Study on the Interaction of a Peptide Targeting Specific G-Quadruplex Structures Based on Chromatographic Retention Behavior
Abstract
1. Introduction
2. Results and Discussion
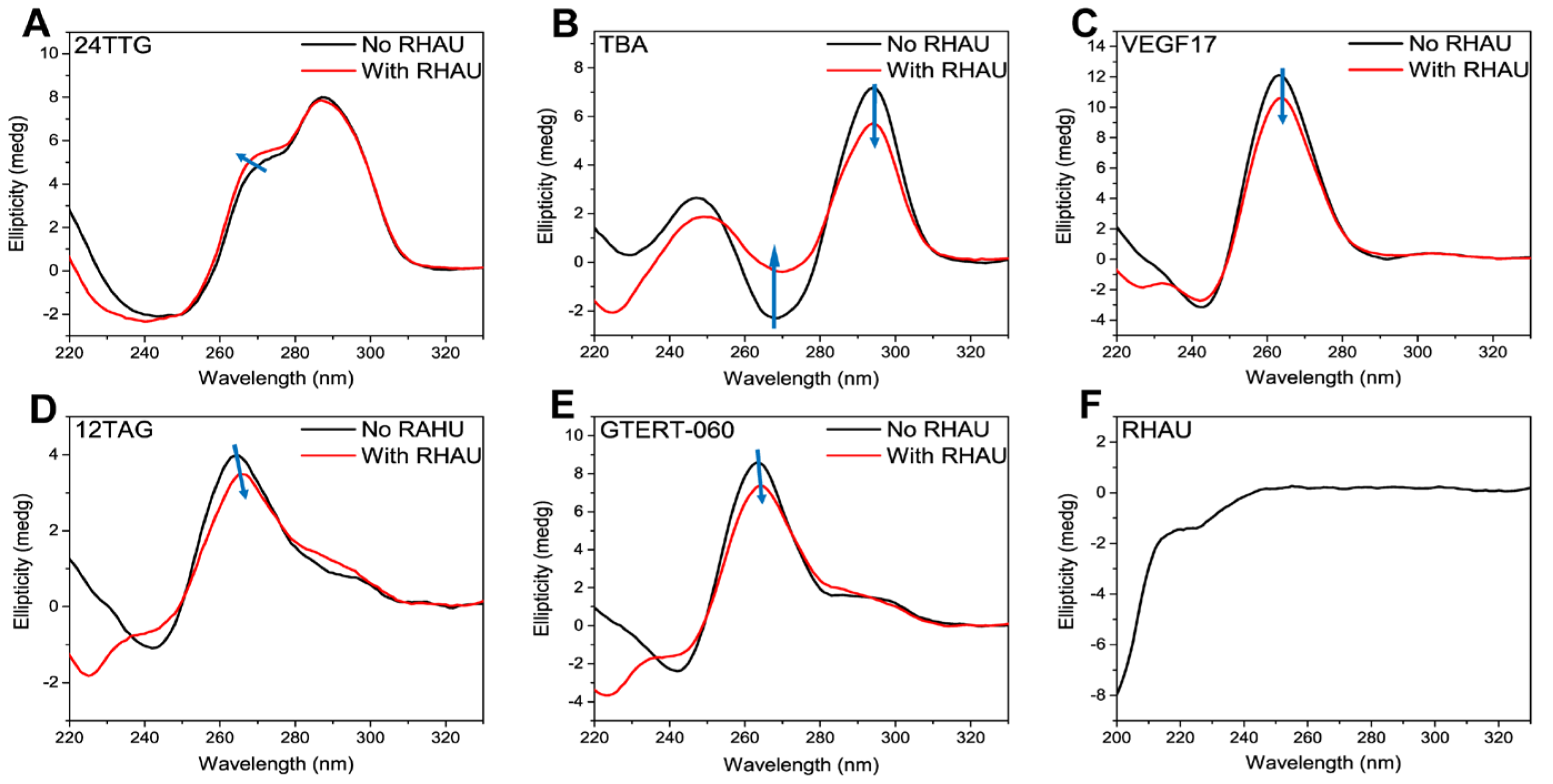
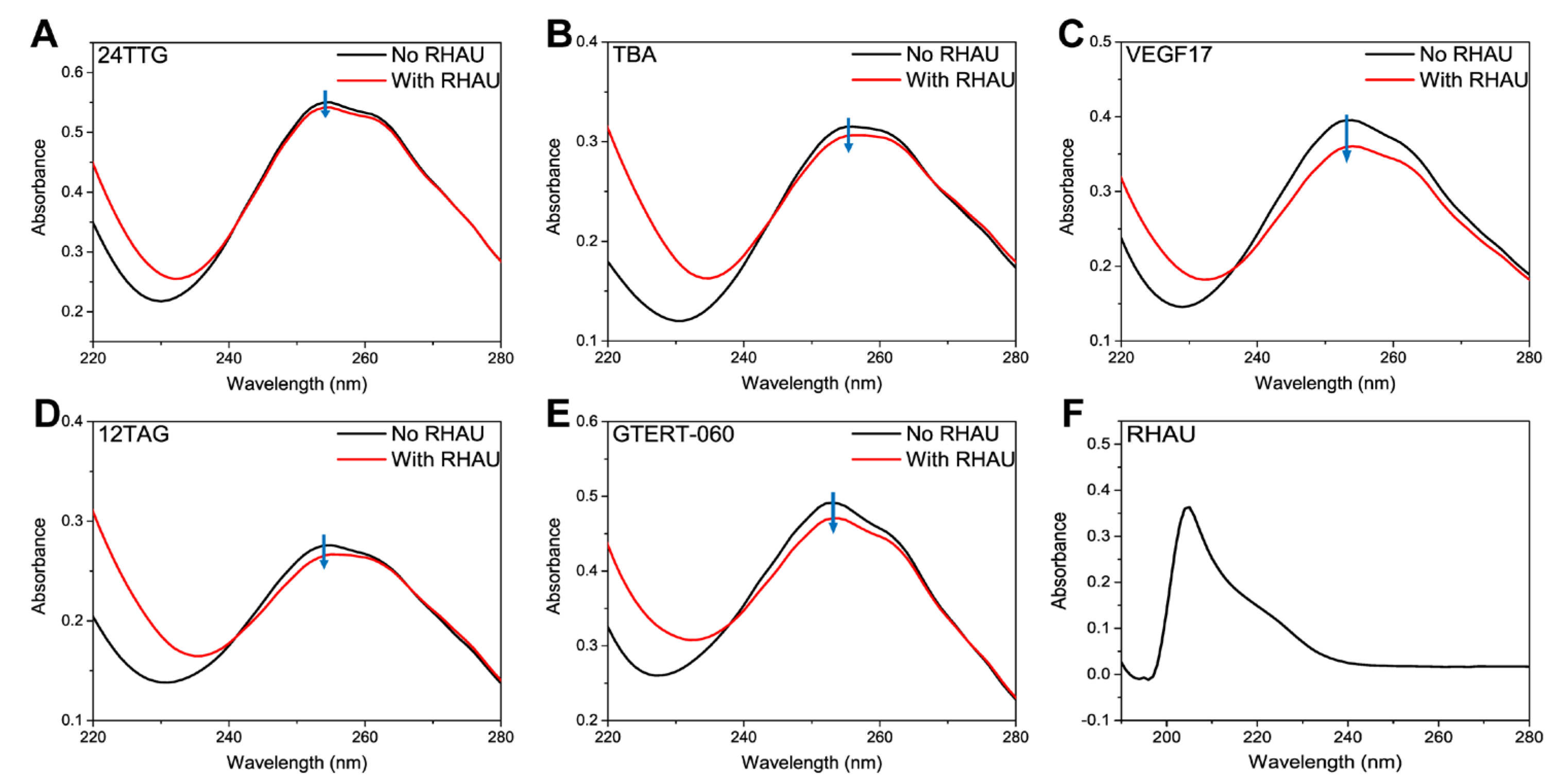
2.1. CD and UV-Vis Analysis
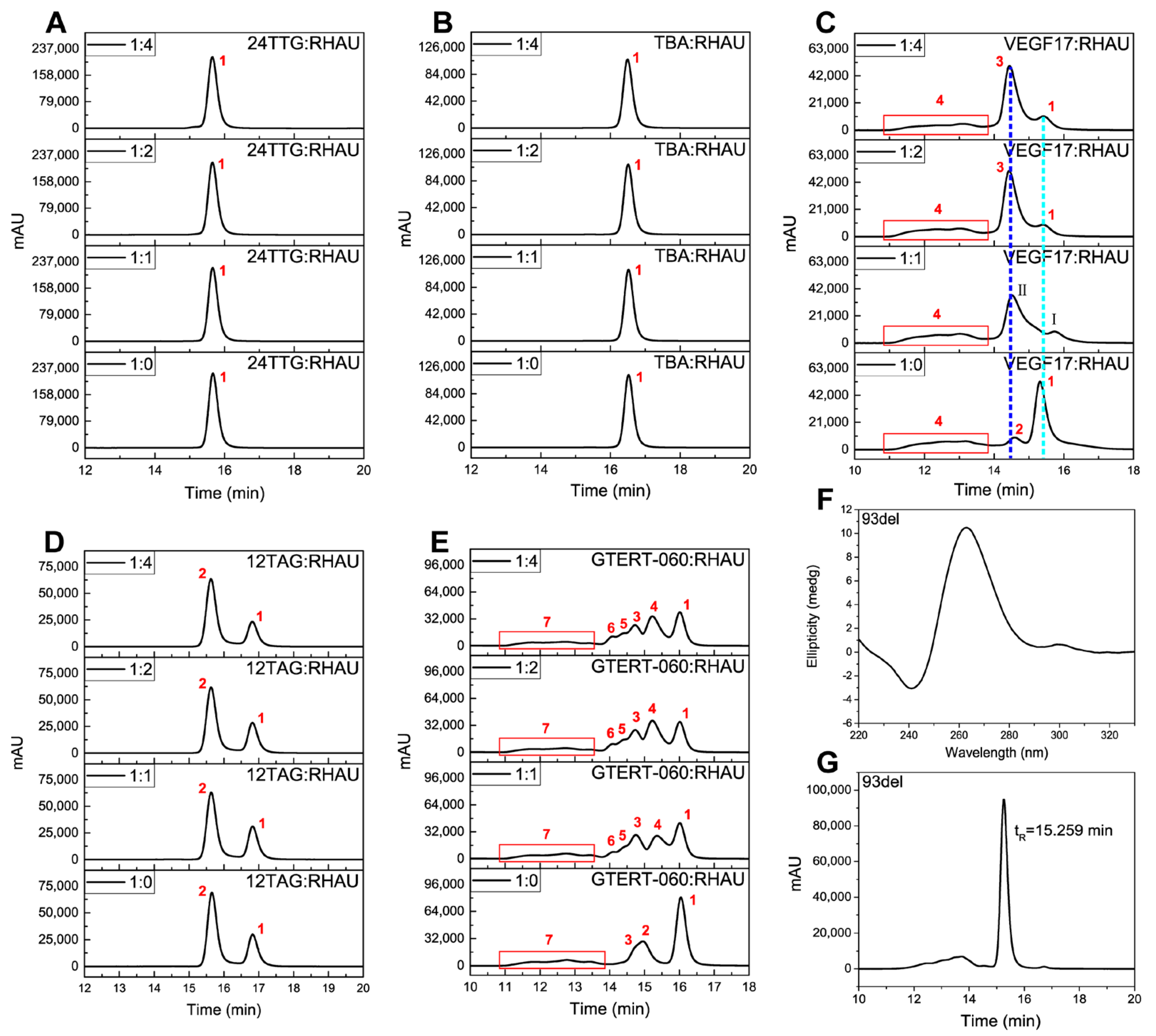
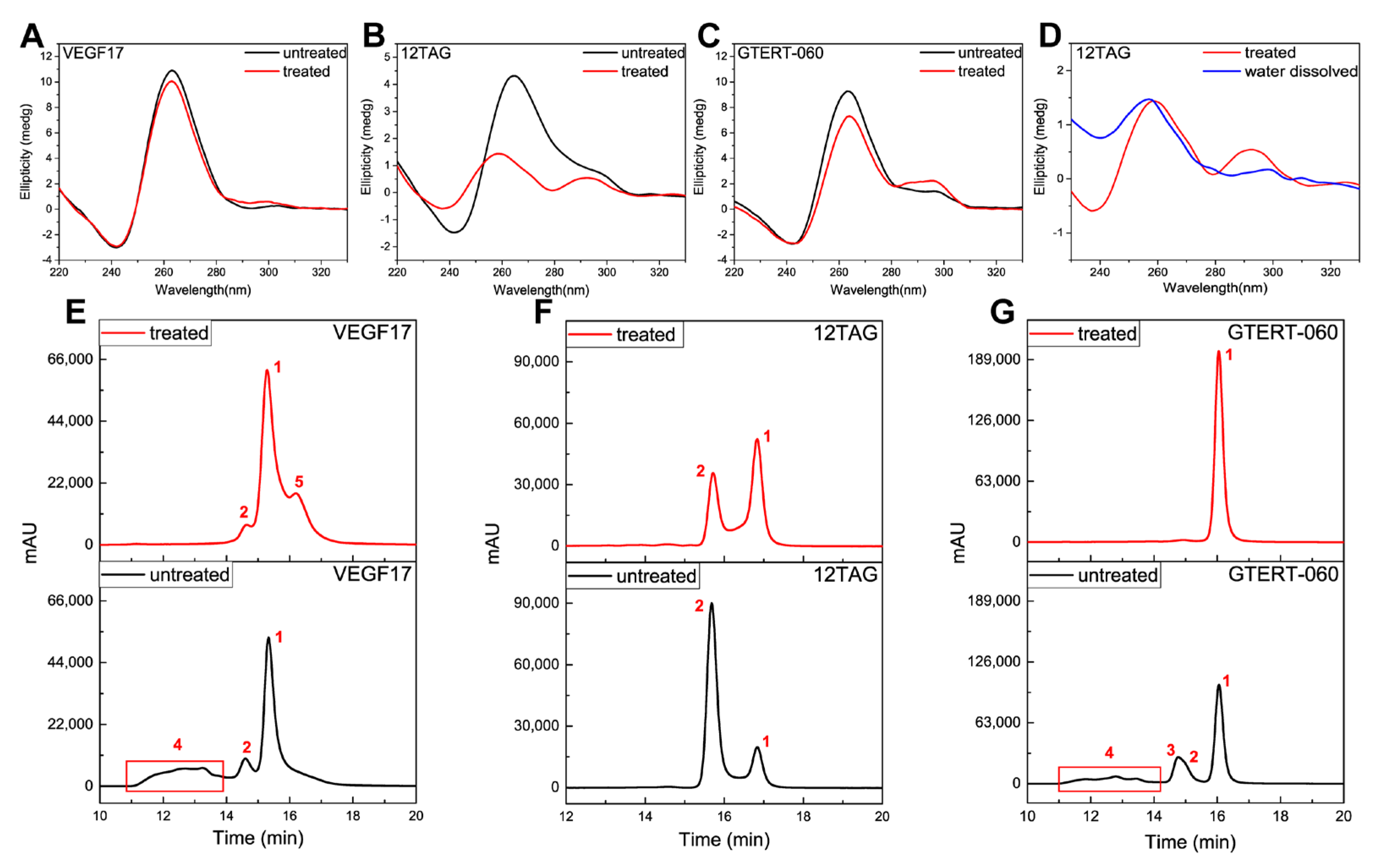
2.2. SEC Analysis
| G4s | Molecular Weight (Mw) | Peaks | tR (min) | Corresponding Substances |
|---|---|---|---|---|
| 24TTG | 7563.97 | Peak 1 | 15.668 | hybrid monomer |
| TBA | 4805.10 | Peak 1 | 16.522 | antiparallel monomer |
| VEGF17 | 5522.5 | Peak 1 | 15.316 | parallel dimer |
| Peak 2 | 14.586 | parallel trimer | ||
| Peak 3 | 14.434 | parallel dimer + RHAU complex | ||
| Peak 4 | 11.1–14.0 | parallel multimers | ||
| 12TAG | 3835.49 | Peak 1 | 16.822 | ssDNA |
| Peak 2 | 15.655 | bimolecular monomers mixture (parallel and antiparallel) | ||
| GTERT-060 | 6448.14 | Peak 1 | 16.045 | monomers mixture (parallel and hybrid) |
| Peak 2 | 14.949 | parallel dimer | ||
| Peak 3 | 14.721 | parallel dimer | ||
| Peak 4 | 15.356 | parallel monomer + RHAU complex | ||
| Peak 5 | 14.422 | parallel dimer + RHAU complex | ||
| Peak 6 | 14.107 | parallel dimer + RHAU complex | ||
| Peak 7 | 11.1–13.8 | parallel multimers |
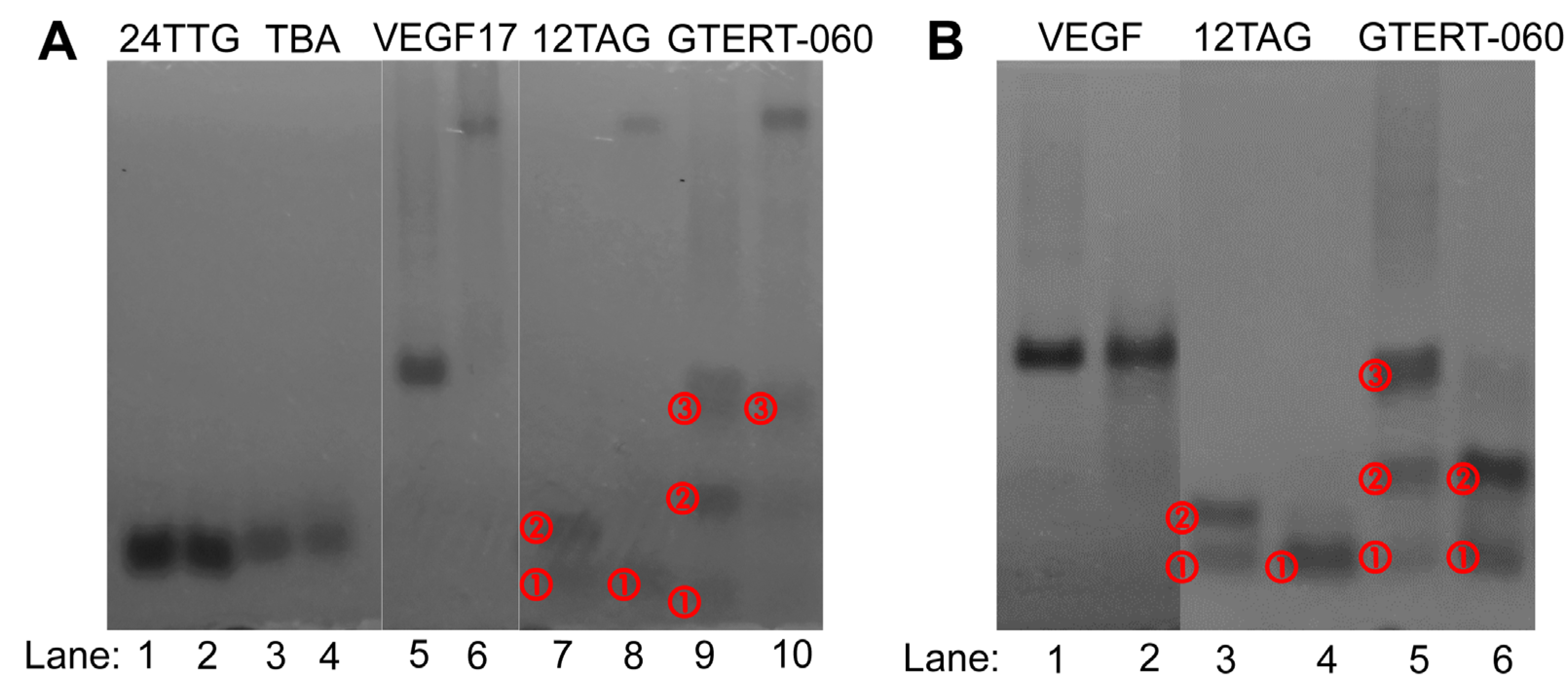
2.3. Native-PAGE Analysis
3. Materials and Methods
3.1. Materials and Reagents
3.2. Sample Preparation
3.3. CD Experiments and UV-Vis Absorption Spectroscopy
3.4. SEC Conditions
3.5. Native-PAGE Experiments
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huppert, J.L. Four-stranded nucleic acids: Structure, function and targeting of G-quadruplexes. Chem. Soc. Rev. 2008, 37, 1375–1384. [Google Scholar] [CrossRef] [PubMed]
- Assi, H.A.; Garavís, M.; González, C.; Damha, M.J. i-Motif DNA: Structural features and significance to cell biology. Nucleic Acids Res. 2018, 46, 8038–8056. [Google Scholar] [CrossRef] [PubMed]
- Phan, A.T.; Kuryavyi, V.; Patel, D.J. DNA architecture: From G to Z. Curr. Opin. Struct. Biol. 2006, 16, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Phan, A.T.; Patel, D.J. Two-repeat human telomeric d(TAGGGTTAGGGT) sequence forms interconverting parallel and antiparallel G-quadruplexes in solution: Distinct topologies, thermodynamic properties, and folding/unfolding kinetics. J. Am. Chem. Soc. 2003, 125, 15021–15027. [Google Scholar] [CrossRef]
