. Specialized metabolites: Physiological and biochemical role in stress resistance, strategies to improve their accumulation, and new applications in crop breeding and management. Plant Physiol. Biochem. 2022, 172, 48–55. [Google Scholar] [CrossRef]
- Bharti, P.; Mahajan, M.; Vishwakarma, A.K.; Bhardwaj, J.; Yadav, S.K. AtROS1 overexpression provides evidence for epigenetic regulation of genes encoding enzymes of flavonoid biosynthesis and antioxidant pathways during salt stress in transgenic tobacco. J. Exp. Bot. 2015, 66, 5959–5969. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Saito, K. Integrated metabolomics for abiotic stress responses in plants. Curr. Opin. Plant Biol. 2015, 24, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Pourcel, L.; Routaboul, J.-M.; Cheynier, V.; Lepiniec, L.; Debeaujon, I. Flavonoid oxidation in plants: From biochemical properties to physiological functions. Trends Plant Sci. 2007, 12, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Ilk, N.; Ding, J.; Ihnatowicz, A.; Koornneef, M.; Reymond, M. Natural variation for anthocyanin accumulation under high-light and low-temperature stress is attributable to the ENHANCER OF AG-4 2 (HUA 2) locus in combination with PRODUCTION OF ANTHOCYANIN PIGMENT 1 (PAP 1) and PAP 2. New Phytol. 2015, 206, 422–435. [Google Scholar] [CrossRef]
- Kusano, M.; Tohge, T.; Fukushima, A.; Kobayashi, M.; Hayashi, N.; Otsuki, H.; Kondou, Y.; Goto, H.; Kawashima, M.; Matsuda, F. Metabolomics reveals comprehensive reprogramming involving two independent metabolic responses of Arabidopsis to UV-B light. Plant J. 2011, 67, 354–369. [Google Scholar] [CrossRef] [PubMed]
- Lotkowska, M.E.; Tohge, T.; Fernie, A.R.; Xue, G.-P.; Balazadeh, S.; Mueller-Roeber, B. The Arabidopsis transcription factor myb112 promotes anthocyanin formation during salinity and under high light stress. Plant Physiol. 2015, 169, 1862–1880. [Google Scholar] [CrossRef]
- Mahmood, K.; Xu, Z.; El-Kereamy, A.; Casaretto, J.A.; Rothstein, S.J. The Arabidopsis transcription factor ANAC032 represses anthocyanin biosynthesis in response to high sucrose and oxidative and abiotic stresses. Front. Plant Sci. 2016, 7, 1548. [Google Scholar] [CrossRef]
- Zhang, C.; Jia, H.; Wu, W.; Wang, X.; Fang, J.; Wang, C. Functional conservation analysis and expression modes of grape anthocyanin synthesis genes responsive to low temperature stress. Gene 2015, 574, 168–177. [Google Scholar] [CrossRef]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef]
- Li, N.; Wang, X.; Ma, B.; Wu, Z.; Zheng, L.; Qi, Z.; Wang, Y. A leucoanthocyanidin dioxygenase gene (RtLDOX2) from the feral forage plant Reaumuria trigyna promotes the accumulation of flavonoids and improves tolerance to abiotic stresses. J. Plant Res. 2021, 134, 1121–1138. [Google Scholar] [CrossRef]
- Xu, C.; Wei, L.; Huang, S.; Yang, C.; Wang, Y.; Yuan, H.; Xu, Q.; Zhang, W.; Wang, M.; Zeng, X. Drought resistance in Qingke involves a reprogramming of the Phenylpropanoid Pathway and UDP-Glucosyltransferase regulation of abiotic stress tolerance targeting Flavonoid Biosynthesis. J. Agric. Food Chem. 2021, 69, 3992–4005. [Google Scholar] [CrossRef] [PubMed]
- Onkokesung, N.; Reichelt, M.; van Doorn, A.; Schuurink, R.C.; van Loon, J.J.; Dicke, M. Modulation of flavonoid metabolites in Arabidopsis thaliana through overexpression of the MYB75 transcription factor: Role of kaempferol-3, 7-dirhamnoside in resistance to the specialist insect herbivore Pieris brassicae. J. Exp. Bot. 2014, 65, 2203–2217. [Google Scholar] [CrossRef] [PubMed]