- Lim, K.W.; Lacroix, L.; Yue, D.J.E.; Lim, J.K.C.; Lim, J.M.W.; Phan, A.T. Coexistence of two distinct G-quadruplex conformations in the hTERT Promoter. J. Am. Chem. Soc. 2010, 132, 12331–12342. [Google Scholar] [CrossRef]
- Phan, A.T.; Kuryavyi, V.; Luu, K.N.; Patel, D.J. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007, 35, 6517–6525. [Google Scholar] [CrossRef]
- Luu, K.N.; Phan, A.T.; Kuryavyi, V.; Lacroix, L.; Patel, D.J. Structure of the human telomere in K+ solution: An intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006, 128, 9963–9970. [Google Scholar] [CrossRef]
- Kolesnikova, S.; Curtis, E.A. Structure and function of multimeric G-quadruplexes. Molecules 2019, 24, 3074. [Google Scholar] [CrossRef]
- Di Antonio, M.; Ponjavic, A.; Radzevičius, A.; Ranasinghe, R.T.; Catalano, M.; Zhang, X.; Shen, J.; Needham, L.-M.; Lee, S.F.; Klenerman, D.; et al. Single-molecule visualization of DNA G-quadruplex formation in live cells. Nat. Chem. 2020, 12, 832–837. [Google Scholar] [CrossRef]
- Müller, S.; Kumari, S.; Rodriguez, R.; Balasubramanian, S. Small-molecule-mediated G-quadruplex isolation from human cells. Nat. Chem. 2010, 2, 1095–1098. [Google Scholar] [CrossRef]
- Zafar, M.K.; Hazeslip, L.; Chauhan, M.Z.; Byrd, A.K. The Expression of human DNA helicase B is affected by G-quadruplexes in the promoter. Biochemistry 2020, 59, 2401–2409. [Google Scholar] [CrossRef]
- Herviou, P.; Le Bras, M.; Dumas, L.; Hieblot, C.; Gilhodes, J.; Cioci, G.; Hugnot, J.-P.; Ameadan, A.; Guillonneau, F.; Dassi, E.; et al. hnRNP H/F drive RNA G-quadruplex-mediated translation linked to genomic instability and therapy resistance in glioblastoma. Nat. Commun. 2020, 11, 2661. [Google Scholar] [CrossRef]
- O’Hagan, M.P.; Ramos-Soriano, J.; Haldar, S.; Sheikh, S.; Morales, J.C.; Mulholland, A.J.; Galan, M.C. Visible-light photoswitching of ligand binding mode suggests G-quadruplex DNA as a target for photopharmacology. Chem. Commun. 2020, 56, 5186–5189. [Google Scholar] [CrossRef]
- Greco, M.L.; Kotar, A.; Rigo, R.; Cristofari, C.; Plavec, J.; Sissi, C. Coexistence of two main folded G-quadruplexes within a single G-rich domain in the EGFR promoter. Nucleic Acids Res. 2017, 45, 10132–10142. [Google Scholar] [CrossRef]
- Šket, P.; Pohleven, J.; Kovanda, A.; Štalekar, M.; Župunski, V.; Zalar, M.; Plavec, J.; Rogelj, B. Characterization of DNA G-quadruplex species forming from C9ORF72 G4C2-expanded repeats associated with amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Neurobiol. Aging 2015, 36, 1091–1096. [Google Scholar] [CrossRef]
- Phan, A.T. Human telomeric G-quadruplex: Structures of DNA and RNA sequences. FEBS J. 2010, 277, 1107–1117. [Google Scholar] [CrossRef]
- Grande, V.; Doria, F.; Freccero, M.; Würthner, F. An aggregating amphiphilic squaraine: A light-up probe that discriminates parallel G-quadruplexes. Angew. Chem. Int. Ed. 2017, 56, 7520–7524. [Google Scholar] [CrossRef]
- Jin, B.; Zhang, X.; Zheng, W.; Liu, X.; Zhou, J.; Zhang, N.; Wang, F.; Shangguan, D. Dicyanomethylene-functionalized squaraine as a highly selective probe for parallel G-quadruplexes. Anal. Chem. 2014, 86, 7063–7070. [Google Scholar] [CrossRef]
- Hu, M.-H.; Chen, S.-B.; Wang, Y.-Q.; Zeng, Y.-M.; Ou, T.-M.; Li, D.; Gu, L.-Q.; Huang, Z.-S.; Tan, J.-H. Accurate high-throughput identification of parallel G-quadruplex topology by a new tetraaryl-substituted imidazole. Biosens. Bioelectron. 2016, 83, 77–84. [Google Scholar] [CrossRef]
- Nicoludis, J.; Barrett, S.P.; Mergny, J.-L.; Yatsunyk, L.A. Interaction of human telomeric DNA with N-methyl mesoporphyrin IX. Nucleic Acids Res. 2012, 40, 5432–5447. [Google Scholar] [CrossRef]
- Zhang, L.; Er, J.C.; Ghosh, K.K.; Chung, W.J.; Yoo, J.; Xu, W.; Zhao, W.; Phan, A.T.; Chang, Y.-T. Discovery of a structural-element specific G-quadruplex “light-up” probe. Sci. Rep. 2014, 4, 3776. [Google Scholar] [CrossRef] [PubMed]
- Zuffo, M.; Guédin, A.; Leriche, E.-D.; Doria, F.; Pirota, V.; Gabelica, V.; Mergny, J.-L.; Freccero, M. More is not always better: Finding the right trade-off between affinity and selectivity of a G-quadruplex ligand. Nucleic Acids Res. 2018, 46, e115. [Google Scholar] [CrossRef]
- Li, T.; Wang, E.; Dong, S. Parallel G-quadruplex-specific fluorescent probe for monitoring DNA structural changes and label-free detection of potassium ion. Anal. Chem. 2010, 82, 7576–7580. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Sun, H.; Chai, Y.; Zhang, S.; Guan, A.; Li, Q.; Yao, L.; Tang, Y. Insulin-like growth factor type I selectively binds to G-quadruplex structures. Biochim. Biophys. Acta (BBA) Gen. Subj. 2018, 1863, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Carle, C.M.; Zaher, H.S.; Chalker, D.L. A parallel G-quadruplex-binding protein regulates the boundaries of DNA elimination events of tetrahymena thermophila. PLoS Genet. 2016, 12, e1005842. [Google Scholar] [CrossRef]