- Kiani, R.; Arzani, A.; Mirmohammady Maibody, S.A.M. Polyphenols, flavonoids, and antioxidant activity involved in salt tolerance in wheat, Aegilops cylindrica and their amphidiploids. Front. Plant Sci. 2021, 12, 646221. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Tan, J.; Zhou, C.; Yang, X.; Yang, F.; Zhang, S.; Sun, S.; Miao, X.; Shi, Z. The OsmiR396–Os GRF 8–OsF3H-flavonoid pathway mediates resistance to the brown planthopper in rice (Oryza sativa). Plant Biotechnol. J. 2019, 17, 1657–1669. [Google Scholar] [CrossRef]
- Bai, Q.; Duan, B.; Ma, J.; Fen, Y.; Sun, S.; Long, Q.; Lv, J.; Wan, D. Coexpression of PalbHLH1 and PalMYB90 genes from Populus alba enhances pathogen resistance in poplar by increasing the flavonoid content. Front. Plant Sci. 2020, 10, 1772. [Google Scholar] [CrossRef] [PubMed]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI reference sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005, 33, 501–504. [Google Scholar] [CrossRef]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Hall, T.; Biosciences, I.; Carlsbad, C. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Chow, C.-N.; Lee, T.-Y.; Hung, Y.-C.; Li, G.-Z.; Tseng, K.-C.; Liu, Y.-H.; Kuo, P.-L.; Zheng, H.-Q.; Chang, W.-C. PlantPAN3. 0: PlantPAN3. 0: A new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Res. 2019, 47, 1155–1163. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ Preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Ru, M.; Wang, K.; Bai, Z.; Peng, L.; He, S.; Pei, T.; Jia, Y.; Li, H.; Liang, Z. Molecular cloning and characterisation of two enzymes involved in the rosmarinic acid biosynthesis pathway of prunella vulgaris L. Plant Cell Tissue Organ Cult. 2017, 128, 381–390. [Google Scholar] [CrossRef]
- Saini, R.P.; Raman, V.; Dhandapani, G.; Malhotra, E.V.; Sreevathsa, R.; Kumar, P.A.; Sharma, T.R.; Pattanayak, D. Silencing of HaAce1 gene by host-delivered artificial microRNA disrupts growth and development of Helicoverpa armigera. PLoS ONE 2018, 13, e0194150. [Google Scholar] [CrossRef] [PubMed]
- Mellors, A.; Tappel, A.L. The inhibition of mitochondrial peroxidation by ubiquinone and ubiquinol. J. Biol. Chem. 1966, 241, 4353–4356. [Google Scholar] [CrossRef] [PubMed]
- Tohidi, B.; Rahimmalek, M.; Arzani, A. Essential oil composition, total phenolic, flavonoid contents, and antioxidant activity of thymus species collected from different regions of Iran. Food Chem. 2017, 220, 153–161. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tyagi, S.; Rathinam, M.; Dokka, N.; Chaudhary, N.; Satish, L.; Dash, P.K.; Shasany, A.K.; Sreevathsa, R. Cajanus platycarpus Flavonoid 3′5′ Hydroxylase_2 (CpF3′5′H_2) Confers Resistance to Helicoverpa armigera by Modulating Total Polyphenols and Flavonoids in Transgenic Tobacco. Int. J. Mol. Sci. 2023, 24, 1755. https://doi.org/10.3390/ijms24021755
Tyagi S, Rathinam M, Dokka N, Chaudhary N, Satish L, Dash PK, Shasany AK, Sreevathsa R. Cajanus platycarpus Flavonoid 3′5′ Hydroxylase_2 (CpF3′5′H_2) Confers Resistance to Helicoverpa armigera by Modulating Total Polyphenols and Flavonoids in Transgenic Tobacco. International Journal of Molecular Sciences. 2023; 24(2):1755. https://doi.org/10.3390/ijms24021755
Chicago/Turabian StyleTyagi, Shaily, Maniraj Rathinam, Narasimham Dokka, Nidhee Chaudhary, Lakkakula Satish, Prasanta K. Dash, Ajit Kumar Shasany, and Rohini Sreevathsa. 2023. "Cajanus platycarpus Flavonoid 3′5′ Hydroxylase_2 (CpF3′5′H_2) Confers Resistance to Helicoverpa armigera by Modulating Total Polyphenols and Flavonoids in Transgenic Tobacco" International Journal of Molecular Sciences 24, no. 2: 1755. https://doi.org/10.3390/ijms24021755
APA StyleTyagi, S., Rathinam, M., Dokka, N., Chaudhary, N., Satish, L., Dash, P. K., Shasany, A. K., & Sreevathsa, R. (2023). Cajanus platycarpus Flavonoid 3′5′ Hydroxylase_2 (CpF3′5′H_2) Confers Resistance to Helicoverpa armigera by Modulating Total Polyphenols and Flavonoids in Transgenic Tobacco. International Journal of Molecular Sciences, 24(2), 1755. https://doi.org/10.3390/ijms24021755