- Heddi, B.; Cheong, V.V.; Martadinata, H.; Phan, A.T. Insights into G-quadruplex specific recognition by the DEAH-box helicase RHAU: Solution structure of a peptide–quadruplex complex. Proc. Natl. Acad. Sci. USA 2015, 112, 9608–9613. [Google Scholar] [CrossRef]
- Roach, R.J.; Garavís, M.; González, C.; Jameson, G.B.; Filichev, V.; Hale, T.K. Heterochromatin protein 1α interacts with parallel RNA and DNA G-quadruplexes. Nucleic Acids Res. 2019, 48, 682–693. [Google Scholar] [CrossRef]
- Booy, E.P.; Meier, M.; Okun, N.; Novakowski, S.K.; Xiong, S.; Stetefeld, J.; McKenna, S.A. The RNA helicase RHAU (DHX36) unwinds a G4-quadruplex in human telomerase RNA and promotes the formation of the P1 helix template boundary. Nucleic Acids Res. 2012, 40, 4110–4124. [Google Scholar] [CrossRef]
- Jiang, M.; Hu, H.; Zhao, K.; Di, R.; Huang, X.; Shi, Y.; Yue, Y.; Nie, J.; Yu, S.; Wang, W.; et al. The G4 resolvase RHAU modulates mRNA translation and stability to sustain postnatal heart function and regeneration. J. Biol. Chem. 2021, 296, 100080. [Google Scholar] [CrossRef]
- Booy, E.P.; Howard, R.; Marushchak, O.; Ariyo, E.O.; Meier, M.; Novakowski, S.K.; Deo, S.R.; Dzananovic, E.; Stetefeld, J.; McKenna, S.A. The RNA helicase RHAU (DHX36) suppresses expression of the transcription factor PITX. Nucleic Acids Res. 2013, 42, 3346–3361. [Google Scholar] [CrossRef]
- Creacy, S.D.; Routh, E.D.; Iwamoto, F.; Nagamine, Y.; Akman, S.A.; Vaughn, J.P. G4 Resolvase 1 binds both DNA and RNA tetramolecular quadruplex with high affinity and is the major source of tetramolecular quadruplex G4-DNA and G4-RNA resolving activity in HeLa cell lysates. J. Biol. Chem. 2008, 283, 34626–34634. [Google Scholar] [CrossRef]
- Vaughn, J.P.; Creacy, S.D.; Routh, E.D.; Joyner-Butt, C.; Jenkins, G.S.; Pauli, S.; Nagamine, Y.; Akman, S.A. The DEXH protein product of the DHX36 gene is the major source of tetramolecular quadruplex G4-DNA resolving activity in HeLa cell lysates. J. Biol. Chem. 2005, 280, 38117–38120. [Google Scholar] [CrossRef]
- Lattmann, S.; Giri, B.; Vaughn, J.P.; Akman, S.A.; Nagamine, Y. Role of the amino terminal RHAU-specific motif in the recognition and resolution of guanine quadruplex-RNA by the DEAH-box RNA helicase RHAU. Nucleic Acids Res. 2010, 38, 6219–6233. [Google Scholar] [CrossRef]
- Dang, D.T.; Phan, A.T. Development of fluorescent protein probes specific for parallel DNA and RNA G-quadruplexes. Chembiochem 2015, 17, 42–45. [Google Scholar] [CrossRef]
- Gueddouda, N.M.; Mendoza, O.; Gomez, D.; Bourdoncle, A.; Mergny, J.-L. G-quadruplexes unfolding by RHAU helicase. Biochim. Biophys. Acta (BBA) Gen. Subj. 2017, 1861, 1382–1388. [Google Scholar] [CrossRef]
- Hossain, K.A.; Jurkowski, M.; Czub, J.; Kogut, M. Mechanism of recognition of parallel G-quadruplexes by DEAH/RHAU helicase DHX36 explored by molecular dynamics simulations. Comput. Struct. Biotechnol. J. 2021, 19, 2526–2536. [Google Scholar] [CrossRef]
- Meier, M.; Patel, T.; Booy, E.P.; Marushchak, O.; Okun, N.; Deo, S.; Howard, R.; McEleney, K.; Harding, S.E.; Stetefeld, J.; et al. Binding of G-quadruplexes to the N-terminal recognition domain of the RNA helicase associated with AU-rich element (RHAU). J. Biol. Chem. 2013, 288, 35014–35027. [Google Scholar] [CrossRef]
- Chen, M.C.; Murat, P.; Abecassis, K.; Ferré-D’Amaré, A.R.; Balasubramanian, S. Insights into the mechanism of a G-quadruplex-unwinding DEAH-box helicase. Nucleic Acids Res. 2015, 43, 2223–2231. [Google Scholar] [CrossRef]
- Chen, M.C.; Tippana, R.; Demeshkina, N.A.; Murat, P.; Balasubramanian, S.; Myong, S.; Ferré-D’Amaré, A.R. Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX. Nature 2018, 558, 465–469. [Google Scholar] [CrossRef]
- Shishkova, E.; Hebert, A.S.; Coon, J.J. Now, more than ever, proteomics needs better chromatography. Cell Syst. 2016, 3, 321–324. [Google Scholar] [CrossRef]
- Qiao, J.-Q.; Liang, C.; Zhu, Z.-Y.; Cao, Z.-M.; Zheng, W.-J.; Lian, H.-Z. Monolithic alkylsilane column: A promising separation medium for oligonucleotides by ion-pair reversed-phase liquid chromatography. J. Chromatogr. A 2018, 1569, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Cai, P.; Meng, S.-S.; Yang, Y.; Zeng, D.-S.; Zhang, Q.-H.; Gu, L.; Zhou, H.-C.; Gu, Z.-Y. Linker scissoring strategy enables precise shaping of metal-organic frameworks for chromatographic separation. Angew. Chem. Int. Ed. 2022, 134, e202207786. [Google Scholar] [CrossRef]
- Yang, J.; Li, J.-Y.; Qiao, J.-Q.; Lian, H.-Z.; Chen, H.-Y. Solid phase extraction of magnetic carbon doped Fe3O4 nanoparticles. J. Chromatogr. A 2014, 1325, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.-Q.; Cao, Z.-M.; Liang, C.; Chen, H.-J.; Zheng, W.-J.; Lian, H.-Z. Study on the polymorphism of G-quadruplexes by reversed-phase HPLC and LC–MS. J. Chromatogr. A 2018, 1542, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.; Liu, X.; Cheng, X.; Qi, C.; Mei, H.; Shangguan, D. Selective isolation of G-quadruplexes by affinity chromatography. J. Chromatogr. A 2012, 1246, 62–68. [Google Scholar] [CrossRef]
- Ferreira, J.; Santos, T.; Pereira, P.; Corvo, M.C.; Queiroz, J.A.; Sousa, F.; Cruz, C. Naphthalene amine support for G-quadruplex isolation. Analyst 2017, 142, 2982–2994. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Geng, Y.; Liu, C.; Miao, H.; Ren, Y.; Xu, N.; Shi, X.; You, Y.; Lee, T.; Zhu, G. Characterizations of distinct parallel and antiparallel G-quadruplexes formed by two-repeat ALS and FTD related GGGGCC sequence. Sci. Rep. 2018, 8, 2366. [Google Scholar] [CrossRef]
- Miller, M.C.; Le, H.T.; Dean, W.L.; Holt, P.A.; Chaires, J.B.; Trent, J.O. Polymorphism and resolution of oncogene promoter quadruplex-forming sequences. Org. Biomol. Chem. 2011, 9, 7633–7637. [Google Scholar] [CrossRef]
- Marzano, M.; Falanga, A.P.; D’Errico, S.; Pinto, B.; Roviello, G.N.; Piccialli, G.; Oliviero, G.; Borbone, N. New G-quadruplex-forming oligodeoxynucleotides incorporating a bifunctional double-ended linker (DEL): Effects of DEL size and ODNs orientation on the topology, stability, and molecularity of DEL-G-quadruplexes. Molecules 2019, 24, 654. [Google Scholar] [CrossRef]
- Marzano, M.; Falanga, A.P.; Dardano, P.; D’Errico, S.; Rea, I.; Terracciano, M.; De Stefano, L.; Piccialli, G.; Borbone, N.; Oliviero, G. π–π stacked DNA G-wire nanostructures formed by a short G-rich oligonucleotide containing a 3′–3′ inversion of polarity site. Org. Chem. Front. 2020, 7, 2187–2195. [Google Scholar] [CrossRef]
- Benito, S.; Ferrer, A.; Benabou, S.; Aviñó, A.; Eritja, R.; Gargallo, R. Evaluation of the effect of polymorphism on G-quadruplex-ligand interaction by means of spectroscopic and chromatographic techniques. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 196, 185–195. [Google Scholar] [CrossRef]
- Kypr, J.; Kejnovska, I.; Renciuk, D.; Vorlickova, M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009, 37, 1713–1725. [Google Scholar] [CrossRef]
- Rehm, C.; Holder, I.T.; Groß, A.; Wojciechowski, F.; Urban, M.; Sinn, M.; Drescher, M.; Hartig, J.S. A bacterial DNA quadruplex with exceptional K+ selectivity and unique structural polymorphism. Chem. Sci. 2014, 5, 2809–2818. [Google Scholar] [CrossRef]
- Ambrus, A.; Chen, D.; Dai, J.; Bialis, T.; Jones, R.A.; Yang, D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006, 34, 2723–2735. [Google Scholar] [CrossRef]
- Sun, D.; Guo, K.; Rusche, J.J.; Hurley, L.H. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005, 33, 6070–6080. [Google Scholar] [CrossRef]
- Macaya, R.F.; Schultze, P.; Smith, F.W.; Roe, J.A.; Feigon, J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. USA 1993, 90, 3745–3749. [Google Scholar] [CrossRef]
- Trizna, L.; Janovec, L.; Halaganová, A.; Víglaský, V. Rhodamine 6G-ligand influencing G-quadruplex stability and topology. Int. J. Mol. Sci. 2021, 22, 7639. [Google Scholar] [CrossRef]
- Lai, H.; Xiao, Y.; Yan, S.; Tian, F.; Zhong, C.; Liu, Y.; Weng, X.; Zhou, X. Symmetric cyanovinyl-pyridinium triphenylamine: A novel fluorescent switch-on probe for an antiparallel G-quadruplex. Analyst 2014, 139, 1834–1838. [Google Scholar] [CrossRef]
- Honisch, C.; Ragazzi, E.; Hussain, R.; Brazier, J.; Siligardi, G.; Ruzza, P. Interaction of a short peptide with G-quadruplex-forming sequences: An SRCD and CD study. Pharmaceutics 2021, 13, 1104. [Google Scholar] [CrossRef]
- Li, L.; Lu, J.; Zhou, W.; Li, H.; Yang, X. Studies on the interaction mechanism of aminopyrene derivatives with human tumor-related DNA. J. Photochem. Photobiol. B Biol. 2013, 123, 32–40. [Google Scholar] [CrossRef]
- Marzano, M.; Falanga, A.P.; Marasco, D.; Borbone, N.; D’Errico, S.; Piccialli, G.; Roviello, G.N.; Oliviero, G. Evaluation of an analogue of the marine ε-PLL peptide as a ligand of G-quadruplex DNA structures. Mar. Drugs 2020, 18, 49. [Google Scholar] [CrossRef] [PubMed]
- Phan, A.T.; Kuryavyi, V.; Ma, J.B.; Faure, A.; Andréola, M.L.; Patel, D.J. An interlocked dimeric parallel-stranded DNA quadruplex: A potent inhibitor of HIV-1 integrase. Proc. Natl. Acad. Sci. USA 2005, 102, 634–639. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Liu, X.; Bing, T.; Cao, Z.; Shangguan, D. General peroxidase activity of G-quadruplex−hemin complexes and its application in ligand screening. Biochemistry 2009, 48, 7817–7823. [Google Scholar] [CrossRef] [PubMed]
- Rauser, V.; Weinhold, E.G. Quantitative Formation of monomeric G-quadruplex DNA from multimeric structures of c-Myc promoter sequence. ChemBioChem 2020, 21, 2445–2448. [Google Scholar] [CrossRef]
| Names | DNA Sequences (5′ to 3′) | Description | G4 Conformations |
|---|---|---|---|
| 24TTG | TTG GGT TAG GGT TAG GGT TAG GGA | Human telomere | Hybrid monomer [7] |
| TBA | GGT TGG TGT GGT TGG | Thrombin-binding aptamer | Antiparallel monomer [56] |
| VEGF17 | GGG AGG GTT GGG GTG GG | Human vascular endothelial growth factor proximal promoter | Parallel [55] |
| 12TAG | TAG GGT TAG GGT | Human telomere | Double-stranded parallel and antiparallel [4] |
| GTERT-060 | AGG GGA GGG GCT GGG AGG GC | Human telomerase, hTERT promoter | Parallel and hybrid [5] |
| 93del | GGG GTG GGA GGA GGG T | Aptamer, inhibitor of HIV-1 integrase | Interlocked bimolecular parallel [62] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Qiao, J.; Zheng, W.; Lian, H. Study on the Interaction of a Peptide Targeting Specific G-Quadruplex Structures Based on Chromatographic Retention Behavior. Int. J. Mol. Sci. 2023, 24, 1438. https://doi.org/10.3390/ijms24021438
Wang J, Qiao J, Zheng W, Lian H. Study on the Interaction of a Peptide Targeting Specific G-Quadruplex Structures Based on Chromatographic Retention Behavior. International Journal of Molecular Sciences. 2023; 24(2):1438. https://doi.org/10.3390/ijms24021438
Chicago/Turabian StyleWang, Ju, Junqin Qiao, Weijuan Zheng, and Hongzhen Lian. 2023. "Study on the Interaction of a Peptide Targeting Specific G-Quadruplex Structures Based on Chromatographic Retention Behavior" International Journal of Molecular Sciences 24, no. 2: 1438. https://doi.org/10.3390/ijms24021438
APA StyleWang, J., Qiao, J., Zheng, W., & Lian, H. (2023). Study on the Interaction of a Peptide Targeting Specific G-Quadruplex Structures Based on Chromatographic Retention Behavior. International Journal of Molecular Sciences, 24(2), 1438. https://doi.org/10.3390/ijms24021438






